RORα Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation
Abstract
:1. Introduction
2. Results
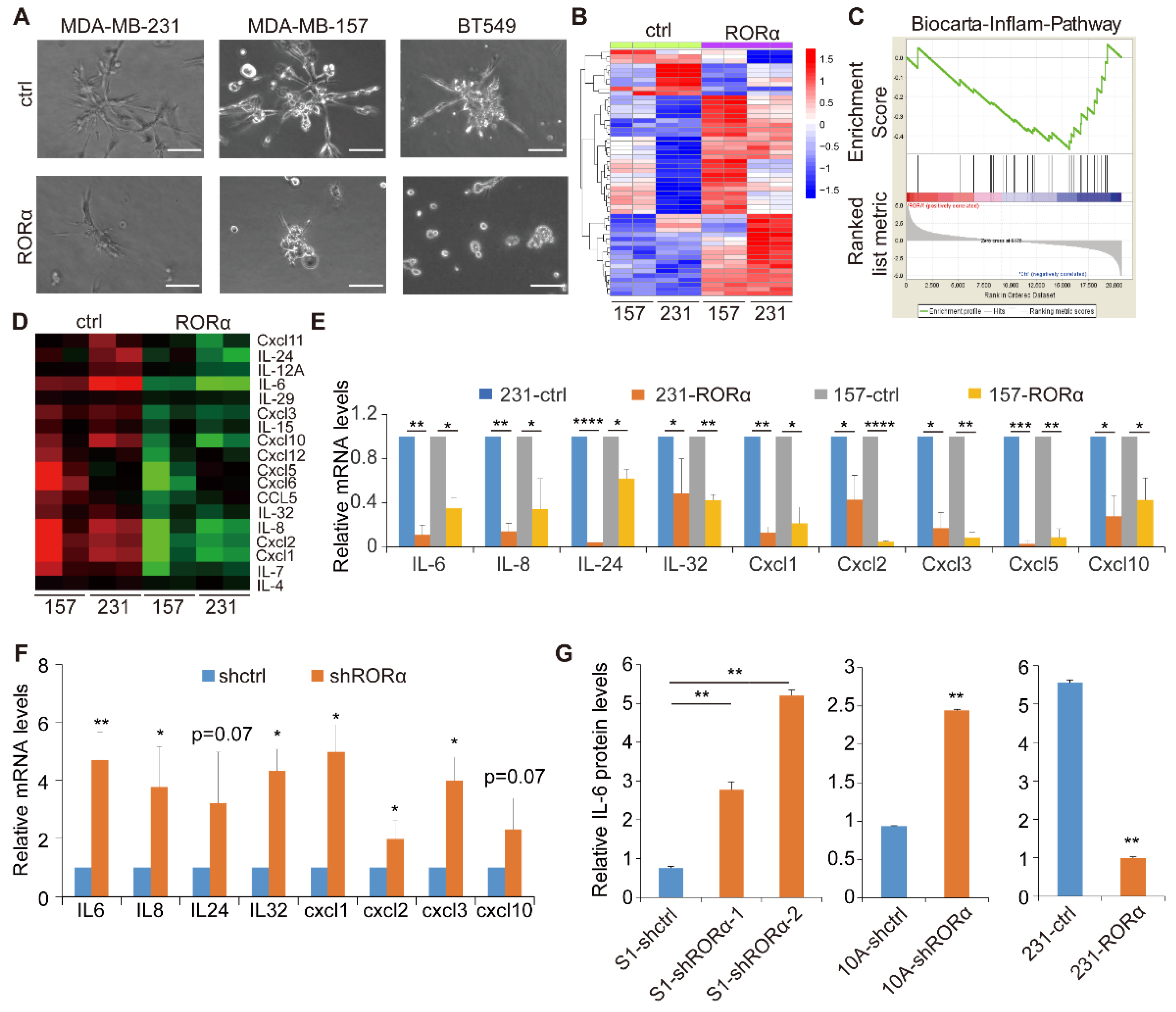
2.1. RORα Inhibits Activation of Inflammation Gene Signature and Expression of Cytokines in Breast Cancer Cells
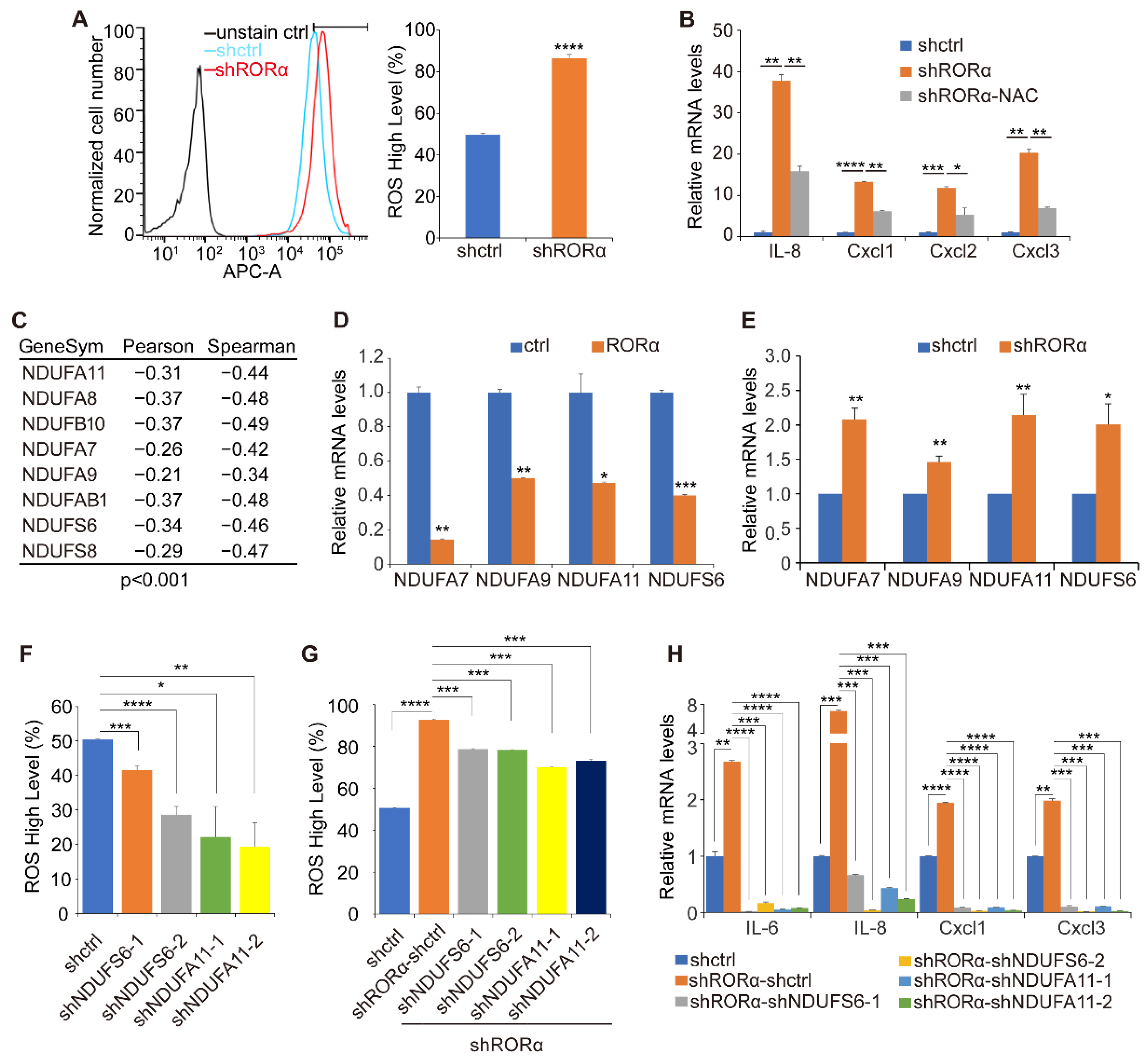
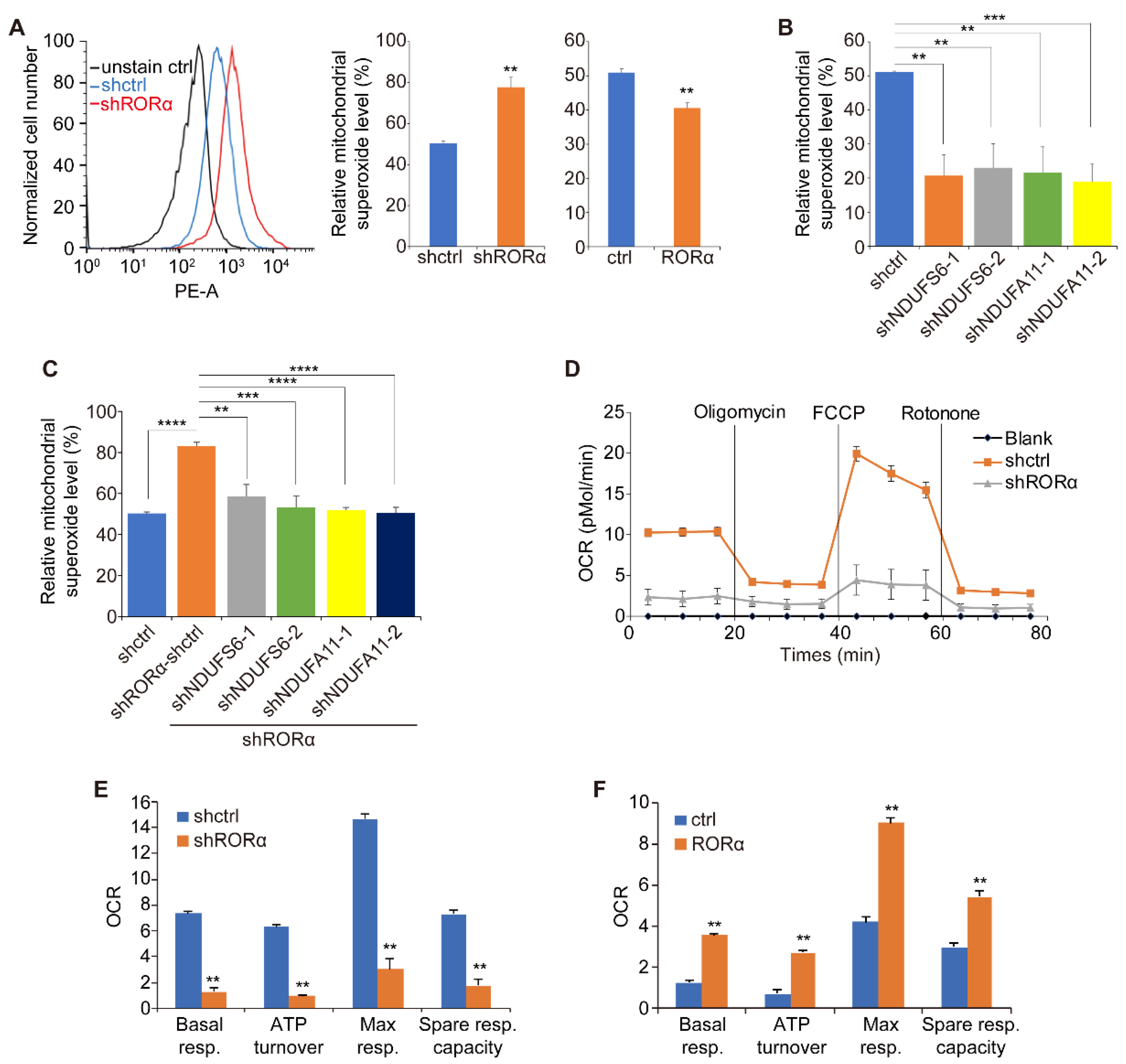
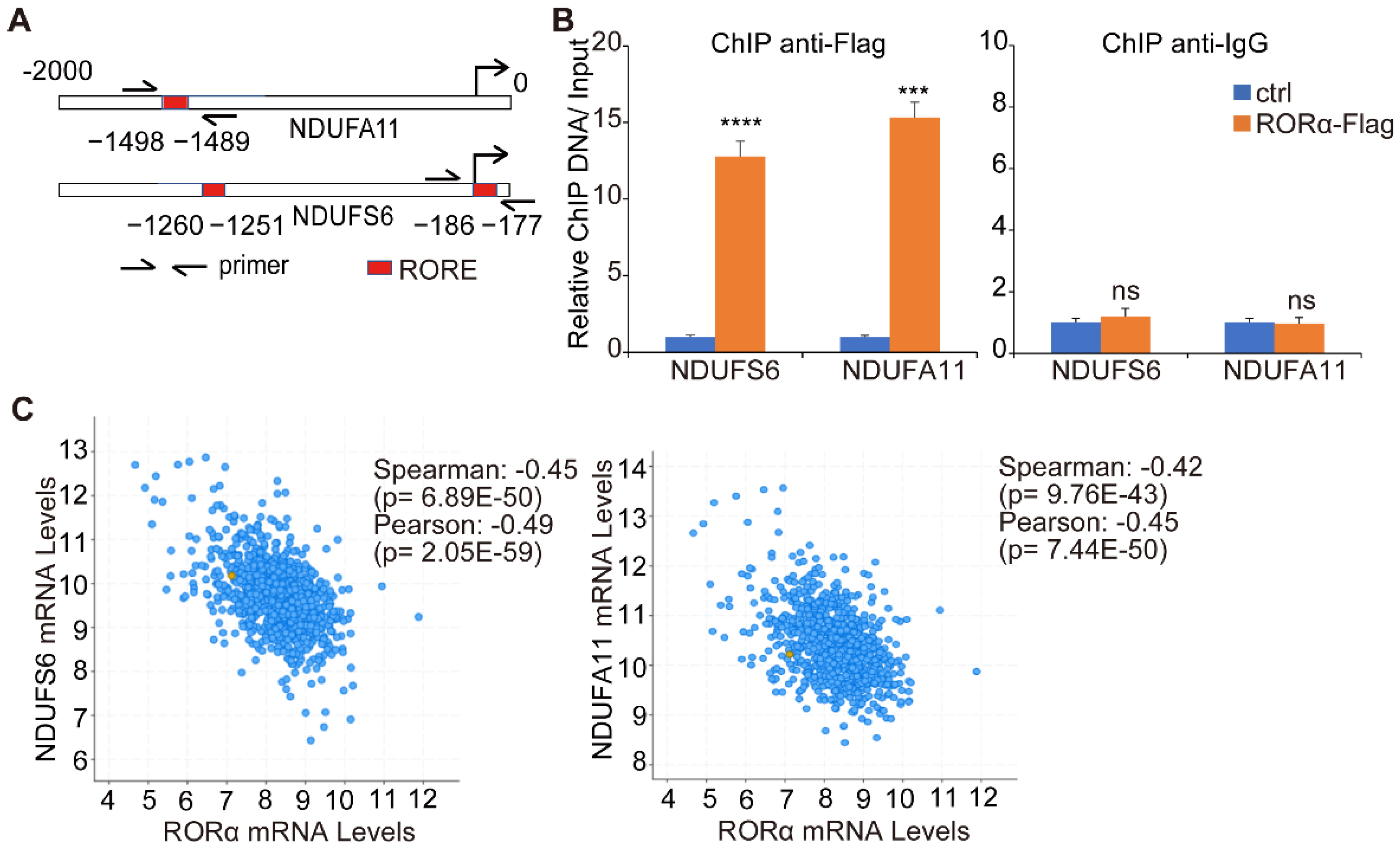
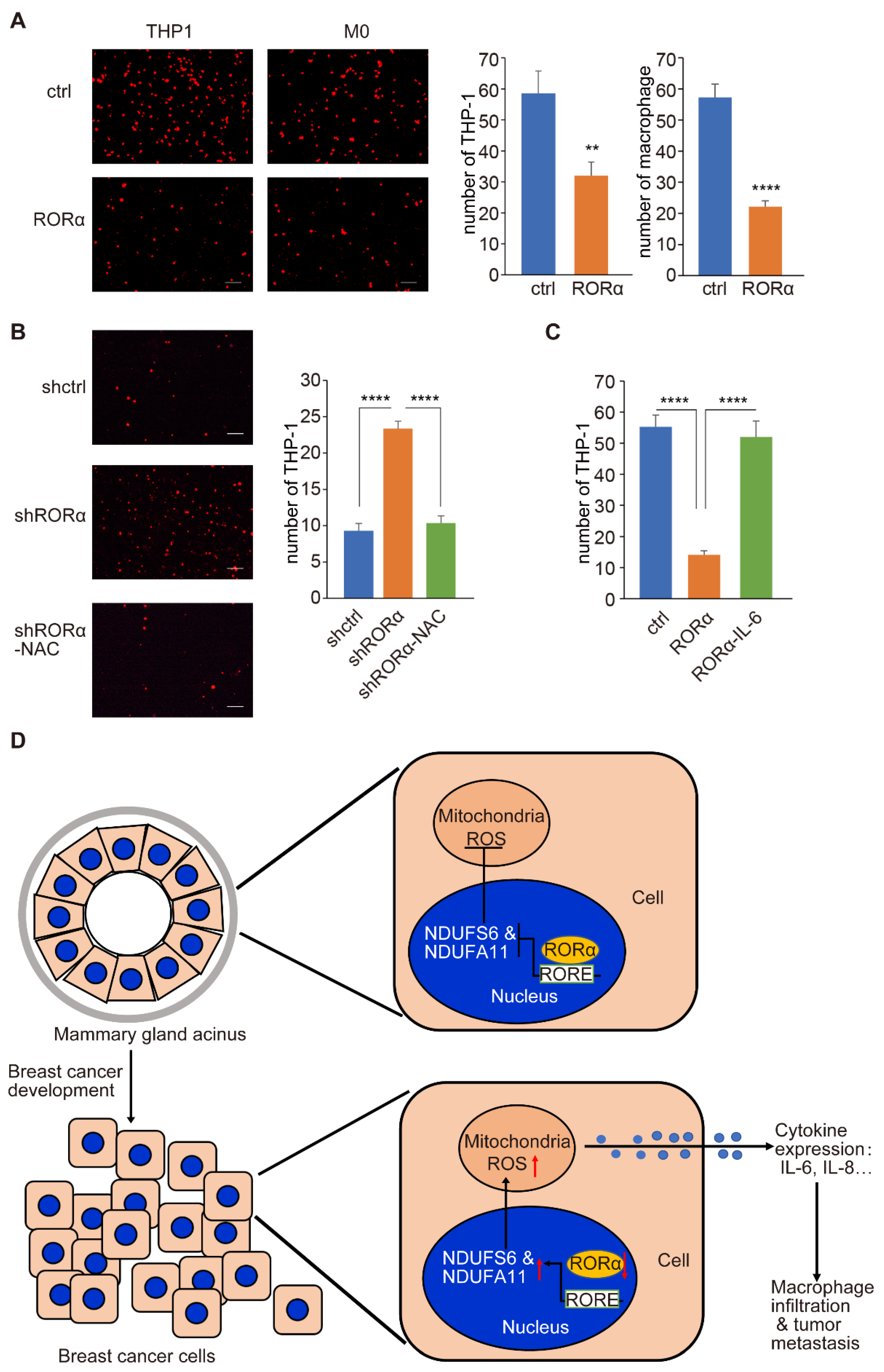
2.2. RORα Inhibits ROS Production by Repressing the Expression of Complex I Genes
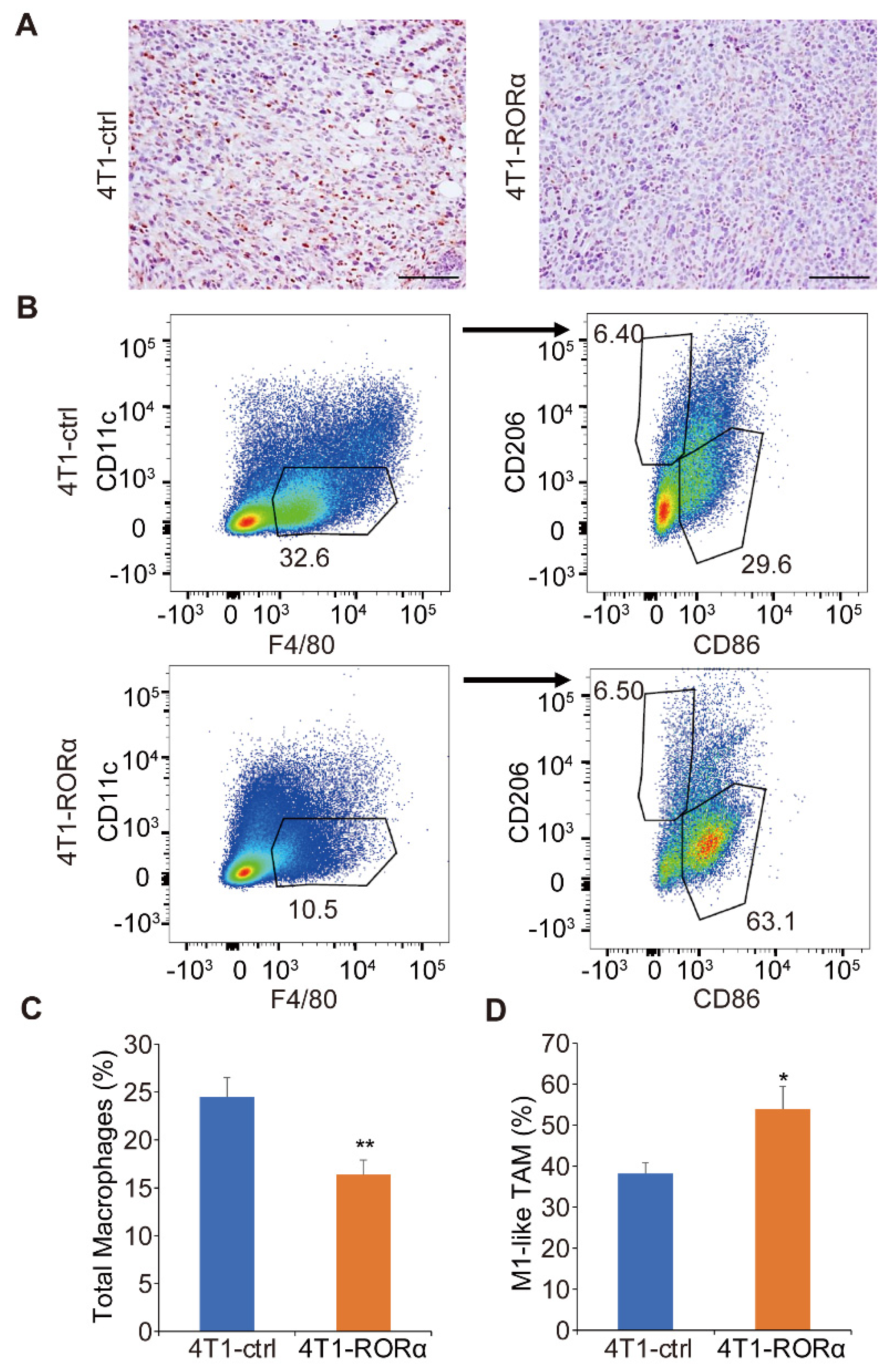
2.3. RORα Expression Inhibits Mammary Tumor Metastasis and Macrophage Infiltration in Tumor Tissue
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Lines and Culture Conditions
4.3. 3D Culture
4.4. Co-Culture Assay
4.5. Microarray Analysis and Quantitative RT-PCR
4.6. Seahorse Assay
4.7. ROS and Superoxide FACS Analysis
4.8. Chromatin Immunoprecipitation (ChIP) Assay
4.9. Mouse Experiments
4.10. Tumor Infiltrating Lymphocytes Isolation and FACS Analysis
4.11. Kaplan-Meier Survival Analysis and Other Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. The Yin-Yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol. Rev. 2008, 222, 155–161. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Brennan, D.J.; Rexhepaj, E.; Ruffell, B.; Shiao, S.L.; Madden, S.F.; Gallagher, W.M.; Wadhwani, N.; Keil, S.D.; Junaid, S.A.; et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011, 1, 54–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, S.; Liu, Q.; Chen, J.; Chen, J.; Chen, F.; He, C.; Huang, D.; Wu, W.; Lin, L.; Huang, W.; et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell 2014, 25, 605–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Gonzalez-Junca, A.; Zheng, Y.; Ouyang, H.; Illa-Bochaca, I.; Horst, K.C.; Krings, G.; Wang, Y.; Fernandez-Garcia, I.; Chou, W.; et al. Inflammation Mediates the Development of Aggressive Breast Cancer Following Radiotherapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 1778–1791. [Google Scholar] [CrossRef]
- Sharma, M.; Beck, A.H.; Webster, J.A.; Espinosa, I.; Montgomery, K.; Varma, S.; van de Rijn, M.; Jensen, K.C.; West, R.B. Analysis of stromal signatures in the tumor microenvironment of ductal carcinoma in situ. Breast Cancer Res. Treat. 2010, 123, 397–404. [Google Scholar] [CrossRef]
- Wang, H.H.; Ducatman, B.S.; Eick, D. Comparative features of ductal carcinoma in situ and infiltrating ductal carcinoma of the breast on fine-needle aspiration biopsy. Am. J. Clin. Pathol. 1989, 92, 736–740. [Google Scholar] [CrossRef]
- Muthuswamy, S.K.; Xue, B. Cell polarity as a regulator of cancer cell behavior plasticity. Annu. Rev. Cell Dev. Biol. 2012, 28, 599–625. [Google Scholar] [CrossRef] [Green Version]
- Boghaert, E.; Radisky, D.C.; Nelson, C.M. Lattice-based model of ductal carcinoma in situ suggests rules for breast cancer progression to an invasive state. PLoS Comput. Biol. 2014, 10, e1003997. [Google Scholar] [CrossRef]
- Li, L.; Chen, J.; Xiong, G.; St Clair, D.K.; Xu, W.; Xu, R. Increased ROS production in non-polarized mammary epithelial cells induces monocyte infiltration in 3D culture. J. Cell Sci. 2017, 130, 190–202. [Google Scholar] [CrossRef] [Green Version]
- Kamp, D.W.; Shacter, E.; Weitzman, S.A. Chronic inflammation and cancer: The role of the mitochondria. Oncology 2011, 25, 400–410, 413. [Google Scholar]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Radisky, D.C.; Levy, D.D.; Littlepage, L.E.; Liu, H.; Nelson, C.M.; Fata, J.E.; Leake, D.; Godden, E.L.; Albertson, D.G.; Nieto, M.A.; et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 2005, 436, 123–127. [Google Scholar] [CrossRef] [Green Version]
- Chance, B.; Sies, H.; Boveris, A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979, 59, 527–605. [Google Scholar] [CrossRef]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of reactive oxygen species by mitochondria: Central role of complex III. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Chen, Y.; St Clair, D.K. ROS and p53: A versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, M.J.; Liu, Z.G. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Jetten, A.M.; Kurebayashi, S.; Ueda, E. The ROR nuclear orphan receptor subfamily: Critical regulators of multiple biological processes. Prog. Nucleic Acid Res. Mol. Biol. 2001, 69, 205–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; McAvoy, S.; Kuhn, R.; Smith, D.I. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene 2006, 25, 2901–2908. [Google Scholar] [CrossRef] [Green Version]
- Xiong, G.; Wang, C.; Evers, B.M.; Zhou, B.P.; Xu, R. RORα suppresses breast tumor invasion by inducing SEMA3F expression. Cancer Res. 2012, 72, 1728–1739. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Kim, I.S.; Kim, H.; Lee, J.S.; Kim, K.; Yim, H.Y.; Jeong, J.; Kim, J.H.; Kim, J.Y.; Lee, H.; et al. RORalpha attenuates Wnt/beta-catenin signaling by PKCalpha-dependent phosphorylation in colon cancer. Mol. Cell 2010, 37, 183–195. [Google Scholar] [CrossRef]
- Stapleton, C.M.; Jaradat, M.; Dixon, D.; Kang, H.S.; Kim, S.C.; Liao, G.; Carey, M.A.; Cristiano, J.; Moorman, M.P.; Jetten, A.M. Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005, 289, L144–L152. [Google Scholar] [CrossRef] [PubMed]
- Schreck, R.; Rieber, P.; Baeuerle, P.A. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991, 10, 2247–2258. [Google Scholar] [CrossRef]
- Karin, M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a000141. [Google Scholar] [CrossRef]
- Delerive, P.; Monté, D.; Dubois, G.; Trottein, F.; Fruchart-Najib, J.; Mariani, J.; Fruchart, J.C.; Staels, B. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001, 2, 42–48. [Google Scholar] [CrossRef]
- Journiac, N.; Jolly, S.; Jarvis, C.; Gautheron, V.; Rogard, M.; Trembleau, A.; Blondeau, J.P.; Mariani, J.; Vernet-der Garabedian, B. The nuclear receptor ROR(alpha) exerts a bi-directional regulation of IL-6 in resting and reactive astrocytes. Proc. Natl. Acad. Sci. USA 2009, 106, 21365–21370. [Google Scholar] [CrossRef] [Green Version]
- Mookerjee, S.A.; Divakaruni, A.S.; Jastroch, M.; Brand, M.D. Mitochondrial uncoupling and lifespan. Mech. Ageing Dev. 2010, 131, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Grivennikova, V.G.; Vinogradov, A.D. Generation of superoxide by the mitochondrial Complex I. Biochim. Biophys. Acta 2006, 1757, 553–561. [Google Scholar] [CrossRef] [Green Version]
- Hanukoglu, I.; Rapoport, R.; Weiner, L.; Sklan, D. Electron leakage from the mitochondrial NADPH-adrenodoxin reductase-adrenodoxin-P450scc (cholesterol side chain cleavage) system. Arch. Biochem. Biophys. 1993, 305, 489–498. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, Y.; Wu, Y.; Wang, L.; Wang, X.; Du, J. Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J. Biol. Chem. 2013, 288, 1489–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; He, Z.; Huang, M.; Liu, T.; Wang, Y.; Xu, H.; Duan, H.; Ma, P.; Zhang, L.; Zamvil, S.S.; et al. Vascular niche IL-6 induces alternative macrophage activation in glioblastoma through HIF-2α. Nat. Commun. 2018, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Xu, R. RORalpha, a potential tumor suppressor and therapeutic target of breast cancer. Int. J. Mol. Sci. 2012, 13, 15755–15766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohrer, L.R.; Schwertfeger, K.L. Macrophages promote fibroblast growth factor receptor-driven tumor cell migration and invasion in a CXCR2-dependent manner. Mol. Cancer Res. MCR 2012, 10, 1294–1305. [Google Scholar] [CrossRef] [Green Version]
- Ingman, W.V.; Wyckoff, J.; Gouon-Evans, V.; Condeelis, J.; Pollard, J.W. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006, 235, 3222–3229. [Google Scholar] [CrossRef]
- Gouon-Evans, V.; Rothenberg, M.E.; Pollard, J.W. Postnatal mammary gland development requires macrophages and eosinophils. Development 2000, 127, 2269–2282. [Google Scholar] [CrossRef]
- Ewald, A.J.; Huebner, R.J.; Palsdottir, H.; Lee, J.K.; Perez, M.J.; Jorgens, D.M.; Tauscher, A.N.; Cheung, K.J.; Werb, Z.; Auer, M. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J. Cell Sci. 2012, 125, 2638–2654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rejon, C.; McCaffrey, L. Cell Polarity in Mammary Gland Morphogenesis and Breast Cancer. In Cell Polarity 2: Role in Development and Disease; Ebnet, K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 187–207. [Google Scholar]
- Xiong, G.; Xu, R. RORα binds to E2F1 to inhibit cell proliferation and regulate mammary gland branching morphogenesis. Mol. Cell. Biol. 2014, 34, 3066–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orrenius, S.; Gogvadze, V.; Zhivotovsky, B. Mitochondrial oxidative stress: Implications for cell death. Annu. Rev. Pharm. Toxicol. 2007, 47, 143–183. [Google Scholar] [CrossRef] [PubMed]
- Urra, F.A.; Munoz, F.; Lovy, A.; Cardenas, C. The Mitochondrial Complex(I)ty of Cancer. Front. Oncol. 2017, 7, 118. [Google Scholar] [CrossRef]
- Han, Y.H.; Kim, H.J.; Kim, E.J.; Kim, K.S.; Hong, S.; Park, H.G.; Lee, M.O. RORα decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid. Redox Signal. 2014, 21, 2083–2094. [Google Scholar] [CrossRef] [Green Version]
- Santidrian, A.F.; Matsuno-Yagi, A.; Ritland, M.; Seo, B.B.; LeBoeuf, S.E.; Gay, L.J.; Yagi, T.; Felding-Habermann, B. Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J. Clin. Investig. 2013, 123, 1068–1081. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Fabuel, I.; Le Douce, J.; Logan, A.; James, A.M.; Bonvento, G.; Murphy, M.P.; Almeida, A.; Bolaños, J.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA 2016, 113, 13063–13068. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.K.; Kim, D.; Kim, K.; Boo, K.; Yu, Y.S.; Kim, I.S.; Jeon, Y.; Im, S.K.; Lee, S.H.; Lee, J.M.; et al. RORalpha is crucial for attenuated inflammatory response to maintain intestinal homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 21140–21149. [Google Scholar] [CrossRef] [Green Version]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Condeelis, J.; Pollard, J.W. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006, 124, 263–266. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, N.J.; Sasser, A.K.; Axel, A.E.; Vesuna, F.; Raman, V.; Ramirez, N.; Oberyszyn, T.M.; Hall, B.M. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 2009, 28, 2940–2947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Spencer, V.A.; Bissell, M.J. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J. Biol. Chem. 2007, 282, 14992–14999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, G.; Deng, L.; Zhu, J.; Rychahou, P.G.; Xu, R. Prolyl-4-hydroxylase alpha subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 2014, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Zhou, H.; Krueger, J.; Kaplan, C.; Lee, S.H.; Dolman, C.; Markowitz, D.; Wu, W.; Liu, C.; Reisfeld, R.A.; et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J. Clin. Investig. 2006, 116, 2132–2141. [Google Scholar] [CrossRef] [Green Version]
- Jezequel, P.; Campone, M.; Gouraud, W.; Guerin-Charbonnel, C.; Leux, C.; Ricolleau, G.; Campion, L. bc-GenExMiner: An easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res. Treat. 2012, 131, 765–775. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, W.; Xiong, G.; Wu, Y.; Wang, C.; St. Clair, D.; Li, J.-D.; Xu, R. RORα Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation. Int. J. Mol. Sci. 2021, 22, 10665. https://doi.org/10.3390/ijms221910665
Mao W, Xiong G, Wu Y, Wang C, St. Clair D, Li J-D, Xu R. RORα Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation. International Journal of Molecular Sciences. 2021; 22(19):10665. https://doi.org/10.3390/ijms221910665
Chicago/Turabian StyleMao, Wei, Gaofeng Xiong, Yuanyuan Wu, Chi Wang, Daret St. Clair, Jia-Da Li, and Ren Xu. 2021. "RORα Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation" International Journal of Molecular Sciences 22, no. 19: 10665. https://doi.org/10.3390/ijms221910665
APA StyleMao, W., Xiong, G., Wu, Y., Wang, C., St. Clair, D., Li, J.-D., & Xu, R. (2021). RORα Suppresses Cancer-Associated Inflammation by Repressing Respiratory Complex I-Dependent ROS Generation. International Journal of Molecular Sciences, 22(19), 10665. https://doi.org/10.3390/ijms221910665





