In Silico Study of the RSH (RelA/SpoT Homologs) Gene Family and Expression Analysis in Response to PGPR Bacteria and Salinity in Brassica napus
Abstract
:1. Introduction
2. Results and Discussion
2.1. In Silico Analysis of RSH Genes and Proteins in B. napus and Selected Close Relatives from the Brassicaceae Family
2.1.1. Characteristics of Selected Brassicaceae RSH Genes
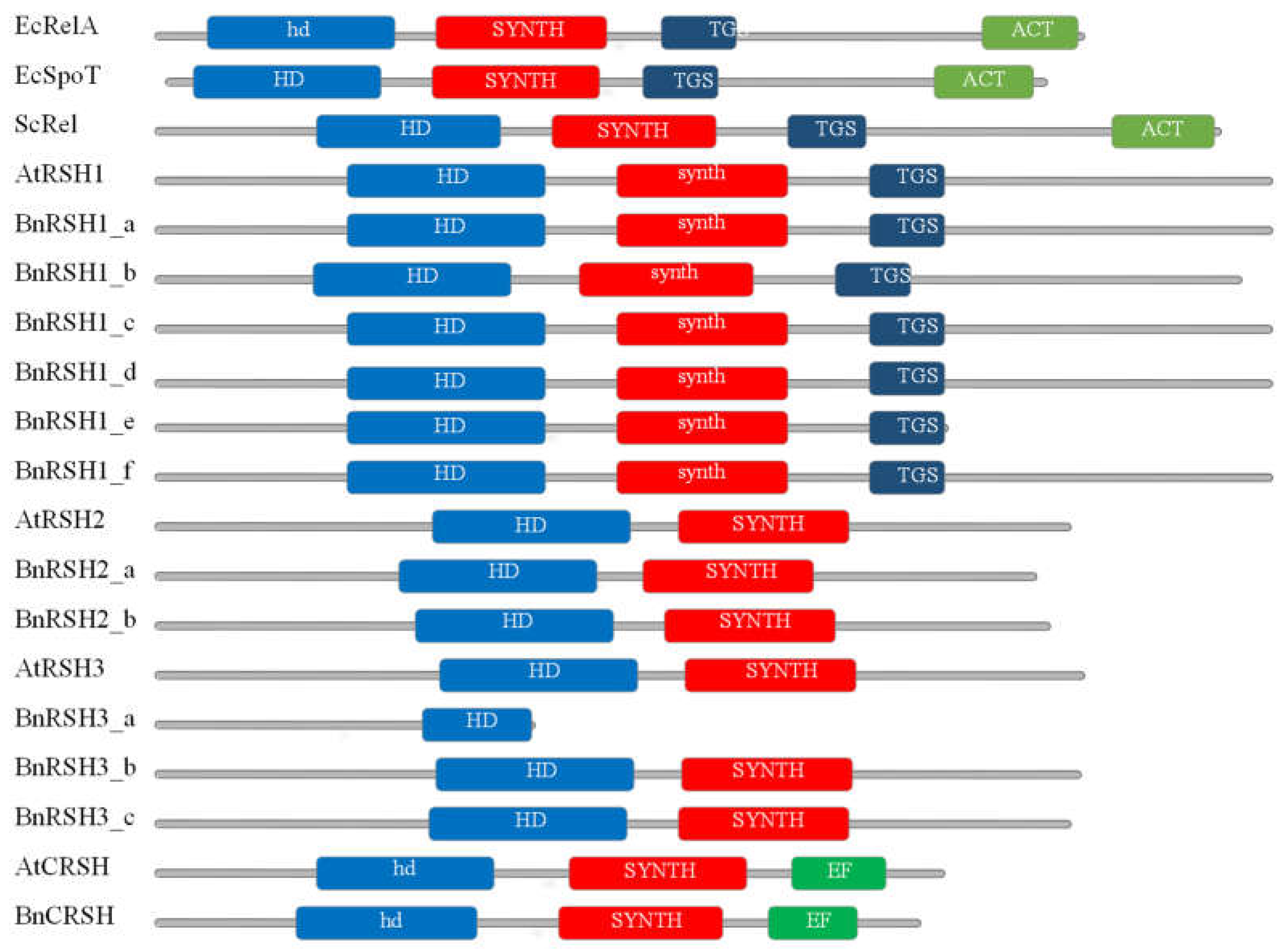
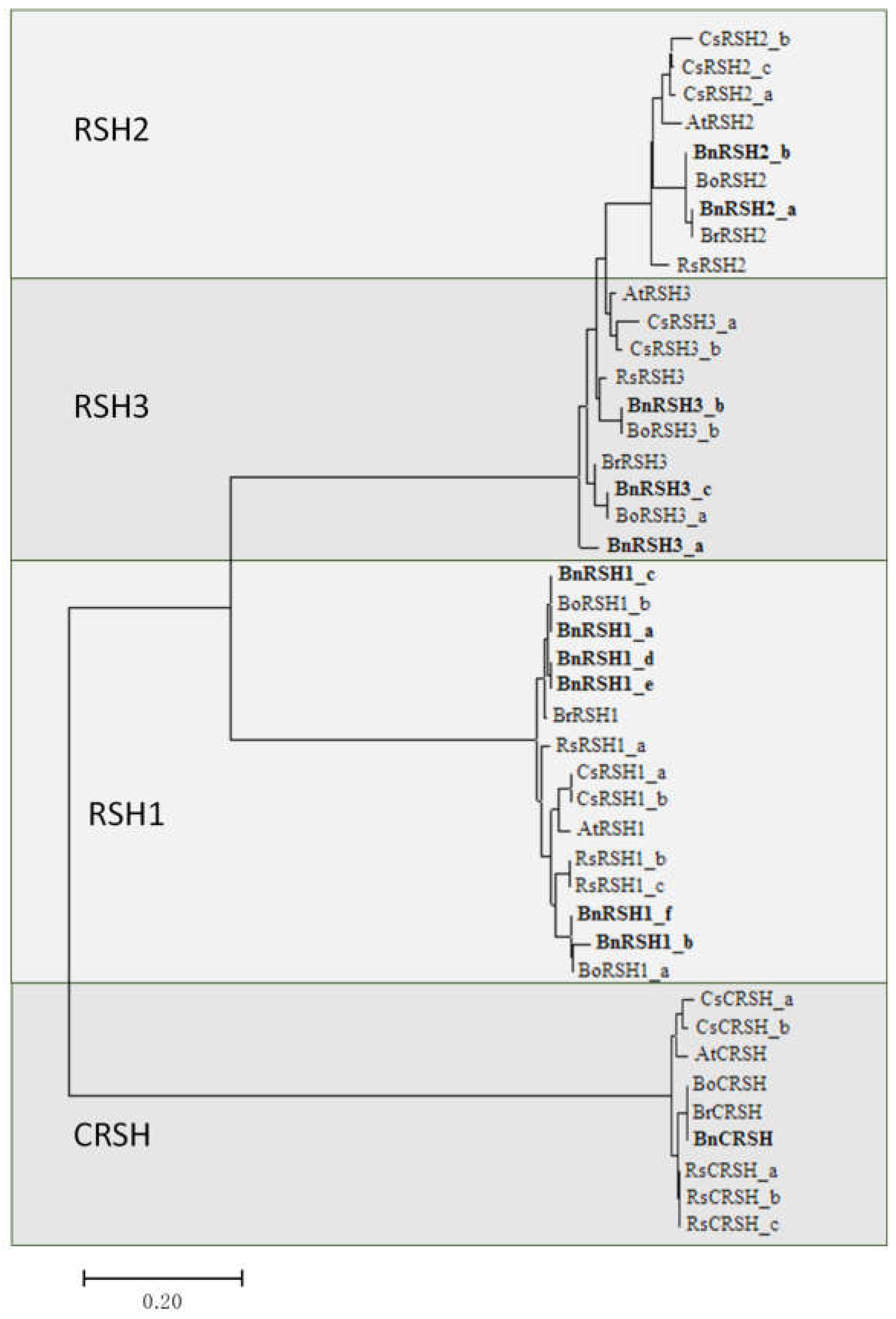
2.1.2. Characteristic of Selected Brassicaeae RSH Proteins
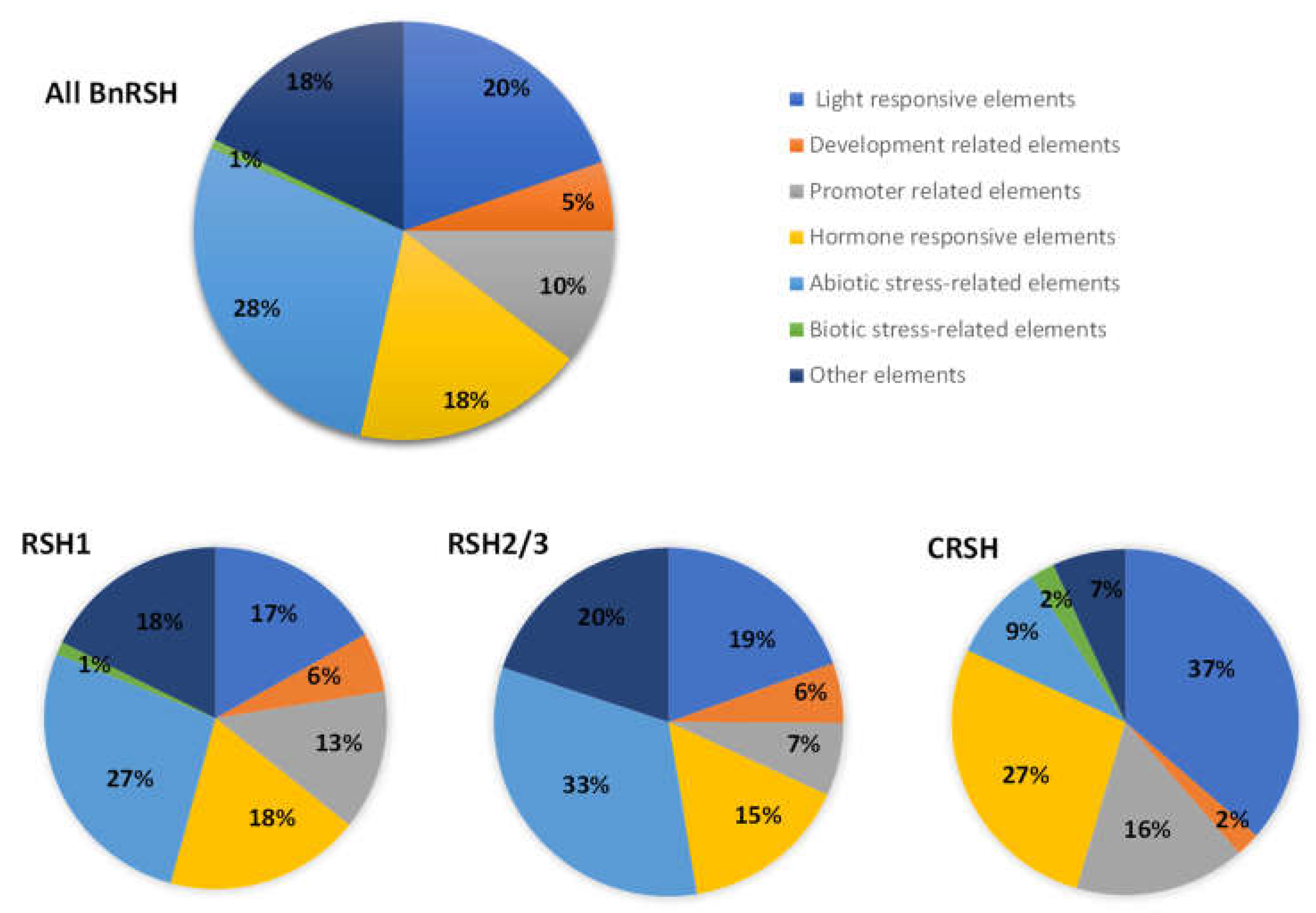
2.2. Regulatory Elements Present in Promoter Regions of B. napus RSH Genes
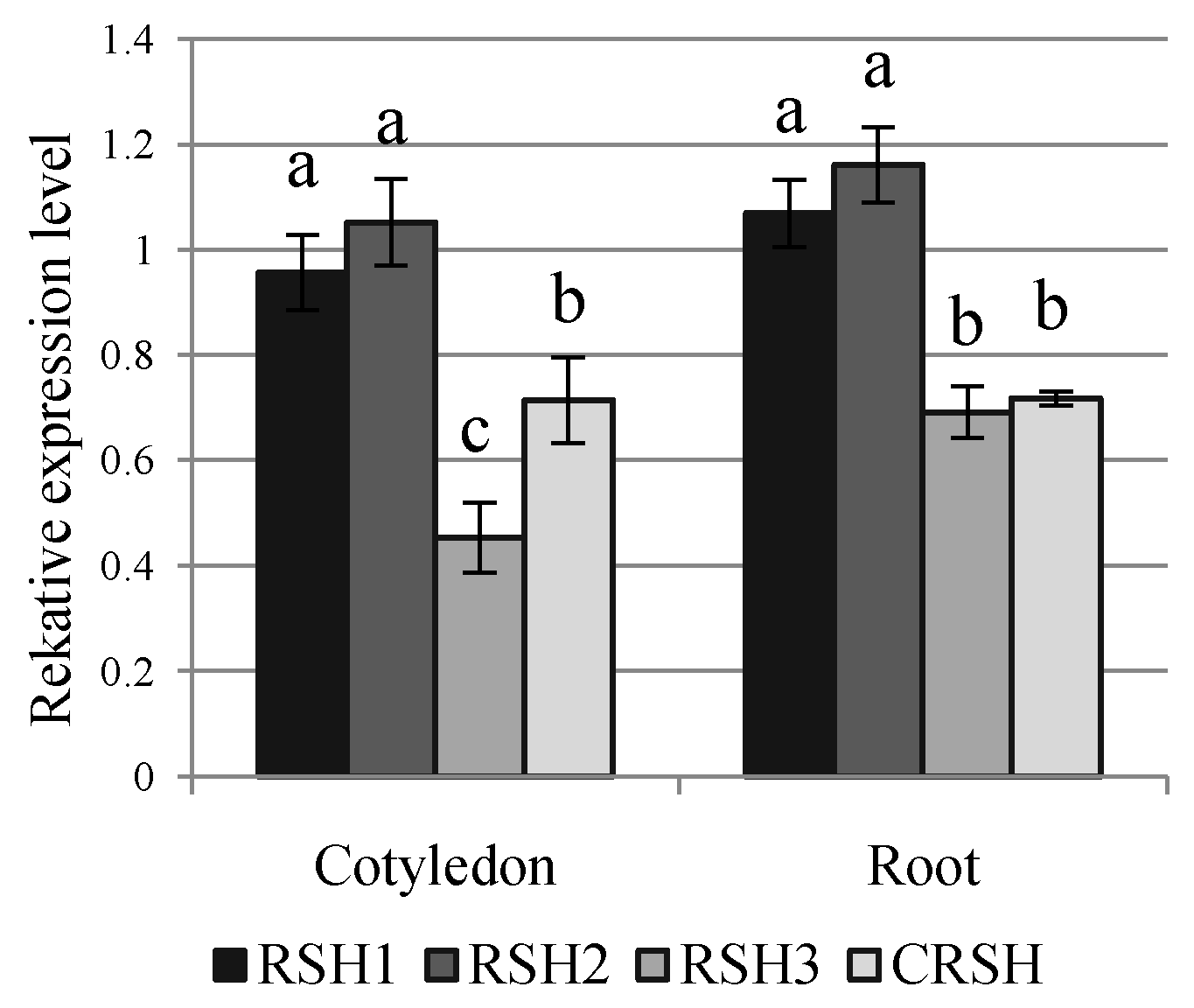
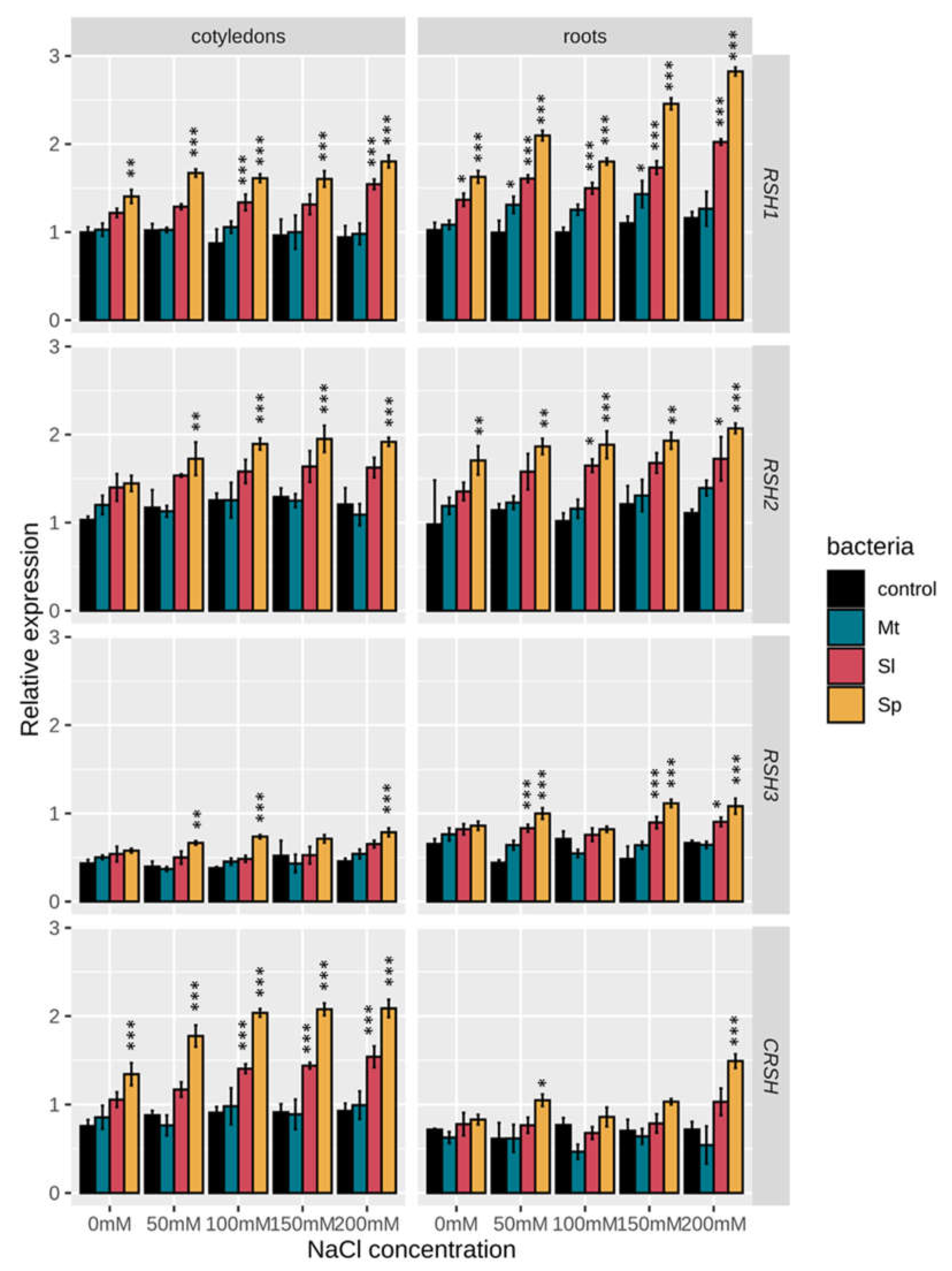
2.3. Effect of Salinity and Rhizobacteria on the Expression of BnRSH Genes
3. Materials and Methods
3.1. In Silico Analysis of B. napus, B. olearacea, B. rapa, C. sativa, and R. sativus RSH Genes and Proteins
3.2. Bacterial Strains
3.3. Plant Material
3.4. Expression Analysis of BnRSH Genes
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, S.K. Breeding Oilseed Crops for Sustainable Production: Opportunities and Constraints; Elsevier Inc.: Amsterdam, The Netherlands, 2015; ISBN 9780128014691. [Google Scholar]
- Warner, D.J.; Lewis, K.A. Evaluation of the risks of contaminating low erucic acid rapeseed with high erucic rapeseed and identification of mitigation strategies. Agriculture 2019, 9, 190. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Liang, Z.; Yan, T.; Xu, Y.; Xuan, L.; Tang, J.; Zhou, G.; Lohwasser, U.; Hua, S.; Wang, H.; et al. Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant 2019, 12, 30–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szymańska, S.; Dąbrowska, G.B.; Tyburski, J.; Niedojadło, K.; Piernik, A.; Hrynkiewicz, K. Boosting the Brassica napus L. tolerance to salinity by the halotolerant strain Pseudomonas stutzeri ISE12. Environ. Exp. Bot. 2019, 163, 55–68. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alzahrani, S.M.; Ali, H.M.; Alayafi, A.A.; Ahmad, M. Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 2018, 19, 3310. [Google Scholar] [CrossRef] [Green Version]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Dąbrowska, G.B.; Zdziechowska, E.; Hrynkiewicz, K. Evaluation of potential suitability of Rhizobacteria for phytodesalination of soils. Environ. Pollut. Control 2016, 38, 9–14. [Google Scholar]
- Shilev, S. Plant-growth-promoting bacteria mitigating soil salinity stress in plants. Appl. Sci. 2020, 10, 7326. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Kohler, J.; Hernández, J.A.; Caravaca, F.; Roldán, A. Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ. Exp. Bot. 2009, 65, 245–252. [Google Scholar] [CrossRef]
- Cordero, I.; Balaguer, L.; Rincón, A.; Pueyo, J.J. Inoculation of tomato plants with selected PGPR represents a feasible alternative to chemical fertilization under salt stress. J. Plant Nutr. Soil Sci. 2018, 181, 694–703. [Google Scholar] [CrossRef]
- Cashel, M.; Gallant, J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 1969, 224, 177–178. [Google Scholar] [CrossRef]
- Cashel, M.; Gentry, D.; VJ, H.; Vinella, D. The stringent response in: Escherichia coli and Salmonella typhimurium. Cell. Mol. Biol. 1996, 2, 1458–1496. [Google Scholar]
- Dabrowska, G.; Prusinska, J.; Goc, A. The stringent response-the mechanism of bacterial adaptive response to stress conditions. Adv. Biochem. 2006, 52, 87–93. [Google Scholar]
- Xiao, H.; Kalman, M.; Ikehara, K.; Zemel, S.; Glaser, G.; Cashel, M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991, 266, 5980–5990. [Google Scholar] [CrossRef]
- Berdychowska, J.; Boniecka, J.; Dąbrowska, G.B. The stringent response and its involvement in the actions of bacterial cells to stress. Adv. Microbiol. 2019, 58, 127–142. [Google Scholar] [CrossRef] [Green Version]
- Irving, S.E.; Choudhury, N.R.; Corrigan, R.M. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 2020, 19, 256–271. [Google Scholar] [CrossRef]
- Mittenhuber, G. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 2001, 3, 585–600. [Google Scholar]
- Heizmann, P.; Howell, S.H. Synthesis of ppGpp and chloroplast ribosomal RNA in Chlamydomonas reinhardi. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 1978, 517, 115–124. [Google Scholar] [CrossRef]
- Van der Biezen, E.A. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. USA 2000, 97, 3747–3752. [Google Scholar] [CrossRef]
- Dąbrowska, G.; Prusińska, J.; Goc, A. Identification of RSH gene cDNA (RelA/SpoT homolog) involved in Pharbitis nil response to stress condition. Adv. Agric. Sci. Probl. Issues 2006, 509, 333–341. [Google Scholar]
- Dabrowska, G.; Prusińska, J.; Goc, A. Plant mechanism of an adaptive stress response homologous to bacterial stringent response. Adv. Biochem. 2006, 52, 94–100. [Google Scholar]
- Boniecka, J.; Prusińska, J.; Dąbrowska, G.B.; Goc, A. Within and beyond the stringent response-RSH and (p)ppGpp in plants. Planta 2017, 246, 817–842. [Google Scholar] [CrossRef] [Green Version]
- Prusińska, J.M.; Boniecka, J.; Dąbrowska, G.B.; Goc, A. Identification and characterization of the Ipomoea nil RelA/SpoT Homologs (InRSHs) and potential directions of their transcriptional regulation. Plant Sci. 2019, 284, 161–176. [Google Scholar] [CrossRef]
- Mizusawa, K.; Masuda, S.; Ohta, H. Expression profiling of four RelA/SpoT-like proteins, homologues of bacterial stringent factors, in Arabidopsis thaliana. Planta 2008, 228, 553–562. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Aravind, L.; Grishin, N.V.; Koonin, E.V. Evolution of aminoacyl-tRNA synthetases-analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999, 9, 689–710. [Google Scholar] [CrossRef]
- Brown, A.; Fernández, I.S.; Gordiyenko, Y.; Ramakrishnan, V. Ribosome-dependent activation of stringent control. Nature 2016, 534, 277–280. [Google Scholar] [CrossRef] [Green Version]
- Loveland, A.B.; Bah, E.; Madireddy, R.; Zhang, Y.; Brilot, A.F.; Grigorieff, N.; Korostelev, A.A. Ribosome-RelA structures reveal the mechanism of stringent response activation. eLife 2016, 5, 1–23. [Google Scholar] [CrossRef]
- Ito, D.; Ihara, Y.; Nishihara, H.; Masuda, S. Phylogenetic analysis of proteins involved in the stringent response in plant cells. J. Plant Res. 2017, 130, 625–634. [Google Scholar] [CrossRef]
- Tozawa, Y.; Nozawa, A.; Kanno, T.; Narisawa, T.; Masuda, S.; Kasai, K.; Nanamiya, H. Calcium-activated (p)ppGpp synthetase in chloroplasts of land plants. J. Biol. Chem. 2007, 282, 35536–35545. [Google Scholar] [CrossRef] [Green Version]
- Ono, S.; Suzuki, S.; Ito, D.; Tagawa, S.; Shiina, T.; Masuda, S. Plastidial (p)ppGpp synthesis by the Ca2+-dependent RelA–SpoT homolog regulates the adaptation of chloroplast gene expression to darkness in Arabidopsis. Plant Cell Physiol. 2020, 61, 2077–2086. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lee, G.; Lee, J.; Kim, H.; Rhee, H.; Park, S.; Kim, K.; Kim, Y.; Kim, B.; Hong, J.; et al. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 2010, 17, 1188–1194. [Google Scholar] [CrossRef] [PubMed]
- Ito, D.; Kawamura, H.; Oikawa, A.; Ihara, Y.; Shibata, T.; Nakamura, N.; Asano, T.; Kawabata, S.-I.; Suzuki, T.; Masuda, S. ppGpp functions as an alarmone in metazoa. Commun. Biol. 2020, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Masuda, S. The stringent response in phototrophs. In Advances in Photosynthesis—Fundamental Aspects.; Najfpour, M., Ed.; InTech: Shanghai, China, 2012; pp. 487–500. ISBN 978-953-307-928-8. [Google Scholar]
- Atkinson, G.C.; Tenson, T.; Hauryliuk, V. The RelA/SpoT Homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 2011, 6, e23479. [Google Scholar] [CrossRef] [Green Version]
- Dabrowska, G.; Mierek-Adamska, A.; Goc, A. Plant metallothioneins: Putative functions identified by promoter analysis in silico. Acta Biol. Crac. Ser. Bot. 2012, 54, 109–120. [Google Scholar] [CrossRef]
- Pan, Y.; Zhu, M.; Wang, S.; Ma, G.; Huang, X.; Qiao, C.; Wang, R.; Xu, X.; Liang, Y.; Lu, K.; et al. Genome-wide characterization and analysis of metallothionein family genes that function in metal stress tolerance in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2181. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-wide identification, classification and expression analysis of the HSP gene superfamily in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. [Google Scholar] [CrossRef] [Green Version]
- Yuan, D.; Li, W.; Hua, Y.; King, G.J.; Xu, F.; Shi, L. Genome-wide identification and characterization of the aquaporin gene family and transcriptional responses to boron deficiency in Brassica napus. Front. Plant Sci. 2017, 8, 1336. [Google Scholar] [CrossRef] [Green Version]
- Sugliani, M.; Abdelkefi, H.; Ke, H.; Bouveret, E.; Robaglia, C.; Caffarri, S.; Field, B. An ancient bacterial signaling pathway regulates chloroplast function to influence growth and development in Arabidopsis. Plant Cell 2016, 28, 661–679. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Lee, M.W.; Qi, M.; Yang, Y. Identification of defense-related rice genes by suppression subtractive hybridization and differential screening. Mol. Plant Microbe Interact. 2001, 14, 685–692. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.H.; Ok, S.H.; Kim, D.; Suh, S.C.; Byun, M.O.; Shin, J.S. Molecular characterization of a biotic and abiotic stress resistance-related gene RelA/SpoT homologue (PepRSH) from pepper. Plant Sci. 2009, 176, 635–642. [Google Scholar] [CrossRef]
- Givens, R.M.; Lin, M.H.; Taylor, D.J.; Mechold, U.; Berry, J.O.; Hernandez, V.J. Inducible expression, enzymatic activity, and origin of higher plant homologues of bacterial RelA/SpoT stress proteins in Nicotiana tabacum. J. Biol. Chem. 2004, 279, 7495–7504. [Google Scholar] [CrossRef] [Green Version]
- Yamada, A.; Tsutsumi, K.; Tanimoto, S.; Ozeki, Y. Plant RelA/SpoT homolog confers salt tolerance in Escherichia coli and Saccharomyces cerevisiae. Plant Cell Physiol. 2003, 44, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Morello, L.; Breviario, D. Plant spliceosomal introns: Not only cut and paste. Curr. Genom. 2008, 9, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Frey, K.; Pucker, B. Animal, fungi, and plant genome sequences harbor different non-canonical splice sites. Cells 2020, 9, 458. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.-B.; Brendel, V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef] [Green Version]
- Jo, B.-S.; Choi, S.S. Introns: The functional benefits of introns in genomes. Genom. Inform. 2015, 13, 112. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.Y.; Vorst, O.; Fiers, M.W.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Masuda, S.; Mizusawa, K.; Narisawa, T.; Tozawa, Y.; Ohta, H.; Takamiya, K.I. The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 2008, 49, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, M.; Honoki, R.; Ihara, Y.; Sato, R.; Oikawa, A.; Kanno, Y.; Ohta, H.; Seo, M.; Saito, K.; Masuda, S. Impact of the plastidial stringent response in plant growth and stress responses. Nat. Plants 2015, 1, 1–7. [Google Scholar] [CrossRef]
- Sato, M.; Takahashi, T.; Ochi, K.; Matsuura, H.; Nabeta, K.; Takahashi, K. Overexpression of RelA/SpoT homologs, PpRSH2a and PpRSH2b, induces the growth suppression of the moss Physcomitrella patens. Biosci. Biotechnol. Biochem. 2015, 79, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Kasai, K.; Ochi, K. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. USA 2004, 101, 4320–4324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, B. Green magic: Regulation of the chloroplast stress response by (p)ppGpp in plants and algae. J. Exp. Bot. 2018, 69, 2797–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamburenko, M.V.; Zubo, Y.O.; Börner, T. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: Repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J. 2015, 82, 1030–1041. [Google Scholar] [CrossRef]
- Ito, D.; Kato, T.; Maruta, T.; Tamoi, M.; Yoshimura, K.; Shigeoka, S. Enzymatic and molecular characterization of Arabidopsis ppGpp pyrophosphohydrolase, AtNUDX26. Biosci. Biotechnol. Biochem. 2012, 76, 2236–2241. [Google Scholar] [CrossRef] [Green Version]
- Abdelkefi, H.; Sugliani, M.; Ke, H.; Harchouni, S.; Soubigou-Taconnat, L.; Citerne, S.; Mouille, G.; Fakhfakh, H.; Robaglia, C.; Field, B. Guanosine tetraphosphate modulates salicylic acid signalling and the resistance of Arabidopsis thaliana to Turnip mosaic virus. Mol. Plant Pathol. 2018, 19, 634–646. [Google Scholar] [CrossRef] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.-C.; Lee, T.-Y.; Huang, H.-D.; Huang, H.-Y.; Pan, R.-L. PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genom. 2008, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Ihara, Y.; Ohta, H.; Masuda, S. A highly sensitive quantification method for the accumulation of alarmone ppGpp in Arabidopsis thaliana using UPLC-ESI-qMS/MS. J. Plant Res. 2015, 128, 511–518. [Google Scholar] [CrossRef]
- Etesami, H.; Noori, F. Soil Salinity as a Challenge for Sustainable Agriculture and Bacterial-Mediated Alleviation of Salinity Stress in Crop Plants BT-Saline Soil-based Agriculture by Halotolerant Microorganisms; Kumar, M., Etesami, H., Kumar, V., Eds.; Springer Singapore: Singapore, 2019; pp. 1–22. ISBN 978-981-13-8335-9. [Google Scholar]
- Ashraf, M.; McNeilly, T. Salinity tolerance in Brassica oilseeds. Crit. Rev. Plant Sci. 2004, 23, 157–174. [Google Scholar] [CrossRef]
- Chen, J.; Bang, W.Y.; Lee, Y.; Kim, S.; Lee, K.W.; Kim, S.W.; Son, Y.S.; Kim, D.W.; Akhter, S.; Bahk, J.D. AtObgC-AtRSH1 interaction may play a vital role in stress response signal transduction in Arabidopsis. Plant Physiol. Biochem. 2014, 74, 176–184. [Google Scholar] [CrossRef]
- Brouwer, C.; Goffeau, A.; Heibloem, M. Irrigation Water Management: Training Manual No. 1—Introduction to Irrigation; FAO: Rome, Italy, 1985. [Google Scholar]
- Nawaz, A.; Shahbaz, M.; Asadullah; Imran, A.; Marghoob, M.U.; Imtiaz, M.; Mubeen, F. Potential of salt tolerant PGPR in growth and yield augmentation of wheat (Triticum aestivum L.) under saline conditions. Front. Microbiol. 2020, 11, 2019. [Google Scholar] [CrossRef]
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Al-Huqail, A.A.; Wirth, S.; Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 2016, 7, 1089. [Google Scholar] [CrossRef] [Green Version]
- Janczak, K.; Hrynkiewicz, K.; Znajewska, Z.; Dąbrowska, G. Use of rhizosphere microorganisms in the biodegradation of PLA and PET polymers in compost soil. Int. Biodeterior. Biodegrad. 2018, 130, 65–75. [Google Scholar] [CrossRef]
- Janczak, K.; Dąbrowska, G.B.; Raszkowska-Kaczor, A.; Kaczor, D.; Hrynkiewicz, K.; Richert, A. Biodegradation of the plastics PLA and PET in cultivated soil with the participation of microorganisms and plants. Int. Biodeterior. Biodegrad. 2020, 155, 105087. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd-Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Miguel, P.S.B.; de Oliveira, M.N.V.; Delvaux, J.C.; de Jesus, G.L.; Borges, A.C.; Tótola, M.R.; Neves, J.C.L.; Costa, M.D. Diversity and distribution of the endophytic bacterial community at different stages of Eucalyptus growth. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2016, 109, 755–771. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Ru, Y.; Brendel, V.P. Common introns within orthologous genes: Software and application to plants. Brief. Bioinform. 2009, 10, 631–644. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; Von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [Green Version]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Hrynkiewicz, K.; Złoch, M.; Kowalkowski, T.; Baum, C.; Niedojadło, K.; Buszewski, B. Strain-specific bioaccumulation and intracellular distribution of Cd2+ in bacteria isolated from the rhizosphere, ectomycorrhizae, and fruitbodies of ectomycorrhizal fungi. Environ. Sci. Pollut. Res. 2015, 22, 3055–3067. [Google Scholar] [CrossRef] [Green Version]
- Hrynkiewicz, K.; Baum, C.; Leinweber, P. Density, metabolic activity, and identity of cultivable rhizosphere bacteria on Salix viminalis in disturbed arable and landfill soils. J. Plant Nutr. Soil Sci. 2010, 173, 747–756. [Google Scholar] [CrossRef]
- Janczak, K.; Dąbrowska, G. Bacteria able to polylactide and polycaprolactone biodegradation. Przem. Chem. 2018, 97, 435–438. [Google Scholar] [CrossRef]
- Mierek-Adamska, A.; Kotowicz, K.; Goc, A.; Boniecka, J.; Berdychowska, J.; Dąbrowska, G.B. Potential involvement of rapeseed (Brassica napus L.) metallothioneins in the hydrogen peroxide-induced regulation of seed vigour. J. Agron. Crop Sci. 2019, 205, 598–607. [Google Scholar] [CrossRef]
- Marone, M.; Mozzetti, S.; De Ritis, D.; Pierelli, L.; Scambia, G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcriptsfrom the same sample. Biol. Proced. Online 2001, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Species | Genes | Gene ID | Transcript ID | CDS (bp) | Chromosome Location | Protein ID | AA | pI | Mw (kD) | Introns | Exons | Predicted Transfer Peptide (Probability) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. thaliana | RSH1 | 828096 | NM_116459.4 | 2655 | 4 | NP_567226.1 | 883 | 6.65 | 98.58 | 23 | 24 | cTP (0.455), mTP (0.0002), tlTP (0.0051), other (0.5393) |
| RSH2 | 820619 | NM_112259.5 | 2130 | 3 | NP_188021.1 | 709 | 6.89 | 79.05 | 5 | 6 | cTP (0.6081), mTP (0.0003), tlTP (0.0846), other (0.3047) | |
| RSH3 | 841853 | NM_104291.8 | 2148 | 1 | NP_564652.2 | 715 | 6.66 | 79.72 | 5 | 6 | cTP (0.7887), mTP (0.0024), tlTP (0.0397), other (0.1669) | |
| CRSH | 821012 | NM_001338291.1 | 1752 | 3 | NP_001327079.1 | 598 | 6.14 | 68.28 | 3 | 4 | cTP (0.0708), mTP (0.1949), tlTP (0.0002), other (0.7341) | |
| B. napus | RSH1_a | 106345251 | XM_013784481.2 | 2652 | unknown | XP_013639935.1 | 883 | 6.64 | 98.56 | 23 | 24 | cTP (0.3786), mTP (0.0002), tlTP (0.0059), other (0.6149) |
| RSH1_b * | 106399012 | XM_013839498.2 | 2565 | A5 | XP_013694952.1 | 854 | 6.60 | 95.88 | 22 | 23 | cTP (0.4978), mTP (0.0025), tlTP(0.004), other (0.4957) | |
| RSH1_c | 106436227 | XM_013877186.2 | 2652 | A8 | XP_013732640.1 | 883 | 6.64 | 98.58 | 23 | 24 | cTP (0.3786), mTP (0.0002), tlTP(0.0059), other (0.6149) | |
| RSH1_d | 106365508 | XM_013804925.2 | 2652 | A9 | XP_013660379.1 | 883 | 6.48 | 98.55 | 23 | 24 | cTP (0.5232), mTP (0.0005), tlTP(0.0171), other (0.459) | |
| RSH1_e | 106362473 | XM_013802370.2 | 1860 | A9 | XP_013657824.1 | 619 | 6.62 | 69.26 | 19 | 20 | cTP (0.5232), mTP (0.0005), tlTP(0.0171), other (0.459) | |
| RSH1_f | 106381614 | XM_013821535.2 | 2640 | C2 | XP_013676989.1 | 879 | 6.60 | 98.44 | 23 | 24 | cTP (0.2021), mTP (0.0011), tlTP(0.0043), other (0.7924) | |
| RSH2_a | 106452255 | XM_013894318.2 | 2055 | A5 | XP_013749772.2 | 684 | 6.67 | 77.13 | 5 | 6 | cTP (0.2425), mTP (0.0001), tlTP(0.0058), other (0.7498) | |
| RSH2_b * | 111206471 | XM_022703426.1 | 2091 | C5 | XP_022559147.1 | 696 | 6.56 | 77.98 | 5 | 6 | cTP (0.4948), mTP (0.0003), tlTP(0.0816), other (0.4229) | |
| RSH3_a * | 106345829 | XM_013785013.2 | 861 | A6 | XP_013640467.1 | 286 | 6.04 | 31.11 | 1 | 2 | cTP (0.6255), mTP (0.0001), tlTP(0.0181), other (0.3557) | |
| RSH3_b | 106431664 | XM_013872470.2 | 2133 | C6 | XP_013727924.1 | 710 | 6.50 | 78.78 | 5 | 6 | cTP (0.7621), mTP (0.0005), tlTP(0.0889), other (0.1473) | |
| RSH3_c | 106348454 | XM_022704818.1 | 2109 | C6 | XP_022560539.1 | 702 | 6.77 | 78.07 | 6 | 7 | cTP (0.5288), mTP (0.0001), tlTP(0.0244), other (0.446) | |
| RSH3 _pseudo | 106345828 | - | - | A6 | - | - | - | - | - | - | - | |
| CRSH * | 106389210 | XM_013829418.2 | 1743 | C3 | XP_013684872.1 | 580 | 6.03 | 65.83 | 3 | 4 | cTP (0.2211), mTP (0.0648), tlTP(0.0095), other (0.7045) | |
| CRSH _pseudo | 106439579 | - | - | A3 | - | - | - | - | - | - | - | |
| B. oleracea | RSH1_a | 106327624 | XM_013765826.1 | 2628 | C2 | XP_013621280.1 | 875 | 6.52 | 97.87 | 23 | 24 | cTP (0.1379), mTP (0.0016), tlTP(0.0142), other (0.8462) |
| RSH1_b | 106318815 | XM_013756949.1 | 2652 | C9 | XP_013612403.1 | 883 | 6.64 | 98.55 | 23 | 24 | cTP (0.3786), mTP (0.0002), tlTP(0.0059), other (0.6149) | |
| RSH2 | 106295267 | XM_013731123.1 | 2091 | C5 | XP_013586577 | 696 | 6.56 | 77.98 | 5 | 6 | cTP (0.4948), mTP (0.0003), tlTP(0.0816), other (0.4229) | |
| RSH3_a | 106300657 | XM_013736852.1 | 2109 | C6 | XP_013592306.1 | 702 | 6.77 | 78.07 | 5 | 6 | cTP (0.5288), mTP (0.0001), tlTP(0.0244), other (0.446) | |
| RSH3_b | 106300381 | XM_013736509.1 | 2133 | C6 | XP_013591963.1 | 710 | 6.50 | 78.78 | 5 | 6 | cTP (0.7621), mTP (0.0005), tlTP(0.0889), other (0.1473) | |
| CRSH | 106334911 | XM_013773298.1 | 1743 | C3 | XP_013628752.1 | 580 | 6.11 | 65.88 | 2 | 3 | cTP (0.2263), mTP (0.0536), tlTP(0.0104), other (0.7096) | |
| B. rapa | RSH1 | 103836764 | XM_033278751.1 | 2685 | A9 | XP_033134642.1 | 894 | 6.38 | 100.02 | 23 | 24 | cTP (0.5255), mTP (0.0005), tlTP(0.0172), other (0.4566) |
| RSH2 | 103870072 | XM_009148172.3 | 2064 | A5 | XP_009146420.1 | 687 | 6.67 | 77.3 | 5 | 6 | cTP (0.2318), mTP (0.0001), tlTP(0.0048), other (0.7612) | |
| RSH3 | 103871068 | XM_009149293.3 | 2091 | A6 | XP_009147541.1 | 696 | 6.30 | 77.87 | 5 | 6 | cTP (0.6833), mTP (0.0001), tlTP(0.0135), other (0.3026) | |
| CRSH | 103859710 | XM_009137283.3 | 1731 | A3 | XP_009135531.1 | 576 | 5.99 | 65.47 | 2 | 3 | cTP (0.3013), mTP (0.0173), tlTP(0.0221), other (0.6593) | |
| C. sativa | RSH1_a | 104747094 | XM_010468670.2 | 2664 | 2 | XP_010466972.1 | 887 | 6.66 | 98.83 | 24 | 25 | cTP (0.5423), mTP (0.0003), tlTP(0.0096), other (0.4475) |
| RSH1_b | 104737555 | XM_010457755.2 | 2655 | 13 | XP_010456057.1 | 884 | 6.56 | 98.56 | 25 | 26 | cTP (0.3518), mTP (0.0012), tlTP(0.0168), other (0.6298) | |
| RSH1 _pseudo | 104707874 | - | - | 8 | - | - | - | - | - | - | - | |
| RSH2_a | 104778842 | XM_010503267.2 | 2154 | 1 | XP_010501569.1 | 717 | 6.57 | 79.77 | 6 | 7 | cTP (0.4377), mTP (0.0002), tlTP(0.0256), other (0.5339) | |
| RSH2_b | 104788263 | XM_010513997.2 | 630 | 5 | XP_010512299.1 | 209 | 7.72 | 23.53 | 3 | 4 | cTP (0), mTP (0), tlTP(0), other (0.9999) | |
| RSH2_c | 104745674 | XM_010466982.2 | 2148 | 15 | XP_010465284.1 | 715 | 6.42 | 79.56 | 6 | 7 | cTP (0.3945), mTP (0.0001), tlTP(0.0761), other (0.5286) | |
| RSH3_a | 104778355 | XM_010502782.2 | 2151 | 3 | XP_010501084.1 | 716 | 6.19 | 80.09 | 5 | 6 | cTP (0.8885), mTP (0.002), tlTP(0.0436), other (0.0634) | |
| RSH3_b | 104758764 | XM_010481702.2 | 2151 | 17 | XP_010480004.1 | 716 | 6.77 | 79.75 | 5 | 6 | cTP (0.7837), mTP (0.0027), tlTP(0.0167), other (0.1939) | |
| RSH3 _pseudo 1 | 104742935 | - | - | 14 | - | - | - | - | - | - | - | |
| RSH3 _pseudo 2 | 104761544 | - | - | 18 | - | - | - | - | - | - | - | |
| CRSH_a | 104782095 | XM_010506922.2 | 1764 | 1 | XP_010505224.1 | 587 | 6.20 | 66.97 | 3 | 4 | cTP (0.2705), mTP (0.0903), tlTP(0.0037), other (0.6353) | |
| CRSH_b | 104765592 | XM_010489335.2 | 1758 | 19 | XP_010487637.1 | 585 | 6.07 | 66.89 | 3 | 4 | cTP (0.0869), mTP (0.1501), tlTP(0.0008), other (0.7621) | |
| R. sativus | RSH1_a | 108828360 | XM_018602017.1 | 2640 | unknown | XP_018457519.1 | 879 | 6.78 | 97.76 | 23 | 24 | cTP (0.6503), mTP (0.0013), tlTP(0.0287), other (0.3196) |
| RSH1_b | 108843457 | XM_018616659.1 | 2601 | unknown | XP_018472161.1 | 866 | 6.96 | 97 | 23 | 24 | cTP (0.1051), mTP (0.0009), tlTP(0.0005), other (0.8934) | |
| RSH1_c | 108834481 | XM_018607822.1 | 1290 | unknown | XP_018463324.1 | 429 | 7.56 | 48.26 | 13 | 14 | cTP (0.1051), mTP (0.0009), tlTP(0.0005), other (0.8934) | |
| RSH2 | 108863143 | XM_018637469.1 | 2037 | unknown | XP_018492971.1 | 678 | 6.55 | 76.31 | 5 | 6 | cTP (0.2086), mTP (0), tlTP(0.0096), other (0.7815) | |
| RSH3 | 108862601 | XM_018636787.1 | 2121 | unknown | XP_018492289.1 | 706 | 6.44 | 78.36 | 6 | 7 | cTP (0.6764), mTP (0.0001), tlTP(0.1784), other (0.1413) | |
| RSH3 _pseudo | 108815328 | - | - | unknown | - | - | - | - | - | - | - | |
| CRSH_a | 108857634 | XM_018631638.1 | 1749 | unknown | XP_018487140.1 | 582 | 6.06 | 66.11 | 3 | 4 | cTP (0.1098), mTP (0.0301), tlTP(0.0031), other (0.857) | |
| CRSH_b | 108857621 | XM_018631622.1 | 1749 | unknown | XP_018487124.1 | 582 | 6.06 | 66.11 | 3 | 4 | cTP (0.1098), mTP (0.0301), tlTP(0.0031), other (0.857) | |
| CRSH_c | 108857284 | XM_018631245.1 | 1749 | unknown | XP_018486747.1 | 582 | 6.06 | 66.08 | 3 | 4 | cTP (0.1098), mTP (0.0301), tlTP(0.0031), other (0.857) | |
| E. coli | RelA | 947244 | - | 2235 | - | NP_417264.1 | 744 | 6.29 | 83.89 | - | - | - |
| SpoT | 948159 | - | 2109 | - | NP_418107.1 | 702 | 8.89 | 79.34 | - | - | - | |
| S. coelicolor | Rel | 1096939 | - | 2544 | - | WP_003977314.1 | 847 | 9.36 | 94.2 | - | - | - |
| Primer Name | Sequence of Primers 5′–3′ | Analysed Gene and Amplicon Length [bp] |
|---|---|---|
| BnRSH1_f BnRSH1_r | GGAGGTTCAGATCAGAACGG CCATTCACCTTCGCTGCTAC | BnRSH1 396 |
| BnRSH2_f BnRSH2_r | GCAAGATGTTGAAGAATCTAACG GCACAGACATCTTGTCATTTTCG | BnRSH2 534 |
| BnRSH3_f BnRSH3_r | CCGAAACTTTCCGATTTCAA TCGTAGTCAACGCACGAGTC | BnRSH3 524 |
| BnCRSH_f BnCRSH_r | AAGTGATGGAGGAGCTTGGA CCATTTACTGGAACGCAACA | BnCRSH 263 |
| BnAc_f BnAc_r | CTCACGCTATCCTCCGTCTC TTGATCTTCATGCTGCTTGG | BnAc 469 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dąbrowska, G.B.; Turkan, S.; Tylman-Mojżeszek, W.; Mierek-Adamska, A. In Silico Study of the RSH (RelA/SpoT Homologs) Gene Family and Expression Analysis in Response to PGPR Bacteria and Salinity in Brassica napus. Int. J. Mol. Sci. 2021, 22, 10666. https://doi.org/10.3390/ijms221910666
Dąbrowska GB, Turkan S, Tylman-Mojżeszek W, Mierek-Adamska A. In Silico Study of the RSH (RelA/SpoT Homologs) Gene Family and Expression Analysis in Response to PGPR Bacteria and Salinity in Brassica napus. International Journal of Molecular Sciences. 2021; 22(19):10666. https://doi.org/10.3390/ijms221910666
Chicago/Turabian StyleDąbrowska, Grażyna B., Sena Turkan, Wioleta Tylman-Mojżeszek, and Agnieszka Mierek-Adamska. 2021. "In Silico Study of the RSH (RelA/SpoT Homologs) Gene Family and Expression Analysis in Response to PGPR Bacteria and Salinity in Brassica napus" International Journal of Molecular Sciences 22, no. 19: 10666. https://doi.org/10.3390/ijms221910666
APA StyleDąbrowska, G. B., Turkan, S., Tylman-Mojżeszek, W., & Mierek-Adamska, A. (2021). In Silico Study of the RSH (RelA/SpoT Homologs) Gene Family and Expression Analysis in Response to PGPR Bacteria and Salinity in Brassica napus. International Journal of Molecular Sciences, 22(19), 10666. https://doi.org/10.3390/ijms221910666






