Identification of Arvicola terrestris scherman Sperm Antigens for Immune Contraceptive Purposes
Abstract
1. Introduction
2. Results
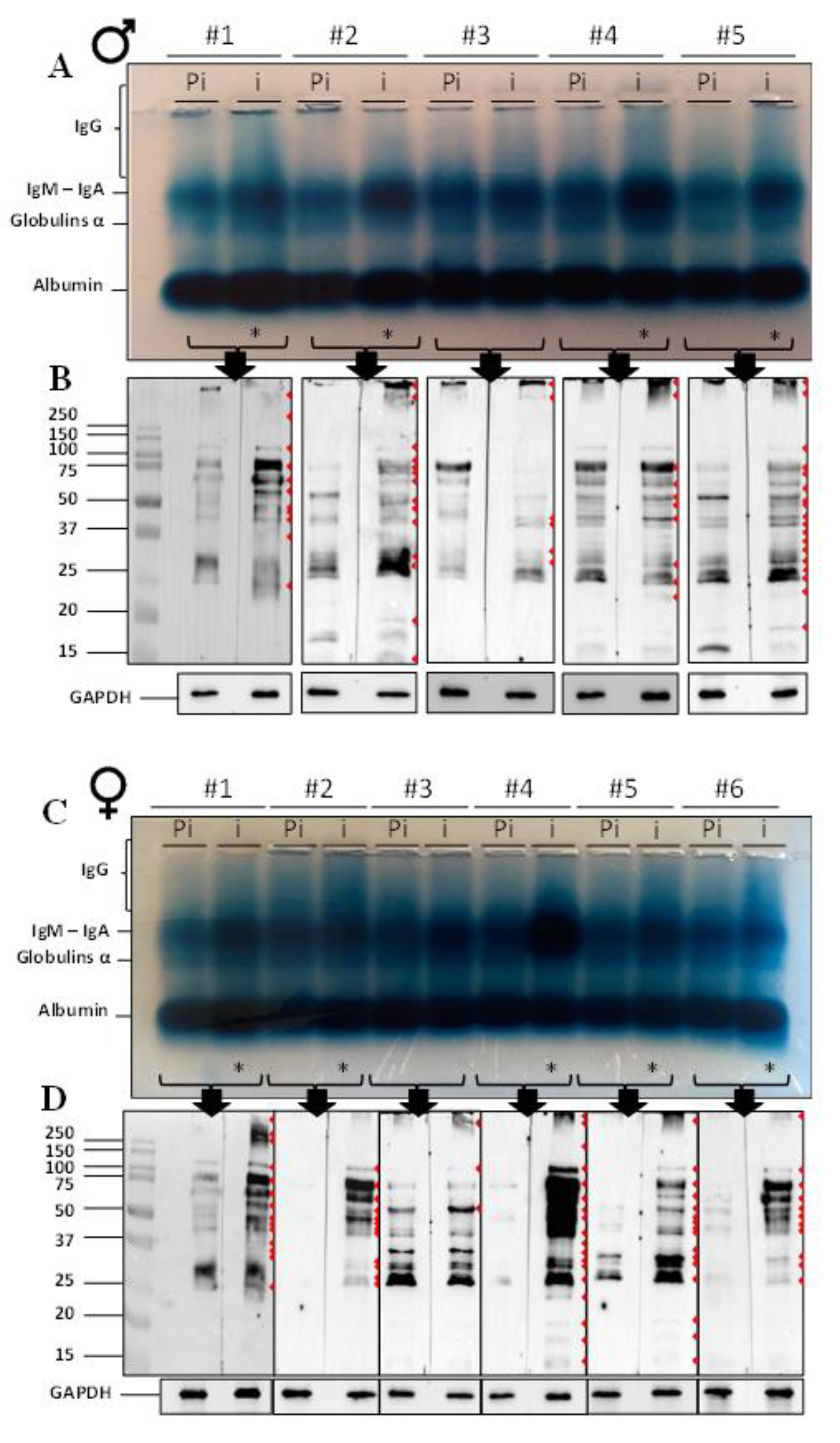
2.1. Immunoglobulin Detection and Western Blot
2.2. ASA Do Target ATS Sperm Surface Proteins
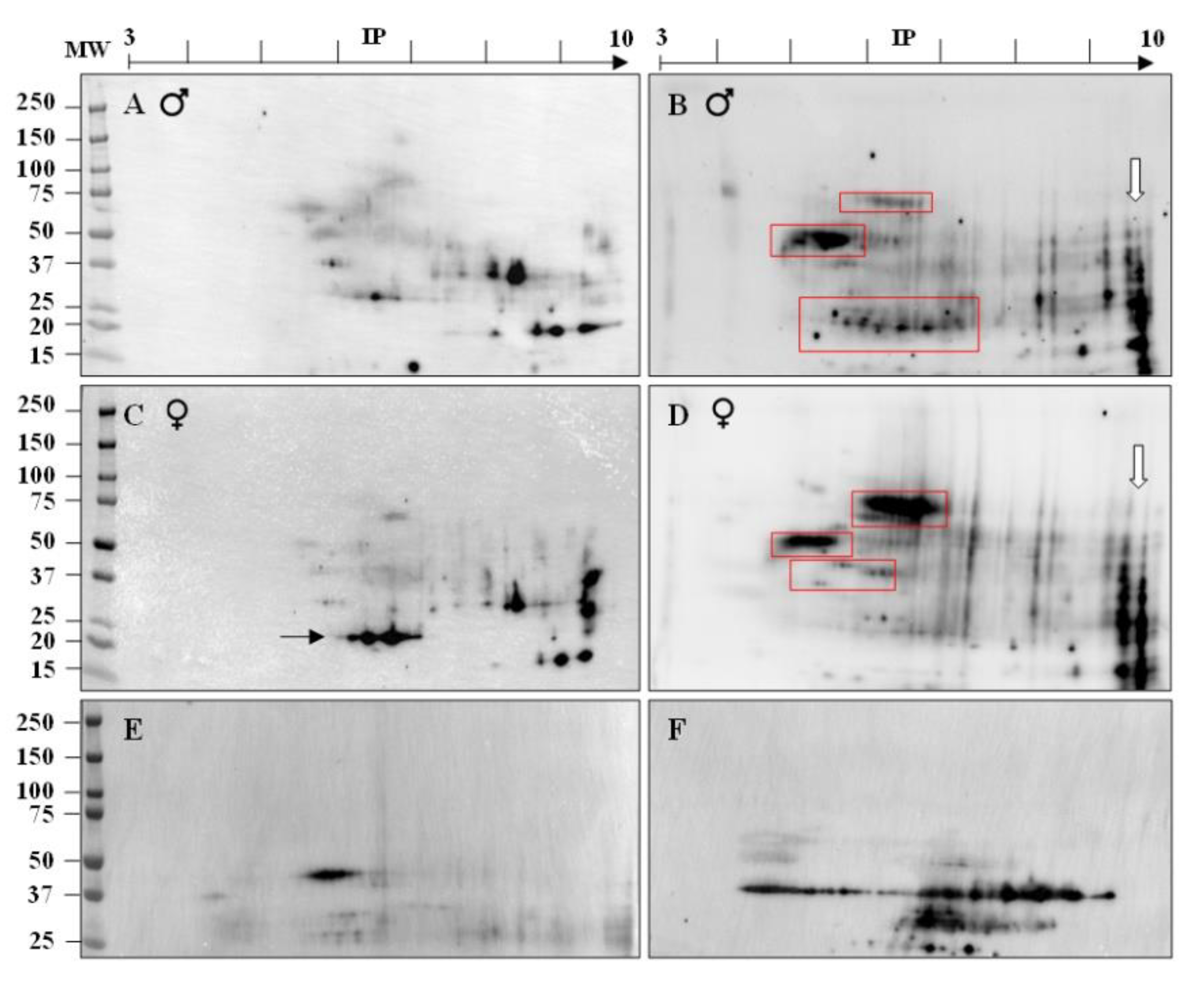
2.3. Identification of Immunogenic ATS Sperm Proteins by Proteomic Analysis
2.4. Immunization of Male ATS with Peptides from Two Candidate Proteins
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Production of Horseradish Peroxidase (HRP)—Conjugated Rabbit Anti-ATS Immunoglobulin G (IgG) Antibodies
4.3. Animal Immunization Procedure
4.4. Sperm Collection
4.5. Extraction of Sperm Proteins
4.6. Serum Globulin Profile with Amido Black Staining
4.7. Western Blot Analysis
4.8. Two-Dimensional Electrophoresis Analysis
4.9. Direct Immunofluorescent Labelling of Sperm Cells
4.10. Confocal Microscopy
4.11. Indirect Immunoprecipitation Assays
4.12. In-Gel Trypsin Digestion
4.13. LC-MS/MS Analysis and Identification of Antigens
4.14. Slot Blot Analysis
4.15. Immunization of Male ATS with Selected Peptides
4.16. Statistical Analysis
4.17. Bioinformatics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Airoldi, J.P.; Altrocchi, R.; Meylan, A. Le comportement fouisseur du Campagnol terrestre Arvicola terrestris Scherman Shaw (Mammalia, Rodentia). Rev. Suisse Zool. 1976, 83, 282–286. [Google Scholar]
- Berthier, K.; Piry, S.; Cosson, J.F.; Giraudoux, P.; Foltête, J.-C.; Defaut, R.; Truchetet, D.; Lambin, X. Dispersal, landscape and travelling waves in cyclic vole populations. Ecol. Lett. 2014, 17, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Saucy, F. Description des cycles pluriannuels d’Arvicola terrestris scherman en Suisse occidentale par la méthode de l’analyse des séries temporelles. EPPO Bull. 1988, 18, 401–413. [Google Scholar] [CrossRef]
- Fichet-Calvet, E.; Pradier, B.; Quéré, J.P.; Giraudoux, P.; Delattre, P. Landscape composition and vole outbreaks: Evidence from an eight year study of Arvicola terrestris. Ecography 2000, 23, 659–668. [Google Scholar] [CrossRef]
- Giraudoux, P.; Delattre, P.; Habert, M.; Quéré, J.P.; Deblay, S.; Defaut, R.; Duhamel, R.; Moissenet, M.F.; Salvi, D.; Truchetet, D. Population dynamics of fossorial water vole (Arvicola terrestris scherman): A land use and landscape perspective. Agric. Ecosyst. Environ. 1997, 66, 47–60. [Google Scholar] [CrossRef]
- Saucy, F. Density dependence in time series of the fossorial form of the water vole, Arvicola terrestris. Oïkos 1994, 74, 381–392. [Google Scholar] [CrossRef]
- Courant, F.; Brunet-Lecomte, P.; Volobouev, V.; Chaline, J.; Quéré, J.P.; Nadachowski, A.; Montuire, S.; Bao, G.; Viriot, L.; Rausch, R.; et al. Karyological and dental identification of Microtus limnophilus in a large focus of alveolar echinococcosis (Gansu, China). Comptes Rendus Acad. Sci. III 1999, 322, 473–480. [Google Scholar] [CrossRef]
- Destrez, A.; Perrot, E.; Granger, S.; Gaillard, C.; Michelin, Y. Les impacts du campagnol terrestre sur les systèmes fourragers: Le cas de l’élevage bovin allaitant en Bourgogne. Fourrages 2014, 220, 291–296. [Google Scholar]
- Viel, J.F.; Giraudoux, P.; Abrial, V.; Bresson-Hadni, S. Water vole (Arvicola terrestris scherman) density as risk factor for human alveolar echinococcosis. Am. J. Trop. Med. Hyg. 1999, 61, 559–565. [Google Scholar] [CrossRef][Green Version]
- Giraudoux, P.; Tremollières, C.; Barbier, B.; Defaut, R.; Rieffel, D.; Bernard, N.; Lucot, É.; Berny, P. Persistence of bromadiolone anticoagulant rodenticide in Arvicola terrestris populations after field control. Environ. Res. 2006, 102, 291–298. [Google Scholar] [CrossRef]
- Berny, P.J.; Buronfosse, T.; Buronfosse, F.; Lamarque, F.; Lorgue, G. Field evidence of secondary poisoning of foxes (Vulpes vulpes) and buzzards (Buteo buteo) by bromadiolone, a 4-year survey. Chemosphere 1997, 35, 1817–1829. [Google Scholar] [CrossRef]
- Fournier-Chambrillon, C.; Berny, P.J.; Coiffier, O.; Barbedienne, P.; Dassé, B.; Delas, G.; Galineau, H.; Mazet, A.; Pouzenc, P.; Rosoux, R.; et al. Evidence of secondary poisoning of free-ranging riparian mustelids by anticoagulant rodenticides in France: Implications for conservation of European mink (Mustela lutreola). J. Wildl. Dis. 2004, 40, 688–695. [Google Scholar] [CrossRef]
- Ruiz-Suarez, N.; Henriquez-Hernandez, L.A.; Valerón, P.F.; Boada, L.D.; Zumbado, M.; Camacho, M.; González, M.A.; Luzardo, O.P. Assessment of anticoagulant rodenticide exposure in six raptor species from the Canary Islands (Spain). Sci. Total Environ. 2014, 485–486, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.K.; Bernoco, M.; Feldman, M. Contraception in mares heteroimmunized with pig zonae pellucidae. J. Reprod. Fertil. 1989, 85, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, J.F.; Turner, J.W., Jr.; Liu, I.K.; Fayrer-Hosken, R. Applications of pig zona pellucida immunocontraception to wildlife fertility control. J. Reprod. Fertil. Suppl. 1996, 50, 183–189. [Google Scholar] [PubMed]
- Miller, L.A.; Johns, B.E.; Killian, G.J. Long-term effects of PZP immunization on reproduction in white-tailed deer. Vaccine 1999, 18, 568–574. [Google Scholar] [CrossRef]
- Moore, H.D.; Jenkins, N.M.; Wong, C. Immunocontraception in rodents: A review of the development of a sperm-based immunocontraceptive vaccine for the grey squirrel (Sciurus carolinensis). Reprod. Fertil. Dev. 1997, 9, 125–129. [Google Scholar] [CrossRef]
- Duckworth, J.A.; Buddle, B.M.; Scobie, S. Fertility of brushtail possums (Trichosurus vulpecula) immunised against sperm. J. Reprod. Immunol. 1998, 37, 125–138. [Google Scholar] [CrossRef]
- Fayrer-Hosken, R.A.; Grobler, D.; van Altena, J.J.; Bertschinger, H.J.; Kirkpatrick, J.F. Immunocontraception of African elephants. Nature 2000, 407, 149. [Google Scholar] [CrossRef]
- Talwar, G.P. Vaccines for control of fertility and hormone-dependent cancers. Immunol. Cell Biol. 1997, 75, 184–189. [Google Scholar] [CrossRef]
- Aitken, R.J.; Paterson, M.; Koothan, P.T. Contraceptive vaccines. Br. Med. Bull. 1993, 49, 88–99. [Google Scholar] [CrossRef]
- Kerr, L.E.; Paterson, M.; Aitken, R.J. Molecular basis of sperm-egg interaction and the prospects for immunocontraception. J. Reprod. Immunol. 1998, 40, 103–118. [Google Scholar] [CrossRef]
- Naz, R.K. Contraceptive vaccines: Success, status, and future perspective. Am. J. Reprod. Immunol. 2011, 66, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Naz, R.K. Antisperm contraceptive vaccines: Where we are and where we are going? Am. J. Reprod. Immunol. 2011, 66, 5–12. [Google Scholar] [CrossRef]
- Alexander, N.J.; Tung, K.S. Immunological and morphological effects of vasectomy in the rabbit. Anat. Rec. 1977, 188, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Ansbacher, R. Sperm-agglutinating and sperm-immobilizing antibodies in vasectomized men. Fertil. Steril. 1971, 22, 629–632. [Google Scholar] [CrossRef]
- Tung, K.S.; Alexander, N.J. Immunopathologic studies on vasectomized guinea pigs. Biol. Reprod. 1977, 17, 241–254. [Google Scholar] [CrossRef]
- Cyr, D.G.; Finsson, K.; Dufresne, J.; Gregory, M. The Epididymis: From Molecules to Clinical Practice; Robaire, B., Hinton, B.T., Eds.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2002; pp. 103–118. [Google Scholar]
- McLaughlin, E.A.; Aitken, R.J. Is there a role for immunocontraception? Mol. Cell Endocrinol. 2011, 335, 78–88. [Google Scholar] [CrossRef]
- Primakoff, P.; Lathrop, W.; Bronson, R. Identification of human sperm surface glycoproteins recognized by autoantisera from immune infertile men, women, and vasectomized men. Biol. Reprod. 1990, 42, 929–942. [Google Scholar] [CrossRef]
- Asquith, K.L.; Kitchener, A.L.; Kay, D.J. Immunisation of the male tammar wallaby (Macropus eugenii) with spermatozoa elicits epididymal antigen-specific antibody secretion and compromised fertilisation rate. J. Reprod. Immunol. 2006, 69, 127–147. [Google Scholar] [CrossRef]
- Tung, K.S.; Goldberg, E.H.; Goldberg, E. Immunobiological consequence of immunization of female mice with homologous spermatozoa: Induction of infertility. J. Reprod. Immunol. 1979, 1, 145–158. [Google Scholar] [CrossRef]
- Castle, P.E.; Whaley, K.J.; Hoen, T.E.; Moench, T.R.; Cone, R.A. Contraceptive effect of sperm-agglutinating monoclonal antibodies in rabbits. Biol. Reprod. 1997, 56, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Grignard, E.; Cadet, R.; Saez, F.; Drevet, J.R.; Vernet, P. Identification of sperm antigens as a first step towards the generation of a contraceptive vaccine to decrease fossorial water vole Arvicola terrestris Scherman proliferations. Theriogenology 2007, 68, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Esmailnejad, A.; Nikahval, B.; Mogheiseh, A.; Karampour, R.; Karami, S. The detection of canine anti-sperm antibody following parenteral immunization of bitches against homogenized whole sperm. Basic Clin. Androl. 2020, 30, 1. [Google Scholar] [CrossRef] [PubMed]
- Naz, R.K.; Phillips, T.M.; Rosenblum, B.B. Characterization of the fertilization antigen 1 for the development of a contraceptive vaccine. Proc. Natl. Acad. Sci. USA 1986, 83, 5713–5717. [Google Scholar] [CrossRef]
- Ellerman, D.A.; Brantúa, V.S.; Martínez, S.P.; Cohen, D.J.; Conesa, D.; Cuasnicú, P.S. Potential contraceptive use of epididymal proteins: Immunization of male rats with epididymal protein DE inhibits sperm fusion ability. Biol. Reprod. 1998, 59, 1029–1036. [Google Scholar] [CrossRef]
- Jagadish, N.; Rana, R.; Mishra, D.; Garg, M.; Selvi, R.; Suri, A. Characterization of immune response in mice to plasmid DNA encoding human sperm associated antigen 9 (SPAG9). Vaccine 2006, 24, 3695–3703. [Google Scholar] [CrossRef]
- Wang, M.; Shi, J.L.; Cheng, G.Y.; Hu, Y.Q.; Xu, C. The antibody against a nuclear autoantigenic sperm protein can result in reproductive failure. Asian J. Androl. 2009, 11, 183–192. [Google Scholar] [CrossRef]
- Flanagan, K.L.; Fink, A.L.; Plebanski, M.; Klein, S.L. Sex and Gender Differences in the Outcomes of Vaccination over the Life Course. Annu. Rev. Cell Dev. Biol. 2017, 33, 577–599. [Google Scholar] [CrossRef]
- Parr, E.L.; Parr, M.B. The effect of sperm immunization in the gastrointestinal tract on anti-sperm antibody production and fertility in female mice. J. Reprod. Immunol. 1986, 9, 49–56. [Google Scholar] [CrossRef]
- Tung, K.S.; Harakal, J.; Qiao, H.; Rival, C.; Li, J.C.; Paul, A.G.; Wheeler, K.; Pramoonjago, P.; Grafer, C.M.; Sun, W.; et al. Egress of sperm autoantigen from seminiferous tubules maintains systemic tolerance. J. Clin. Investig. 2017, 127, 1046–1060. [Google Scholar] [CrossRef]
- Krämer, A.; Sommer, D.; Hahn, E.G.; Riecken, E.O. German experimental hepatitis B vaccine—Influence of variation of dosage schedule, sex and age differences on immunogenicity in health care workers. Klin. Wochenschr. 1986, 64, 688–694. [Google Scholar] [CrossRef][Green Version]
- Peacock, J.W.; Nordone, S.K.; Jackson, S.S.; Liao, H.-X.; Letvin, N.L.; Yafal, A.G.; Gritz, L.; Mazzara, G.P.; Haynes, B.F.; Staats, H.F. Gender differences in human immunodeficiency virus type 1-specific CD8 responses in the reproductive tract and colon following nasal peptide priming and modified vaccinia virus Ankara boosting. J. Virol. 2004, 78, 13163–13172. [Google Scholar] [CrossRef]
- Camus, M.F.; Wolf, J.B.; Morrow, E.H.; Dowling, D.K. Single Nucleotides in the mtDNA Sequence Modify Mitochondrial Molecular Function and Are Associated with Sex-Specific Effects on Fertility and Aging. Curr. Biol. 2015, 25, 2717–2722. [Google Scholar] [CrossRef] [PubMed]
- Ventura-Clapier, R.; Moulin, M.; Piquereau, J.; Lemaire, C.; Mericskay, M.; Veksler, V.; Garnier, A. Mitochondria: A central target for sex differences in pathologies. Clin. Sci. 2017, 131, 803–822. [Google Scholar] [CrossRef] [PubMed]
- Freitas, M.J.; Vijayaraghavan, S.; Fardilha, M. Signaling mechanisms in mammalian sperm motility. Biol. Reprod. 2017, 96, 2–12. [Google Scholar]
- Paradowska, A.; Bohring, C.; Krause, E.; Krause, W. Identification of evolutionary conserved mouse sperm surface antigens by human antisperm antibodies (ASA) from infertile patients. Am. J. Reprod. Immunol. 2006, 55, 321–330. [Google Scholar] [CrossRef]
- Frenette, G.; Girouard, J.; Sullivan, R. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biol. Reprod. 2006, 75, 885–890. [Google Scholar] [CrossRef]
- Girouard, J.; Frenette, G.; Sullivan, R. Comparative proteome and lipid profiles of bovine epididymosomes collected in the intraluminal compartment of the caput and cauda epididymidis. Int. J. Androl. 2011, 34, e475–e486. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.; de Iuliis, G.N.; Hart, H.M.; Zhou, W.; Mathe, A.; Bernstein, I.R.; Anderson, A.L.; Stanger, S.J.; Skerrett-Byrne, D.; Jamaluddin, M.F.B.; et al. Proteomic Profiling of Mouse Epididymosomes Reveals their Contributions to Post-testicular Sperm Maturation. Mol. Cell Proteom. 2019, 18, S91–S108. [Google Scholar] [CrossRef]
- Thimon, V.; Frenette, G.; Saez, F.; Thabet, M.; Sullivan, R. Protein composition of human epididymosomes collected during surgical vasectomy reversal: A proteomic and genomic approach. Hum. Reprod. 2008, 23, 1698–1707. [Google Scholar] [CrossRef]
- Sullivan, R.; Saez, F. Epididymosomes, prostasomes, and liposomes: Their roles in mammalian male reproductive physiology. Reproduction 2013, 146, R21–R35. [Google Scholar] [CrossRef]
- Guiton, R.; Voisin, A.; Henry-Berger, J.; Saez, F.; Drevet, J.R. Of vessels and cells: The spatial organization of the epididymal immune system. Andrology 2019, 7, 712–718. [Google Scholar] [CrossRef]
- Hinton, B.T.; Howards, S.S. Permeability characteristics of the epithelium in the rat caput epididymidis. J. Reprod. Fertil. 1981, 63, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Myers, K.; Khatra, B.; Vijayaraghavan, S. Protein 14-3-3zeta binds to protein phosphatase PP1gamma2 in bovine epididymal spermatozoa. Biol. Reprod. 2004, 71, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Saez, F.; Whitfield, M.; Drevet, J.R. Impairment of sperm maturation and capacitation due to diet-dependent cholesterol overload. Andrology 2019, 7, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Kawase, O.; Jimbo, M. Detection of sperm-reactive antibodies in wild sika deer and identification of the sperm antigens. J. Vet. Med. Sci. 2018, 80, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.M.; Serres, C.; Auer, J. Moonlighting proteins in sperm-egg interactions. Biochem. Soc. Trans. 2014, 42, 1740–1743. [Google Scholar] [CrossRef]
- Fu, J.; Yao, R.; Luo, Y.; Yang, D.; Cao, Y.; Qiu, Y.; Song, W.; Miao, S.; Gu, Y.; Wang, L. Anti-GAPDHS antibodies: A biomarker of immune infertility. Cell Tissue Res. 2016, 364, 199–207. [Google Scholar] [CrossRef]
- Naaby-Hansen, S.; Herr, J.C. Heat shock proteins on the human sperm surface. J. Reprod. Immunol. 2010, 84, 32–40. [Google Scholar] [CrossRef]
- Agarwal, A.; Selvam, M.K.P.; Samanta, L.; Vij, S.C.; Parekh, N.; Sabanegh, E.; Tadros, N.N.; Arafa, M.; Sharma, R. Effect of Antioxidant Supplementation on the Sperm Proteome of Idiopathic Infertile Men. Antioxidants 2019, 8, 488. [Google Scholar] [CrossRef]
- Lin, Y.N.; Roy, A.; Yan, W.; Burns, K.H.; Matzuk, M.M. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol. Cell Biol. 2007, 27, 6794–6805. [Google Scholar] [CrossRef] [PubMed]
- Asquith, K.L.; Harman, A.J.; McLaughlin, E.A.; Nixon, B.; Aitken, R.J. Localization and significance of molecular chaperones, heat shock protein 1, and tumor rejection antigen gp96 in the male reproductive tract and during capacitation and acrosome reaction. Biol. Reprod. 2005, 72, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Nowicka-Bauer, K.; Kamieniczna, M.; Cibulka, J.; Ulcova-Gallova, Z.; Kurpisz, M. Proteomic identification of sperm antigens using serum samples from individuals with and without antisperm antibodies. Andrologia 2016, 48, 693–701. [Google Scholar] [CrossRef]
- Fiedler, S.E.; Bajpai, M.; Carr, D.W. Identification and characterization of RHOA-interacting proteins in bovine spermatozoa. Biol. Reprod. 2008, 78, 184–192. [Google Scholar] [CrossRef]
- Suri, A. Contraceptive vaccines targeting sperm. Expert Opin. Biol. Ther. 2005, 5, 381–392. [Google Scholar] [CrossRef] [PubMed]



| Protein Name | UniProtKB IDs | MW (Da) | Th.Ip | mFC Male | mFC Female | No. of Peptides (% Coverage) | Mascot Score |
|---|---|---|---|---|---|---|---|
| Sperm–egg interaction | |||||||
| ALDOA | P05064 | 37,817 | 8.30 | 3.75 | 4.33 | 8 (43.20) | 497.32 |
| HSPA1L * | P16627 | 70,981 | 5.76 | 6.83 | 25.86 | 2 (7.80) | 234.52 |
| CCT3 | P80318 | 61,265 | 6.10 | 5.86 | 89.02 | 2 (3.49) | 79.20 |
| CCT8 | P42932 | 60,116 | 5.42 | 1.52 | 1.81 | 2 (4.38) | 123.39 |
| ZPBP * | Q62522 | 42,640 | 9.43 | 2.83 | 6.57 | 7 (15.26) | 386.51 |
| ZPBP2 * | Q6X786 | 42,640 | 8.04 | 2.83 | 6.57 | 7 (22.92) | 580.50 |
| HSPD1 (HSP60) | P63038 | 55,970 | 5.70 | 1.79 | 6.83 | 7 (19.01) | 397.55 |
| Penetration of zona pellucida | |||||||
| HEXB | P20060 | 54,911 | 6.29 | 1.96 | 4.07 | 5 (12.82) | 319.44 |
| ACR * | P23578 | 50,286 | 9.26 | 1.65 | 3.34 | 6 (18.88) | 468.93 |
| Sperm capacitation | |||||||
| PEBP1 | P70296 | 21,024 | 7.01 | 3.10 | 5.77 | 3 (28.33) | 198.37 |
| AKAP3 * | O88987 | 94,787 | 5.84 | 3.52 | 12.72 | 3 (3.20) | 169.17 |
| YWHAZ | P63101 | 27,898 | 4.73 | 1.27 | 2.65 | 7 (30.20) | 503.69 |
| PRKACA | P05132 | 40,713 | 8.84 | 0.95 | 0.49 | 6 (14.29) | 322.06 |
| ROPN1 * | Q9ESG2 | 24,123 | 5.52 | 1.51 | 7.34 | 3 (18.90) | 233.78 |
| Acrosomal reaction | |||||||
| CALR | P14211 | 48,159 | 4.29 | 1.76 | 3.41 | 11 (28.92) | 864.52 |
| PDIA3 | P27773 | 57,052 | 5.98 | 1.94 | 5.29 | 12 (26.14) | 770.66 |
| ACR * | P23578 | 50,286 | 9.26 | 1.65 | 3.34 | 6 (18.88) | 468.93 |
| HEXB | P07686 | 54,911 | 6.29 | 1.96 | 4.07 | 5 (12.82) | 319.44 |
| AKAP3 * | O88987 | 94,787 | 5.84 | 3.52 | 12.72 | 3 (3.20) | 169.17 |
| PRKACA | P05132 | 40,713 | 8.84 | 0.95 | 0.49 | 6 (14.29) | 322.06 |
| Sperm part | |||||||
| ALDOA | P05064 | 37,817 | 8.30 | 3.75 | 4.33 | 8 (43.20) | 497.32 |
| NUP210L * | Q9D2F7 | 202,450 | 7.15 | 5.64 | 6.87 | 33 (17.50) | 2493.12 |
| PRKAR2A | P12367 | 45,817 | 4.96 | 1.52 | 3.41 | 3 (7.37) | 167.54 |
| ODF1 * | Q61999 | 29,282 | 8.46 | 1.57 | 2.01 | 23 (53.31) | 1421.42 |
| GAPDHS * | Q64467 | 35,936 | 8.39 | 1.51 | 2.86 | 8 (9.36) | 178.10 |
| PGK2 * | P09041 | 27,881 | 8.74 | 1.33 | 2.77 | 4 (6.95) | 233.10 |
| OAZ3* | Q9R109 | 26,958 | 6.38 | 8.10 | 20.86 | 2 (7.69) | 202.45 |
| AKAP3 * | O88987 | 94,787 | 5.84 | 3.52 | 12.72 | 3 (3.20) | 169.17 |
| DYNLL2 | Q9D0M5 | 10,457 | 6.81 | 2.79 | 4.45 | 2 (27) | 199.68 |
| ROPN1 * | Q9ESG2 | 24,123 | 5.52 | 1.51 | 7.34 | 3 (18.90) | 233.78 |
| PRKACA | P05132 | 40,713 | 8.84 | 1.06 | 2.05 | 6 (14.29) | 322.06 |
| Carbohydrate metabolism/Glycolysis and Gluconeogenesis | |||||||
| ALDOA | P05064 | 37,817 | 8.30 | 3.75 | 4.33 | 8 (43.20) | 497.32 |
| ACO2 | Q99KI0 | 86,107 | 7.36 | 1.68 | 3.84 | 14 (21.50) | 1013.27 |
| GAPDH | P16858 | 35,936 | 8.57 | 1.51 | 2.86 | 8 (31.23) | 594.17 |
| PGK1 | P09411 | 27,881 | 8.30 | 1.33 | 2.77 | 4 (11.37) | 381.29 |
| PGK2 * | P09041 | 27,881 | 8.74 | 1.33 | 2.77 | 4 (6.95) | 233.10 |
| MDH2 | P08249 | 36,045 | 8.92 | 2.78 | 7.06 | 18 (64) | 1260.95 |
| CS | Q9CZU6 | 52,010 | 8.45 | 3.57 | 7.15 | 8 (21.72) | 699.94 |
| ENO1 | P17182 | 47,602 | 7.01 | 1.86 | 2.40 | 9 (23.04) | 511.79 |
| GPI1 (G6PI) | P06745 | 62,986 | 8.42 | 2.07 | 3.05 | 4 (9.32) | 245.83 |
| SUCLA2 | Q9Z2I9 | 50,481 | 7.05 | 1.32 | 2.22 | 7 (33.81) | 803.25 |
| MTHFD1L | Q3V3R1 | 105,743 | 8.32 | 3.81 | 6.45 | 2 (3.10) | 132.20 |
| LDHC * | P00342 | 28,302 | 7.08 | 1.30 | 0.83 | 3 (17.76) | 323.87 |
| HK3 | Q3TRM8 | 102,184 | 5.23 | 0.39 | 7.61 | 3 (3.15) | 163.23 |
| Energy metabolism | |||||||
| PRDX5 | P99029 | 20,423 | 6.00 | 1.91 | 4.29 | 5 (17.60) | 313.59 |
| CAR1 | P13634 | 28,550 | 6.59 | 1.72 | 3.44 | 9 (52.17) | 686.90 |
| CAR2 | P00920 | 29,294 | 6.87 | 1.11 | 2.21 | 4 (17.31) | 233.50 |
| SDHA | Q8K2B3 | 73,414 | 7.06 | 1.49 | 5.47 | 7 (11) | 393.12 |
| ATP5B | P56480 | 25,127 | 5.26 | 1.27 | 2.67 | 15 (25.39) | 1084.34 |
| ATP5F1 | Q9CQQ7 | 10,909 | 9.37 | 0.97 | 1.87 | 2 (7.11) | 111.97 |
| COX2 | P00405 | 26,025 | 4.67 | 0.96 | 2.01 | 5 (16.30) | 311.67 |
| COX4I1 | P19783 | 10,899 | 9.52 | 0.78 | 1.90 | 6 (41.30) | 357.12 |
| ATP5A1 | Q03265 | 58,502 | 9.16 | 2.47 | 5.22 | 14 (25.28) | 1072.18 |
| Mitochondrial protein | |||||||
| PRDX3 | P20108 | 28,428 | 7.68 | 2.07 | 4.46 | 2 (7.66) | 115.88 |
| MTIF2 | Q91YJ5 | 10,909 | 6.71 | 2.82 | 7.30 | 7 (26.91) | 488.06 |
| COX2 | P00405 | 26,025 | 4.67 | 0.96 | 2.01 | 5 (16.30) | 311.67 |
| SUCLA2 | Q9Z2I9 | 50,481 | 7.05 | 1.32 | 2.22 | 7 (33.81) | 803.25 |
| DIABLO | Q9JIQ3 | 26,705 | 5.68 | 2.95 | 404.15 | 2 (8.09) | 83.35 |
| CHDH | Q8BJ64 | 67020 | 8.57 | 21.01 | 20.77 | 3 (4.86) | 164.31 |
| SDHA | Q8K2B3 | 73,414 | 7.06 | 1.49 | 5.47 | 7 (11) | 393.12 |
| ATP5B | P56480 | 25,127 | 5.26 | 1.27 | 2.67 | 15 (25.39) | 1084.34 |
| ATP5F1 | Q9CQQ7 | 10,909 | 9.37 | 0.97 | 1.87 | 2 (7.11) | 111.97 |
| COX4I1 | P19783 | 10,899 | 9.52 | 0.78 | 1.90 | 6 (41.30) | 357.12 |
| COX6B2 | Q80ZN9 | 10,835 | 9.21 | 0.85 | 1.58 | 2 (23.86) | 100.88 |
| NIPSNAP3B | Q9CQE1 | 28,368 | 9.21 | 0.77 | 1.77 | 3 (22.84) | 213.10 |
| HSPA1L * | P16627 | 70,981 | 5.76 | 6.83 | 25.86 | 2 (7.80) | 234.52 |
| HSPD1 (HSP60) | P63038 | 55,970 | 5.70 | 1.79 | 6.83 | 7 (19.01) | 397.55 |
| ATPIF1 | O35143 | 12,255 | 4.82 | 2.91 | 19.66 | 2 (17.76) | 105.38 |
| CYCT * | P00015 | 11,654 | 9.54 | 2.56 | 10.67 | 2 (27.62) | 776.30 |
| CYCS | P62897 | 11,654 | 9.54 | 2.56 | 10.67 | 2 (8.92) | 250.71 |
| ATP5A1 | Q03265 | 58,502 | 9.16 | 2.47 | 5.22 | 14 (25.28) | 1072.18 |
| MTHFD1L | Q3V3R1 | 105,743 | 8.32 | 3.81 | 6.45 | 2 (3.10) | 132.20 |
| Chaperones | |||||||
| HSPA5 (GR78/BIP) | P20029 | 72,606 | 5.07 | 1.99 | 4.19 | 21 (31.76) | 1701.29 |
| HSC70 (HSPA8) | P63017 | 71,069 | 5.37 | 3.99 | 4.49 | 5 (16.10) | 520.24 |
| HSPA1L * | P16627 | 70,981 | 5.76 | 6.83 | 25.86 | 2 (7.80) | 234.52 |
| HSPD1 (HSP60) | P63038 | 55,970 | 5.70 | 1.79 | 6.83 | 7 (19.01) | 397.55 |
| HSP90B1 | P08113 | 92,865 | 4.76 | 1.19 | 1.95 | 16 (31.49) | 1123.06 |
| Protein Name | UniProtKB IDs | MW (Da) | Th.Ip | mFc Male | mFc Female | No. of Peptides (% Coverage) | Mascot Score |
|---|---|---|---|---|---|---|---|
| Moonlighting proteins | |||||||
| PGK1 | P09411 | 27,881 | 8.30 | 1.33 | 2.77 | 4 (11.37) | 381.29 |
| PGK2 * | P09041 | 27,881 | 8.74 | 1.33 | 2.77 | 4 (6.95) | 233.10 |
| GAPDH | P16858 | 35,936 | 8.57 | 1.51 | 2.86 | 8 (31.23) | 594.17 |
| ACO2 | Q99KI0 | 86,107 | 7.36 | 1.68 | 3.84 | 14 (21.5) | 1013.27 |
| ALDOA | P05064 | 37,817 | 8.30 | 3.75 | 4.33 | 8 (43.20) | 497.32 |
| ODF1 * | Q61999 | 29,282 | 8.46 | 1.57 | 2.01 | 23 (53.31) | 1421.42 |
| HSPA5 | P20029 | 72,606 | 5.07 | 1.99 | 4.19 | 21 (31.76) | 1701.29 |
| ACR * | P23578 | 50,286 | 9.26 | 1.65 | 3.34 | 6 (18.88) | 468.93 |
| ENO1 | P17182 | 47,602 | 7.01 | 1.86 | 2.40 | 9 (23.04) | 511.79 |
| HSPD1 (HSP60) | P63038 | 55,970 | 5.70 | 1.79 | 6.83 | 7 (19.01) | 397.55 |
| Phenotype limited only to sexual organs when the gene expression is invalidated | |||||||
| NUP210L * | Q9D2F7 | 202,450 | 7.15 | 5.64 | 6.87 | 33 (17.50) | 2493.12 |
| PGK1 | P09411 | 27,881 | 8.30 | 1.33 | 2.77 | 4 (11.37) | 381.29 |
| PGK2 * | P09041 | 27,881 | 8.74 | 1.33 | 2.77 | 4 (6.95) | 233.10 |
| ODF1 * | Q61999 | 29,282 | 8.46 | 1.57 | 2.01 | 23 (53.31) | 1421.42 |
| GAPDHS * | Q64467 | 35,936 | 8.39 | 1.51 | 2.86 | 8 (9.36) | 178.10 |
| ACR * | P23578 | 50,286 | 9.26 | 1.65 | 3.34 | 6 (18.88) | 468.93 |
| AKAP3 * | O88987 | 94,787 | 5.84 | 3.52 | 12.72 | 3 (3.20) | 169.17 |
| OAZ3 * | Q9R109 | 26,958 | 6.38 | 8.10 | 20.86 | 2 (7.69) | 202.45 |
| HSPA1L * | P16627 | 70,981 | 5.76 | 6.83 | 25.86 | 2 (7.80) | 234.52 |
| ZPBP * | Q62522 | 42,640 | 9.43 | 2.83 | 6.57 | 7 (15.26) | 386.51 |
| ZPBP2 * | Q6X786 | 42,640 | 8.04 | 2.83 | 6.57 | 7 (22.92) | 580.50 |
| CHDH | Q8BJ64 | 67,020 | 8.57 | 21.01 | 20.77 | 3 (4.86) | 164.31 |
| ROPN1 * | Q9ESG2 | 24,123 | 5.52 | 1.51 | 7.34 | 3 (18.90) | 233.78 |
| PRKACA | P05132 | 40,713 | 8.84 | 0.95 | 0.49 | 6 (14.29) | 322.06 |
| Epididymosomes | |||||||
| HSC70 (HSPA8) | P63017 | 71,069 | 5.37 | 3.99 | 4.49 | 5 (16.10) | 520.24 |
| CAR1 | P13634 | 28,550 | 6.59 | 1.72 | 3.44 | 9 (52.17) | 686.90 |
| PEBP1 | P70296 | 21,024 | 7.01 | 3.10 | 5.77 | 3 (28.33) | 198.37 |
| TUBB4B | P68372 | 50,255 | 4.79 | 2.27 | 2.14 | 15 (63.82) | 2370.87 |
| ALDOA | P05064 | 37,817 | 8.30 | 3.75 | 4.33 | 8 (43.20) | 497.32 |
| CALR | P14211 | 48,159 | 4.29 | 1.76 | 3.41 | 11 (28.92) | 864.52 |
| HSPA5 | P20029 | 72,606 | 5.07 | 1.99 | 4.19 | 21 (31.76) | 1701.29 |
| PRDX5 | P99029 | 20,423 | 6.00 | 1.91 | 4.29 | 5 (17.60) | 313.59 |
| HEXB | P20060 | 54,911 | 6.29 | 1.96 | 4.07 | 5 (12.82) | 319.44 |
| PGK1 | P09411 | 27,881 | 8.30 | 1.33 | 2.77 | 4 (11.37) | 381.29 |
| PRKAR2A | P12367 | 45,817 | 4.96 | 1.52 | 3.41 | 3 (7.37) | 167.54 |
| PDIA3 | P27773 | 57,052 | 5.98 | 1.94 | 5.29 | 12 (26.14) | 770.66 |
| GAPDH | P16858 | 35,936 | 8.57 | 1.51 | 2.86 | 8 (31.23) | 594.17 |
| GAPDHS * | Q64467 | 35,936 | 8.39 | 1.51 | 2.86 | 8 (9.36) | 178.10 |
| PRDX1 | P35700 | 22,309 | 8.27 | 2.21 | 4.55 | 4 (31.30) | 344.45 |
| PRDX2 | Q61171 | 22,163 | 5.66 | 1.50 | 2.93 | 3 (15) | 230.67 |
| PRDX6 | O08709 | 23,741 | 6.00 | 2.75 | 5.31 | 2 (8.93) | 96.41 |
| AKR1B3 | P45376 | 36,056 | 6.52 | 2.09 | 3.59 | 2 (9.05) | 97.03 |
| AKAP3 * | O88987 | 94,787 | 5.84 | 3.52 | 12.72 | 3 (3.20) | 169.17 |
| YWHAZ | P63101 | 27,898 | 4.73 | 1.27 | 2.65 | 7 (30.20) | 503.69 |
| ENO1 | P17182 | 47,602 | 7.01 | 1.86 | 2.40 | 9 (23.04) | 511.79 |
| CCT3 | P80318 | 61,265 | 6.10 | 5.86 | 89.02 | 2 (3.49) | 79.20 |
| HSPD1 (HSP60) | P63038 | 55,970 | 5.70 | 1.79 | 6.83 | 7 (19.01) | 397.55 |
| ZPBP * | Q62522 | 42,640 | 9.43 | 2.83 | 6.57 | 7 (15.26) | 386.51 |
| ZPBP2 * | Q6X786 | 42,640 | 8.04 | 2.83 | 6.57 | 7 (22.92) | 580.50 |
| HSP90B1 | P08113 | 92,865 | 4.76 | 1.20 | 2.09 | 16 (31.49) | 1123.06 |
| PPIA | P17742 | 18,089 | 7.68 | 3.05 | 7.45 | 2 (17.83) | 206.22 |
| PDIA4 | P08003 | 72,914 | 4.96 | 4.75 | 4.94 | 4 (7.01) | 217.37 |
| RAB2A | P53994 | 23,545 | 6.08 | 0.27 | 0.32 | 3 (15.10) | 152.70 |
| Protein Name | UniProtKB IDs | MW (Da) | Th.Ip | mFc Male | mFc Female | No. of Peptides (% Coverage) | Mascot Score |
|---|---|---|---|---|---|---|---|
| DNA/RNA binding | |||||||
| NUP155 | Q99P88 | 156,682 | 5.78 | 7.96 | 3.18 | 5 (3.80) | 297.33 |
| PPIB | P24369 | 23,754 | 9.42 | 1.84 | 5.53 | 3 (15.74) | 212.97 |
| SERPINA3 | Q91WP6 | 46,647 | 5.37 | 1.64 | 2.34 | 3 (11.87) | 103.57 |
| histone H3.1 | P68433 | 15,509 | 10.31 | 2.93 | 3.98 | 5 (22.06) | 285.16 |
| EEF1A1 | P10126 | 50,440 | 9.10 | 0.79 | 1.30 | 5 (10.82) | 311 |
| Histone H4 | P62806 | 11,360 | 11.36 | 0.89 | 7.30 | 5 (55.34) | 396.84 |
| HIST1H1E | P43274 | 21,882 | 11.03 | 3.75 | 1.15 | 5 (23.30) | 484.74 |
| TXN1 | P10639 | 11,952 | 9.59 | 1.26 | 2.07 | 2 (20.95) | 122.51 |
| PPIA | P17742 | 18,089 | 7.68 | 3.05 | 7.45 | 2 (17.83) | 206.22 |
| Hist1h2bc | Q6ZWY9 | 13,898 | 9.68 | 1.67 | 2.60 | 5 (28.57) | 319.82 |
| GRHL3 | Q5FWH3 | 67,554 | 6.40 | 1.30 | 0.97 | 2 (2.66) | 88.29 |
| Complement activation | |||||||
| C3 | P01027 | 173,622 | 6.02 | 1.37 | 2.56 | 26 (48.50) | 7024.70 |
| C1QC | Q02105 | 27,193 | 8.61 | 1.39 | 0.85 | 22 (29.10) | 1752.80 |
| C1QB | P14106 | 27,193 | 8.83 | 1.73 | 0.99 | 54 (40.60) | 4681.20 |
| C4BP | P08607 | 53,951 | 6.83 | 0.85 | 1.05 | 7 (14.50) | 614.73 |
| CPN2 | Q9DBB9 | 61,608 | 5.63 | 0.95 | 3.61 | 3 (4.94) | 191.85 |
| C2 | P21180 | 144,278 | 7.23 | 32.07 | 4.11 | 20 (13.10) | 1593.08 |
| C1QA | P98086 | 26,093 | 9.26 | 1.35 | 0.87 | 17 (29.80) | 1017.74 |
| CFI | Q61129 | 69,853 | 7.72 | 1.47 | 2.11 | 8 (12.67) | 551.94 |
| Cytoskeletal protein | |||||||
| ACTB | P60710 | 41,890 | 5.29 | 1.30 | 2.41 | 6 (14.29) | 322.06 |
| TUBA1C | P68373 | 50,566 | 4.96 | 1.34 | 2.49 | 2 (36.53) | 1498.38 |
| TUBB4B | P68372 | 50,255 | 4.79 | 2.27 | 2.14 | 15 (63.82) | 2370.87 |
| DSP | E9Q557 | 334,465 | 6.44 | 2.25 | 0.08 | 4 (1.25) | 169.10 |
| GFAP | P03995 | 49,987 | 5.42 | 0.70 | 0.26 | 1 (2.56) | 33.58 |
| JUP | Q02257 | 82,479 | 5.75 | 1.34 | 0.31 | 3 (4.56) | 117.12 |
| MSN | P26041 | 67,816 | 6.08 | 0.59 | 0.62 | 2 (2.77) | 106.67 |
| Lipid metabolism | |||||||
| CES1 | Q8VCC2 | 62,577 | 6.15 | 3.75 | 6.59 | 3 (8.85) | 278.86 |
| CRAT | P47934 | 71,289 | 8.63 | 3.52 | 8.07 | 10 (16.45) | 605.83 |
| PTGDS | O09114 | 21,523 | 7.66 | 2.23 | 6.21 | 2 (8.90) | 124.98 |
| NPC2 | Q9Z0J0 | 17,000 | 7.57 | 1.77 | 4.37 | 4 (14.67) | 198.99 |
| EPHX1 | Q9D379 | 52,808 | 6.77 | 4.86 | Infinity | 2 (3.74) | 60.14 |
| GK | Q8C635 | 58,210 | 6.12 | 0.83 | 2.96 | 2 (4.39) | 135.99 |
| GK2 * | Q9WU65 | 58,210 | 5.57 | 0.83 | 2.96 | 2 (4.11) | 127.32 |
| GK3 * (in human) | Q14409 | 58,210 | 6.00 | 0.83 | 2.96 | 2 (4.17) | 129.18 |
| APOE | P08226 | 35,619 | 5.65 | 1.08 | 2.31 | 19 (44.20) | 1278.93 |
| APOA4 | P06728 | 44,043 | 5.28 | 1.03 | 2.23 | 31 (48.90) | 2557.95 |
| APOM | Q9Z1R3 | 21,771 | 5.66 | 4.92 | 1.79 | 2 (8.95) | 109.44 |
| APOC1 | P34928 | 9893 | 8.01 | 1.07 | 6.05 | 2 (20.45) | 104.77 |
| PON1 | P52430 | 39,785 | 5.08 | 0.46 | 0.58 | 6 (23.94) | 458.68 |
| ADIPOQ | Q60994 | 26,435 | 5.42 | 0.48 | 1.01 | 2 (11.11) | 149.13 |
| ECI1 | P42125 | 32,546 | 8.80 | 2.68 | 1.44 | 2 (10.38) | 149.01 |
| Others | |||||||
| RPS27A | P62983 | 12,358 | 9.34 | 1.73 | 3.20 | 9 (40.19) | 503.74 |
| ITIH1 | Q61702 | 101,362 | 6.31 | 1.25 | 3.41 | 11 (17.5) | 1101.71 |
| AMBP | Q07456 | 39,481 | 5.95 | 1.85 | 4.09 | 2 (7.22) | 153.34 |
| TTR | P07309 | 15,833 | 11.13 | 1.27 | 2.33 | 15 (59.90) | 1558.55 |
| ERP44 | Q9D1Q6 | 47,274 | 5.09 | 3.75 | 4.41 | 2 (4.93) | 61.93 |
| CPN2 | Q9DBB9 | 49,092 | 7.61 | 1.05 | 3.61 | 2 (6.87) | 105.60 |
| PROC | P33587 | 52,746 | 5.85 | 1.84 | 3.37 | 2 (4.16) | 84.11 |
| TRF | Q921I1 | 78,371 | 6.81 | 1.09 | 2.13 | 94 (52.9) | 9020.85 |
| KNG1 | O08677 | 38,818 | 6.34 | 0.84 | 1.53 | 3 (21.10) | 872.77 |
| ATRN | Q9WU60 | 162,969 | 7.24 | 0.76 | 2.91 | 2 (1.20) | 80.25 |
| ITIH3 | Q61704 | 78,169 | 5.49 | 0.62 | 1.07 | 2 (13.50) | 896.4 |
| NMES1 | Q810Q5 | 9589 | 9.45 | 0.72 | 1.12 | 2 (11) | 71.69 |
| CTSD | P18242 | 44,768 | 6.10 | 1.63 | 0.75 | 7 (14.2) | 324.5 |
| SERPINA1 | P07758 | 58,642 | 5.37 | 7.14 | 6.26 | 3 (30) | 103.57 |
| GSTA3 | P30115 | 22,658 | 9.21 | 9.08 | 1.81 | 2 (13.30) | 148.63 |
| Il4i1 | O09046 | 72,032 | 8.79 | 2.33 | 4.16 | 5 (9.30) | 236.71 |
| SERPINA3F | Q80X76 | 45,927 | 4.91 | 1.37 | 1.83 | 8 (13.93) | 882.80 |
| PZP | Q61838 | 165,815 | 5.97 | 0.86 | 1.17 | 50 (21.78) | 3570.34 |
| PDIA4 | P08003 | 72,914 | 4.96 | 4.75 | 4.94 | 4 (7.01) | 217.37 |
| RAB2A | P53994 | 23,545 | 6.08 | 0.27 | 0.32 | 3 (15.10) | 152.70 |
| CPB2 | Q9JHH6 | 49,092 | 6.72 | 0.34 | 0.35 | 2 (3.55) | 83.62 |
| Cellular Components | ||||||
|---|---|---|---|---|---|---|
| # Term ID | Term Description | Observed Gene Count | Background Gene Count | Strength | False Discovery Rate | Matching Proteins in Your Network (Labels) |
| GO:0035686 | Sperm fibrous sheath | 2 | 12 | 2.31 | 0.0037 | GAPDHS, AKAP3 |
| GO:0002199 | Zona pellucida receptor complex | 2 | 13 | 2.28 | 0.0037 | CCT3, HSPA1L |
| GO:0097228 | Sperm principal piece | 2 | 29 | 1.93 | 0.0094 | GAPDHS, AKAP3 |
| GO:0043209 | Myelin sheath | 6 | 212 | 1.54 | 1.91e-06 | CCT3, HSPD1, TUBB4B, PEBP1, GPI1, GAPDH |
| GO:0005740 | Mitochondrial envelope | 4 | 590 | 0.92 | 0.0283 | HSPD1, CRAT, PEBP1, DIABLO |
| GO:0031967 | Organelle envelope | 5 | 1015 | 0.78 | 0.0283 | HSPD1, CRAT, PEBP1, DIABLO, GAPDH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chorfa, A.; Goubely, C.; Henry-Berger, J.; Guiton, R.; Drevet, J.R.; Saez, F. Identification of Arvicola terrestris scherman Sperm Antigens for Immune Contraceptive Purposes. Int. J. Mol. Sci. 2021, 22, 9965. https://doi.org/10.3390/ijms22189965
Chorfa A, Goubely C, Henry-Berger J, Guiton R, Drevet JR, Saez F. Identification of Arvicola terrestris scherman Sperm Antigens for Immune Contraceptive Purposes. International Journal of Molecular Sciences. 2021; 22(18):9965. https://doi.org/10.3390/ijms22189965
Chicago/Turabian StyleChorfa, Areski, Chantal Goubely, Joelle Henry-Berger, Rachel Guiton, Joël R. Drevet, and Fabrice Saez. 2021. "Identification of Arvicola terrestris scherman Sperm Antigens for Immune Contraceptive Purposes" International Journal of Molecular Sciences 22, no. 18: 9965. https://doi.org/10.3390/ijms22189965
APA StyleChorfa, A., Goubely, C., Henry-Berger, J., Guiton, R., Drevet, J. R., & Saez, F. (2021). Identification of Arvicola terrestris scherman Sperm Antigens for Immune Contraceptive Purposes. International Journal of Molecular Sciences, 22(18), 9965. https://doi.org/10.3390/ijms22189965







