Differentiation of the High Night Temperature Response in Leaf Segments of Rice Cultivars with Contrasting Tolerance
Abstract
1. Introduction
2. Results
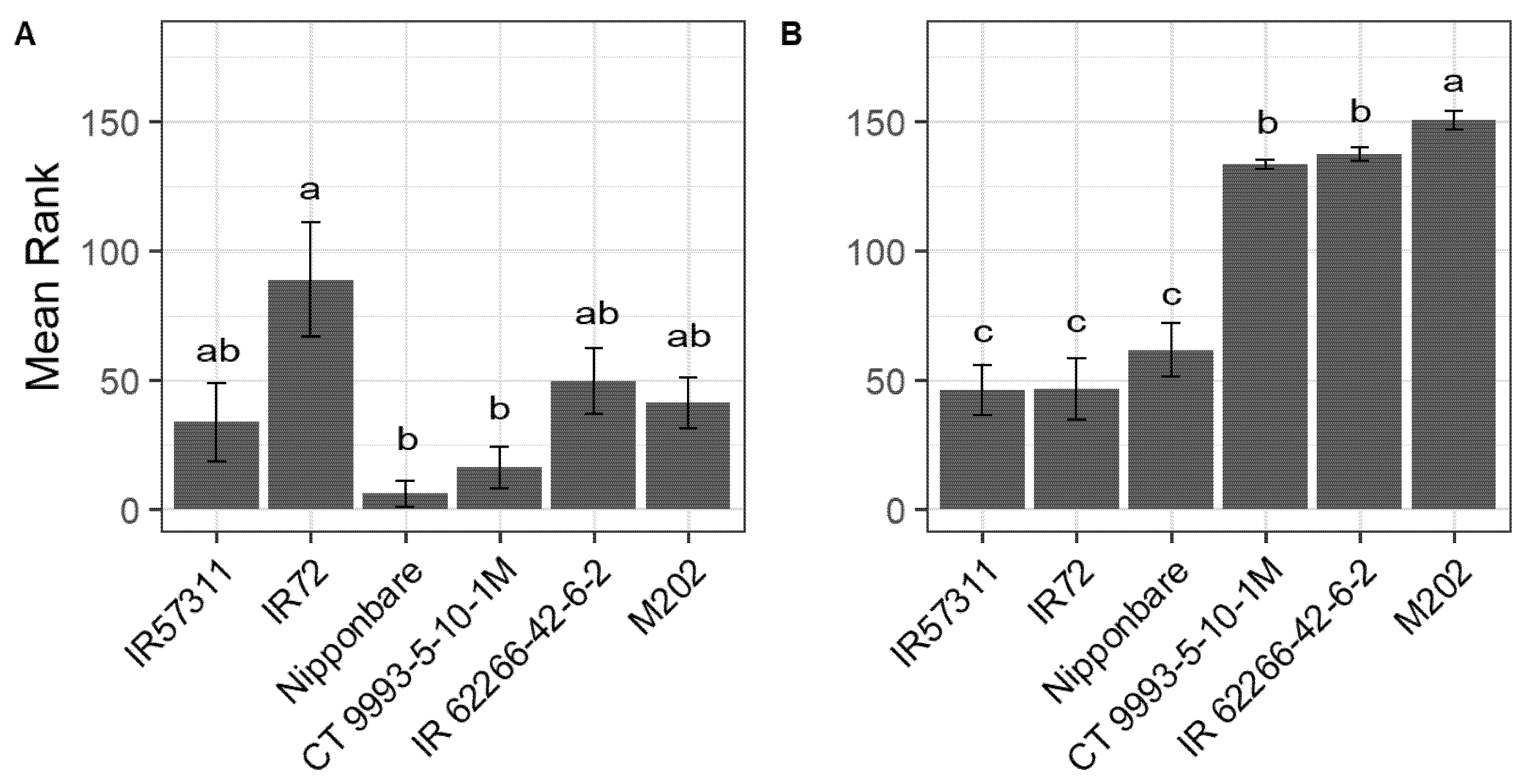
2.1. Sensitive, Intermediate and Tolerant Cultivars Showed Different Phenotypes under HNT
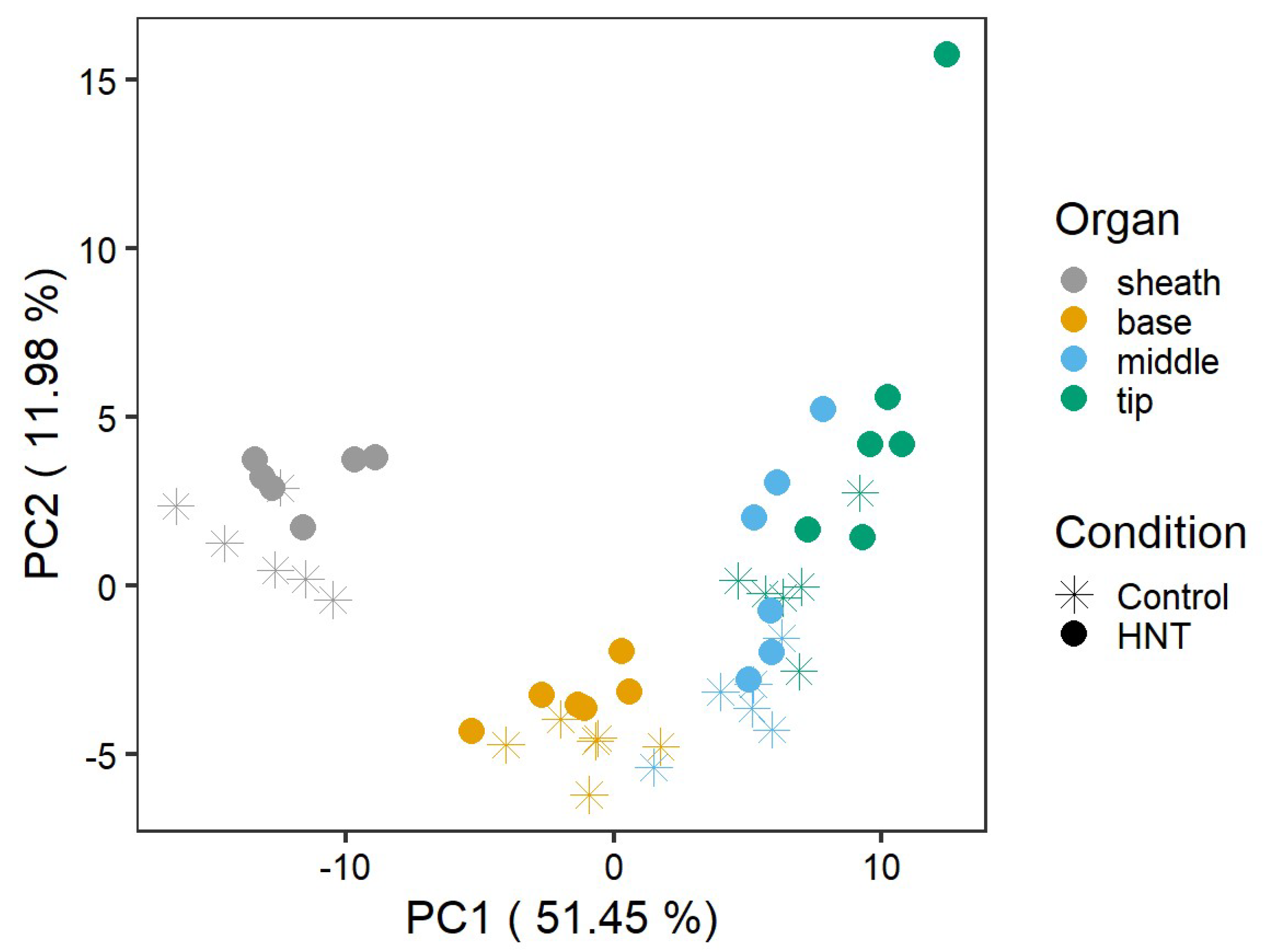
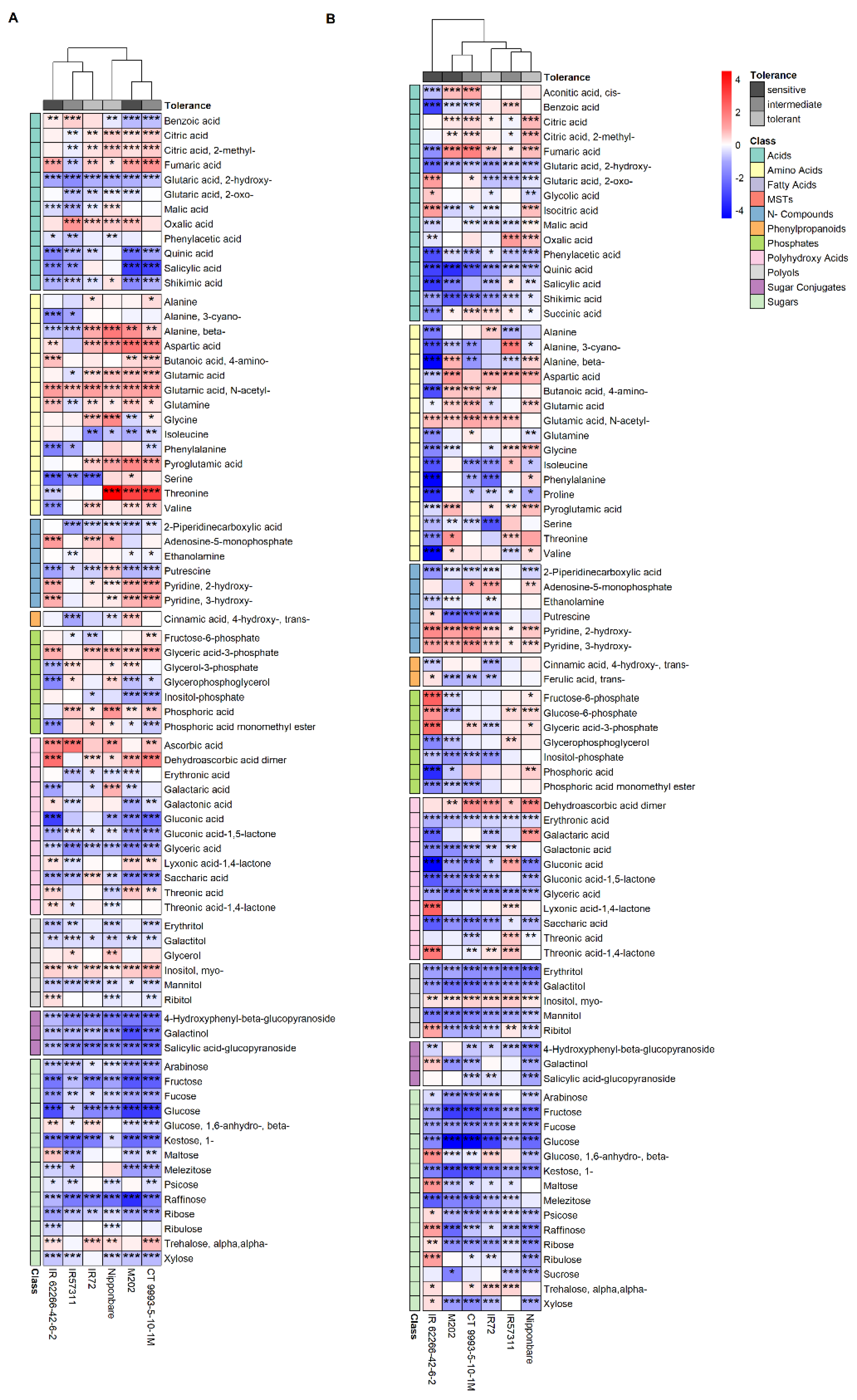
2.2. Metabolite Profile of the Sheath Differed from That of Leaf Blade Segments under Control Conditions
2.3. Leaf Segments Have Distinct Metabolite Profiles under HNT Stress
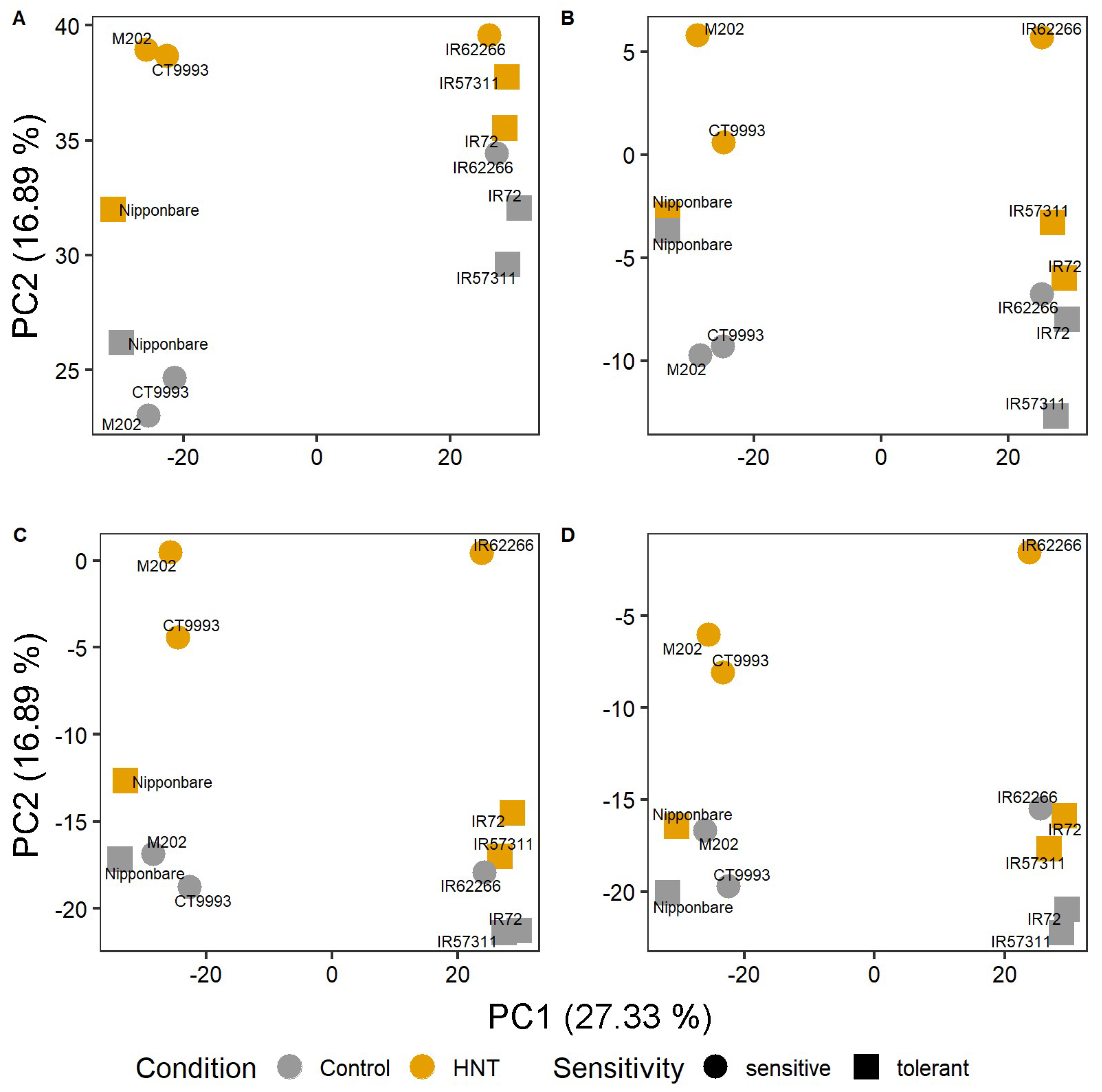
2.4. Gene Expression in the Middle Part of the Leaf and the Sheath of Sensitive Cultivars Is Most Highly Affected by HNT Conditions
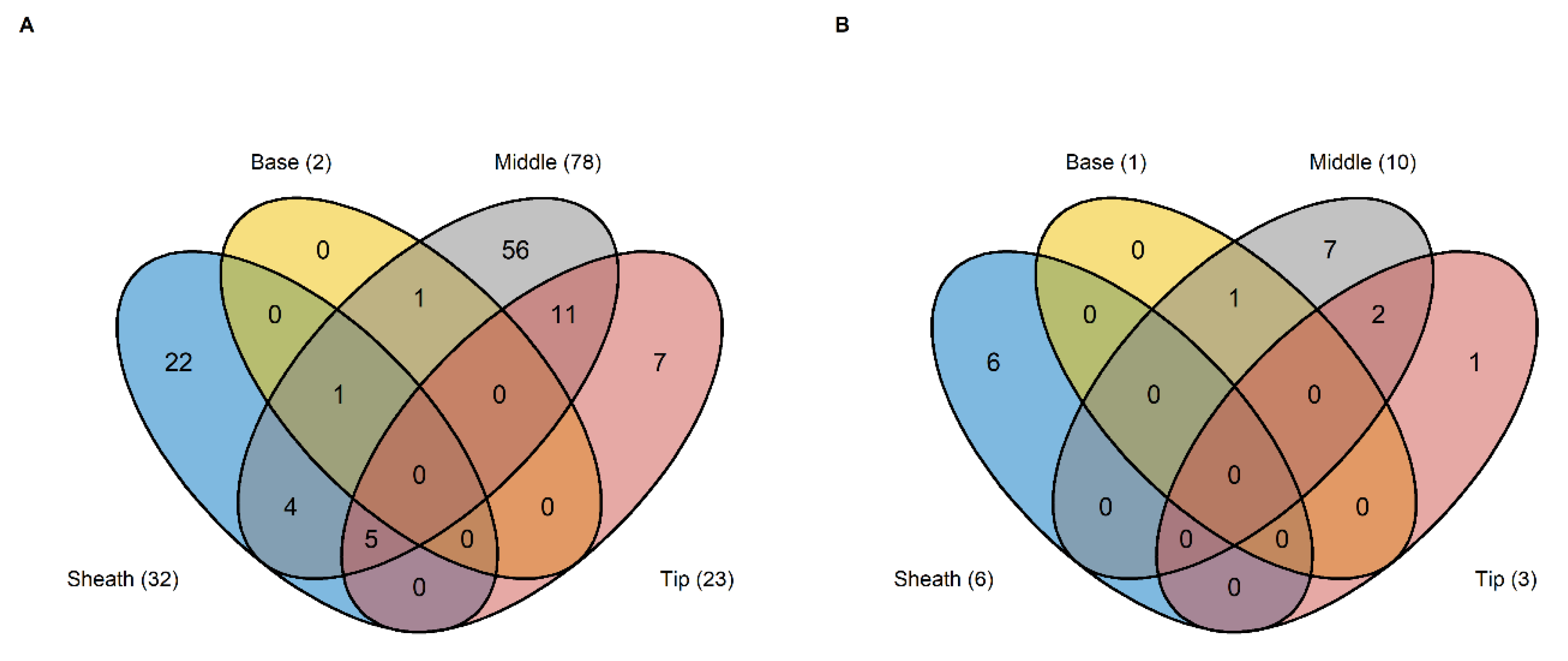
2.5. Identification of Segment Specific Differentially Expressed Genes Regulating HNT Response
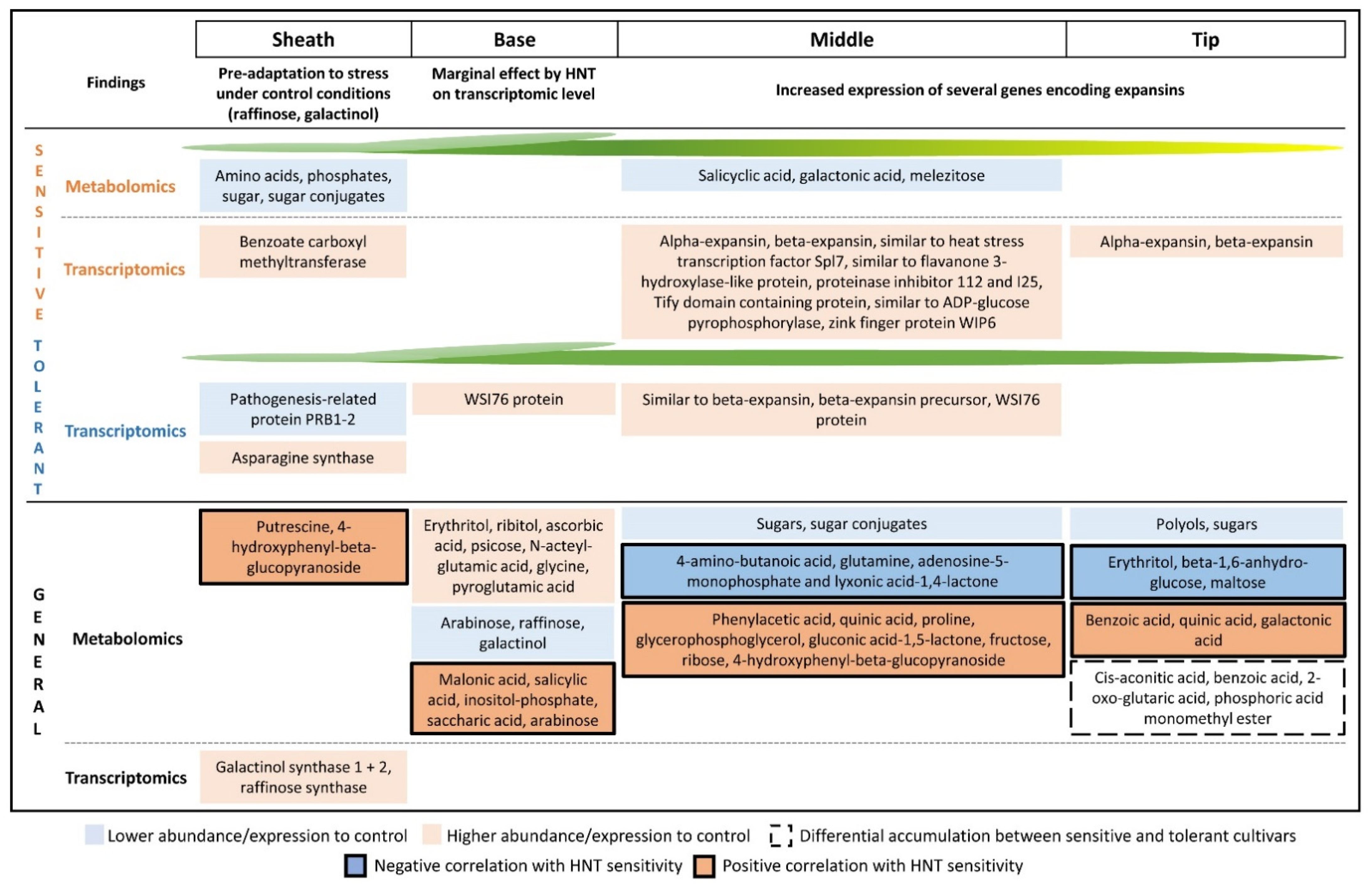
3. Discussion
3.1. Higher Raffinose Abundance in the Sheath Compared to Leaf Blade Segments Reveals a Pre-Adaptation to Abiotic Stress under Control Conditions
3.2. The Leaf Sheath Contributes to HNT Response on Transcriptomic Level
3.3. The Leaf Base Was Marginally Affected by HNT, Especially on Transcriptomic Level
3.4. The Middle Part and the Tip of the Leaf Are Most Highly Affected by HNT on Metabolic and Transcriptomic Level
4. Material and Methods
4.1. Plant Material, Cultivation and Experimental Design of HNT Stress Treatment
4.2. Metabolite Profiling and Data Analysis
4.3. Transcriptome Profiling
4.4. RNA-Seq Data Analysis
4.5. Data Visualization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wassmann, R.; Jagadish, S.V.K.; Sumfleth, K.; Pathak, H.; Howell, G.; Ismail, A.; Serraj, R.; Redona, E.; Singh, R.K.; Heuer, S. Regional vulnerability of climate change impacts on Asian rice production and scope for adaptation. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2009; Volume 102, pp. 91–133. [Google Scholar]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef]
- IPCC. Global Warming of 1.5 °C. An IPCC Special Report on the Impacts of Global Warming of 1.5 °C above Pre-Industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty. Available online: https://www.ipcc.ch/sr15/ (accessed on 1 June 2021).
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland, 2014; p. 151. [Google Scholar]
- Easterling, D.R.; Horton, B.; Jones, P.D.; Peterson, T.C.; Karl, T.R.; Parker, D.E.; Salinger, M.J.; Razuvayev, V.; Plummer, N.; Jamason, P.; et al. Maximum and minimum temperature trends for the globe. Science 1997, 277, 364–367. [Google Scholar] [CrossRef]
- Vose, R.S.; Easterling, D.R.; Gleason, B. Maximum and minimum temperature trends for the globe: An update through 2004. Geophys. Res. Lett. 2005, 32, L23822. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Wang, Y.; Yang, X. On the asymmetry of the urban daily air temperature cycle. J. Geophys. Res. Atmos. 2017, 122, 5625–5635. [Google Scholar] [CrossRef]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef]
- Shi, W.; Muthurajan, R.; Rahman, H.; Selvam, J.; Peng, S.; Zou, Y.; Jagadish, K.S. Source-sink dynamics and proteomic reprogramming under elevated night temperature and their impact on rice yield and grain quality. New Phytol. 2013, 197, 825–837. [Google Scholar] [CrossRef]
- Bahuguna, R.N.; Solis, C.A.; Shi, W.; Jagadish, K.S. Post-flowering night respiration and altered sink activity account for high night temperature-induced grain yield and quality loss in rice (Oryza sativa L.). Physiol. Plant. 2017, 159, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Impa, S.M.; Raju, B.; Hein, N.T.; Sandhu, J.; Prasad, P.V.V.; Walia, H.; Jagadish, S.V.K. High night temperature effects on wheat and rice: Current status and way forward. Plant Cell Environ. 2021, 44, 2049–2065. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Misra, G.; Sreenivasulu, N.; Henry, A. What happens at night? Physiological mechanisms related to maintaining grain yield under high night temperature in rice. Plant Cell Environ. 2021, 44, 2245–2261. [Google Scholar] [CrossRef]
- Cooper, N.T.W.; Siebenmorgen, T.J.; Counce, P.A. Effects of nighttime temperature during kernel development on rice physicochemical properties. Cereal Chem. 2008, 85, 276–282. [Google Scholar] [CrossRef]
- Ambardekar, A.A.; Siebenmorgen, T.J.; Counce, P.A.; Lanning, S.B.; Mauromoustakos, A. Impact of field-scale nighttime air temperatures during kernel development on rice milling quality. Field Crops Res. 2011, 122, 179–185. [Google Scholar] [CrossRef]
- Glaubitz, U.; Li, X.; Köhl, K.I.; van Dongen, J.T.; Hincha, D.K.; Zuther, E. Differential physiological responses of different rice (Oryza sativa) cultivars to elevated night temperature during vegetative growth. Funct. Plant Biol. 2014, 41, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Coast, O.; Ellis, R.H.; Murdoch, A.J.; Quiñones, C.; Jagadish, K.S.V. High night temperature induces contrasting responses for spikelet fertility, spikelet tissue temperature, flowering characteristics and grain quality in rice. Funct. Plant Biol. 2015, 42, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cui, K.; Wang, W.; Li, Q.; Fahad, S.; Hu, Q.; Huang, J.; Nie, L.; Peng, S. Heat-induced phytohormone changes are associated with disrupted early reproductive development and reduced yield in rice. Sci. Rep. 2016, 6, 34978. [Google Scholar] [CrossRef]
- Lanning, S.B.; Siebenmorgen, T.J.; Counce, P.A.; Ambardekar, A.A.; Mauromoustakos, A. Extreme nighttime air temperatures in 2010 impact rice chalkiness and milling quality. Field Crops Res. 2011, 124, 132–136. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, Q.; Peng, S.; Zou, Y.; Chen, S.; Shi, W.; Qin, J.; Laza, M.R.C. Effects of high night temperature on yield and agronomic traits of irrigated rice under field chamber system conditions. Aust. J. Crop Sci. 2013, 7, 7–13. [Google Scholar]
- Shi, W.; Yin, X.; Struik, P.C.; Xie, F.; Schmidt, R.C.; Jagadish, K.S.V. Grain yield and quality responses of tropical hybrid rice to high night-time temperature. Field Crops Res. 2016, 190, 18–25. [Google Scholar] [CrossRef]
- Glaubitz, U.; Erban, A.; Kopka, J.; Hincha, D.K.; Zuther, E. High night temperature strongly impacts TCA cycle, amino acid and polyamine biosynthetic pathways in rice in a sensitivity-dependent manner. J. Exp. Bot. 2015, 66, 6385–6397. [Google Scholar] [CrossRef]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated analysis of rice transcriptomic and metabolomic responses to elevated night temperatures identifies sensitivity- and tolerance-related profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef]
- Schaarschmidt, S.; Lawas, L.M.F.; Kopka, J.; Jagadish, S.V.K.; Zuther, E. Physiological and molecular attributes contribute to high night temperature tolerance in cereals. Plant Cell Environ. 2021, 44, 2034–2048. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2017, 69, 825–844. [Google Scholar] [CrossRef]
- Miya, M.; Yoshikawa, T.; Sato, Y.; Itoh, J.I. Genome-wide analysis of spatiotemporal expression patterns during rice leaf development. BMC Genom. 2021, 22, 169. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Jan, A.; Karibe, H.; Komatsu, S. Identification of phosphoproteins regulated by gibberellin in rice leaf sheath. Plant Mol. Biol. 2005, 58, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Wu, X.; Shi, L.; Zhang, H.; Chen, Y.; Qi, X.; Wang, Z.; Li, X. Transcriptomic and metabolic flux analyses reveal shift of metabolic patterns during rice grain development. BMC Syst. Biol. 2018, 12, 47. [Google Scholar] [CrossRef]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, S.; Tokida, T.; Nakamura, H.; Sakai, H.; Usui, Y.; Okubo, T.; Tago, K.; Hayashi, K.; Sekiyama, Y.; Ono, H.; et al. Characterization of leaf blade- and leaf sheath-associated bacterial communities and assessment of their responses to environmental changes in CO2, temperature, and nitrogen levels under field conditions. Microbes Environ. 2015, 30, 51–62. [Google Scholar] [CrossRef]
- Hirose, T.; Endler, A.; Ohsugi, R. Gene expression of enzymes for starch and sucrose metabolism and transport in leaf sheaths of rice (Oryza sativa L.) during the heading period in relation to the sink to source transition. Plant Prod. Sci. 1999, 2, 178–183. [Google Scholar] [CrossRef]
- Perez, C.M.; Palmiano, E.P.; Baun, L.C.; Juliano, B.O. Starch metabolism in the leaf sheaths and culm of rice. Plant Physiol. 1971, 47, 404–408. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef]
- Kavanová, M.; Lattanzi, F.A.; Grimoldi, A.A.; Schnyder, H. Phosphorus deficiency decreases cell division and elongation in grass leaves. Plant Physiol. 2006, 141, 766–775. [Google Scholar] [CrossRef]
- Schaarschmidt, S.; Lawas, L.M.F.; Glaubitz, U.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Hincha, D.K.; Zuther, E. Season affects yield and metabolic profiles of rice (Oryza sativa) under high night temperature stress in the field. Int. J. Mol. Sci. 2020, 21, 3187. [Google Scholar] [CrossRef]
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 1755–1767. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sengupta, S.; Mukherjee, A.; Basak, P.; Majumder, A.L. Abiotic stress regulates expression of galactinol synthase genes post-transcriptionally through intron retention in rice. Planta 2019, 249, 891–912. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Ishizaki, T.; Valencia, M.; Ogawa, S.; Dedicova, B.; Ogata, T.; Yoshiwara, K.; Maruyama, K.; Kusano, M.; Saito, K.; et al. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 2017, 15, 1465–1477. [Google Scholar] [CrossRef]
- Sadok, W.; Jagadish, S.V.K. The hidden costs of nighttime warming on yields. Trends Plant Sci. 2020, 25, 644–651. [Google Scholar] [CrossRef]
- Xie, Y.; Ravet, K.; Pearce, S. Extensive structural variation in the Bowman-Birk inhibitor family in common wheat (Triticum aestivum L.). BMC Genom. 2021, 22, 218. [Google Scholar] [CrossRef] [PubMed]
- Othman, T.; Bakar, N.T.A.; Abidin, R.Z.; Mahmood, M.; Saidi, N.; Shaharuddin, N.A. Potential of plant’s Bowman-Birk protease inhibitor in combating abiotic stresses: A Mini Review. Bioremediat. Sci. Technol. Res. 2014, 2, 53–61. [Google Scholar]
- Malefo, M.B.; Mathibela, E.O.; Crampton, B.G.; Makgopa, M.E. Investigating the role of Bowman-Birk serine protease inhibitor in Arabidopsis plants under drought stress. Plant Physiol. Biochem. 2020, 149, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Park, S.; Choi, M.J.; Lee, J.Y.; Kim, J.K.; Ha, S.-H.; Lim, S.-H. Molecular and biochemical analysis of two rice flavonoid 3’-hydroxylase to evaluate their roles in flavonoid biosynthesis in rice grain. Int. J. Mol. Sci. 2016, 17, 1549. [Google Scholar] [CrossRef]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112. [Google Scholar] [CrossRef]
- Jones, L.; McQueen-Mason, S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett. 2004, 559, 61–65. [Google Scholar] [CrossRef]
- Li, F.; Han, Y.; Feng, Y.; Xing, S.; Zhao, M.; Chen, Y.; Wang, W. Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 2013, 163, 281–291. [Google Scholar] [CrossRef]
- Xu, J.; Belanger, F.; Huang, B. Differential gene expression in shoots and roots under heat stress for a geothermal and non-thermal Agrostis grass species contrasting in heat tolerance. Environ. Exp. Bot. 2008, 63, 240–247. [Google Scholar] [CrossRef]
- Xu, Q.; Xu, X.; Shi, Y.; Xu, J.; Huang, B. Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress. PLoS ONE 2014, 9, e100792. [Google Scholar] [CrossRef] [PubMed]
- Jadamba, C.; Kang, K.; Paek, N.-C.; Lee, S.I.; Yoo, S.-C. Overexpression of rice expansin7 (Osexpa7) confers enhanced tolerance to salt stress in rice. Int. J. Mol. Sci. 2020, 21, 454. [Google Scholar] [CrossRef]
- Zhao, M.R.; Li, F.; Fang, Y.; Gao, Q.; Wang, W. Expansin-regulated cell elongation is involved in the drought tolerance in wheat. Protoplasma 2011, 248, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, Y.; Yin, S.; Zhang, M.; Wang, W. Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J. Plant Physiol. 2015, 173, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Lü, P.; Kang, M.; Jiang, X.; Dai, F.; Gao, J.; Zhang, C. RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 2013, 237, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Liu, Y.; Deng, X.; Liu, D.; Liu, Y.; Hu, Y.; Yan, Y. Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 101. [Google Scholar] [CrossRef]
- Siahpoosh, M.R.; Sanchez, D.H.; Schlereth, A.; Scofield, G.N.; Furbank, R.T.; van Dongen, J.T.; Kopka, J. Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309. Plant Sci. 2011, 182, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Do, P.T.; Degenkolbe, T.; Erban, A.; Heyer, A.G.; Kopka, J.; Köhl, K.I.; Hincha, D.K.; Zuther, E. Dissecting rice polyamine metabolism under controlled long-term drought stress. PLoS ONE 2013, 8, e60325. [Google Scholar] [CrossRef] [PubMed]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2019, 48, D440–D444. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Zuther, E.; Schulz, E.; Childs, L.H.; Hincha, D.K. Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 2012, 35, 1860–1878. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Andrews, S.; FASTQC. A Quality Control Tool for High Throughput Sequence Data [Online]. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 22 September 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, Version 3.4.2; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.com/ (accessed on 8 June 2021).
- R StudioTeam. RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 8 June 2021).
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods—A Bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]



| Condition | Sheath | Base | Middle | Tip |
|---|---|---|---|---|
| Control | 102 | 130 | 136 | 136 |
| HNT | 108 | 129 | 134 | 133 |
| Tolerance Group | Segment | Up | Down | Total |
|---|---|---|---|---|
| Sensitive | Sheath | 20 | 12 | 32 |
| Base | 0 | 2 | 2 | |
| Middle | 54 | 24 | 78 | |
| Tip | 8 | 15 | 23 | |
| Tolerant | Sheath | 3 | 3 | 6 |
| Base | 1 | 0 | 1 | |
| Middle | 9 | 1 | 10 | |
| Tip | 2 | 1 | 3 |
| Group | Tissue | Gene Identifier | Description | Up/Down |
|---|---|---|---|---|
| Sensitive | Sheath | Os11g0147150 | Hypothetical gene | + |
| Os08g0287200 | Hypothetical conserved gene | + | ||
| Os06g0242000 | Similar to benzoate carboxyl methyltransferase | + | ||
| Middle | Os12g0282000 | Conserved hypothetical protein | - | |
| Os05g0477600 | Alpha-expansin OsEXPA4 | + | ||
| Os04g0344100 | Similar to OSIGBa0106G08.3 protein | + | ||
| Os10g0555600 | Beta-expansin precursor | + | ||
| Os02g0658800 | Beta-expansin | + | ||
| Os04g0418800 | Similar to Hydroxyproline-rich glycoprotein | + | ||
| Os10g0556100 | Similar to beta-expansin EXPB4 | + | ||
| Os04g0350100 | Proteinase inhibitor I25, cystatin domain containing protein | + | ||
| Os02g0236600 | Peroxidase P7 (EC 1.11.1.7) (TP7) | + | ||
| Os02g0112900 | Similar to Viroid RNA-binding protein (Fragment) | + | ||
| Tip | Os04g0493600 | Similar to Lectin-C precursor (PL-C) | + | |
| Tolerant | Sheath | Os01g0382000 | Similar to Pathogenesis-related protein PRB1-2 precursor | - |
| Os03g0291500 | Asparagine synthetase | + | ||
| Os01g0159000 | Similar to cDNA clone: J023049H21 | + | ||
| Os01g0550800 | Protein of unknown function DUF239, plant domain containing protein | + | ||
| Middle | Os10g0555900 | Similar to Beta-expansin | + | |
| Os10g0555600 | Beta-expansin precursor | + |
| #DEGs RNA-Seq | #DEGs Glaubitz et al. 2017 | Overlap | |
|---|---|---|---|
| Sensitive | 85 | 550 | 36 |
| Tolerant | 10 | 29 | 0 |
| ID | Segment | Function | CT9993 | M202 | IR62266 | edgeR | DESeq2 |
|---|---|---|---|---|---|---|---|
| Os04g0689500 | Middle | Conserved hypothetical protein. | NA | 3.87 | 3.59 | 2.80 | 2.80 |
| Os09g0456800 | Middle | Similar to Heat stress transcription factor Spl7 (Heat shock transcription factor). | NA | 4.88 | 3.60 | 3.37 | 3.37 |
| Os05g0444200 | Middle | Similar to T6J4.5 protein (WIP6 protein). | 4.60 | 5.80 | 5.50 | 3.43 | 3.42 |
| Os04g0581000 | Middle | Similar to Flavanone 3-hydroxylase-like protein. | NA | 8.32 | 3.83 | 3.55 | 3.55 |
| Os09g0272600 | Middle | Conserved hypothetical protein. | NA | 3.67 | 4.48 | 3.97 | 3.96 |
| Os01g0124100 | Middle | Proteinase inhibitor I12, Bowman–Birk family protein. | NA | 3.73 | 5.24 | 4.05 | 4.07 |
| Os05g0550300 | Middle | Similar to Lipid transfer protein (Fragment). | 4.31 | 4.64 | 4.40 | 4.14 | 4.14 |
| Os10g0392400 | Middle | Tify domain containing protein. | 5.02 | 5.20 | 6.08 | 4.16 | 4.15 |
| Os05g0580000 | Middle, Tip | Similar to ADP-glucose pyrophosphorylase (EC 2.7.7.27). | NA | 4.29 | 7.41 | 4.65 | 4.64 |
| Os02g0106100 | Middle | Similar to Fructosyltransferase. | 3.99 | 4.97 | 3.67 | 4.70 | 4.69 |
| Os01g0823100 | Middle, Tip | Alpha-expansin OsEXPA2. | NA | 5.61 | 4.36 | 4.84 | 4.85 |
| Os10g0555900 | Middle, Tip | Beta-expansin precursor. | NA | 6.70 | 4.36 | 5.96 | 5.95 |
| Os04g0659300 | Middle, Tip | Protein of unknown function DUF26 domain containing protein. | NA | 6.34 | 6.58 | 7.58 | 7.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schaarschmidt, S.; Glaubitz, U.; Erban, A.; Kopka, J.; Zuther, E. Differentiation of the High Night Temperature Response in Leaf Segments of Rice Cultivars with Contrasting Tolerance. Int. J. Mol. Sci. 2021, 22, 10451. https://doi.org/10.3390/ijms221910451
Schaarschmidt S, Glaubitz U, Erban A, Kopka J, Zuther E. Differentiation of the High Night Temperature Response in Leaf Segments of Rice Cultivars with Contrasting Tolerance. International Journal of Molecular Sciences. 2021; 22(19):10451. https://doi.org/10.3390/ijms221910451
Chicago/Turabian StyleSchaarschmidt, Stephanie, Ulrike Glaubitz, Alexander Erban, Joachim Kopka, and Ellen Zuther. 2021. "Differentiation of the High Night Temperature Response in Leaf Segments of Rice Cultivars with Contrasting Tolerance" International Journal of Molecular Sciences 22, no. 19: 10451. https://doi.org/10.3390/ijms221910451
APA StyleSchaarschmidt, S., Glaubitz, U., Erban, A., Kopka, J., & Zuther, E. (2021). Differentiation of the High Night Temperature Response in Leaf Segments of Rice Cultivars with Contrasting Tolerance. International Journal of Molecular Sciences, 22(19), 10451. https://doi.org/10.3390/ijms221910451







