Points of View on the Tools for Genome/Gene Editing
Abstract
1. Preface
2. Endonucleases for Genome Editing
2.1. Homing Endonuclease and Meganuclease
2.2. Zinc Finger Nuclease (ZFN)
2.3. Transcription Activator-Like Effector Nuclease (TALEN)
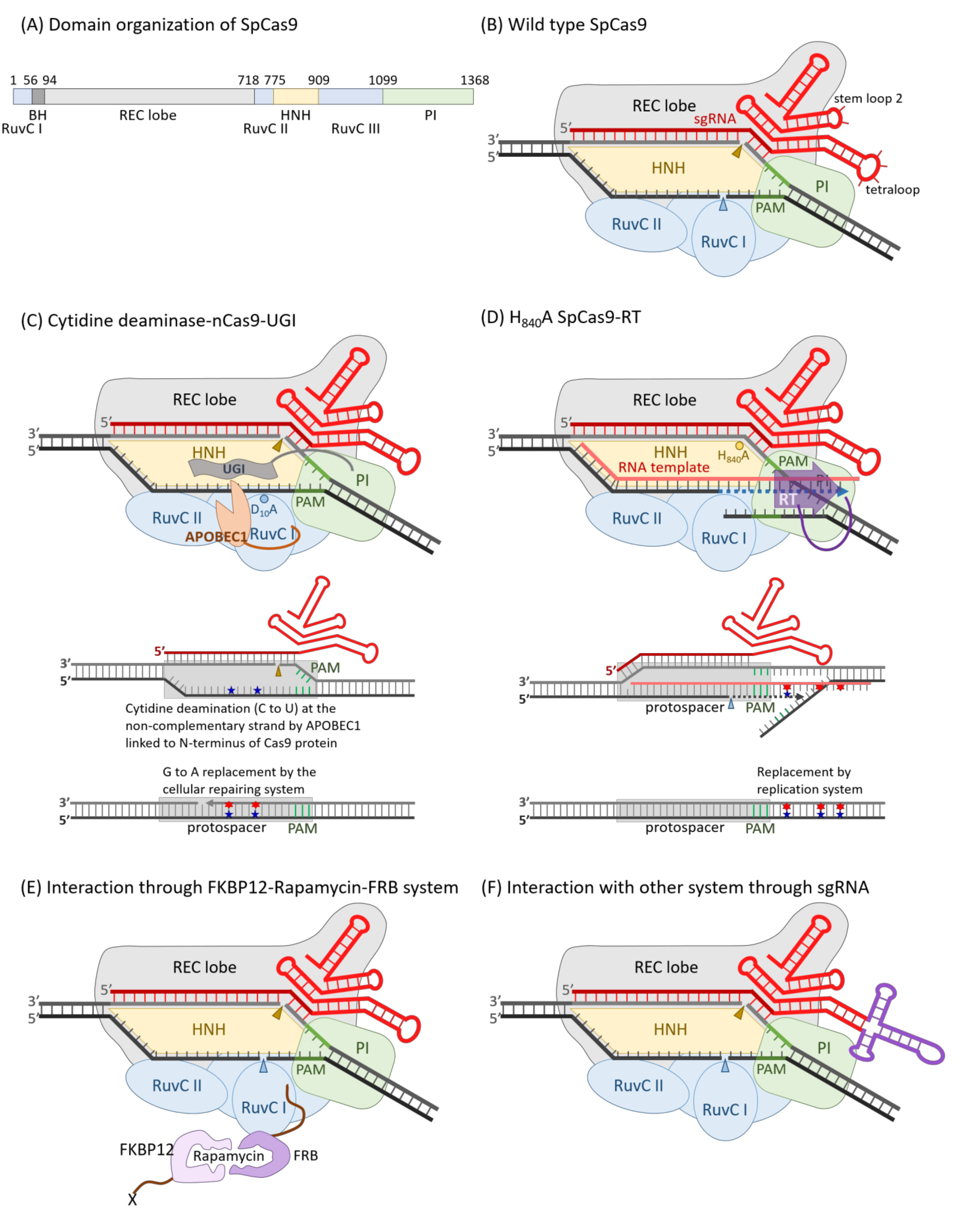
2.4. CRISPR/Cas Nucleases
3. Precise Genome Editing
3.1. With Double-Strand Breaks
3.2. Without Double-Strand Breaks
4. Forms and Delivery Methods of Programmable Site-Specific Endonucleases/DNA Editors
5. Strategies for In Vivo Genome Editing
5.1. Delivery of Cas9/sgRNA RNP with Virus-Like Particles
5.2. Delivery of Cas9/sgRNA RNP with Extracellular Vesicles
6. Concluding Remarks and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Sansbury, B.M.; Kmiec, E.B. On the Origins of Homology Directed Repair in Mammalian Cells. Int. J. Mol. Sci. 2021, 22, 3348. [Google Scholar] [CrossRef]
- Ramirez, C.L.; Certo, M.T.; Mussolino, C.; Goodwin, M.J.; Cradick, T.J.; McCaffrey, A.P.; Cathomen, T.; Scharenberg, A.M.; Joung, J.K. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012, 40, 5560–5568. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Miller, J.C.; Lee, Y.-L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef]
- Gregory, T.R.; Nicol, J.A.; Tamm, H.; Kullman, B.; Kullman, K.; Leitch, I.J.; Murray, B.G.; Kapraun, D.F.; Greilhuber, J.; Bennett, M.D. Eukaryotic genome size databases. Nucleic Acids Res. 2007, 35, D332–D338. [Google Scholar] [CrossRef] [PubMed]
- Redi, C.; Capanna, E. Genome size evolution: Sizing mammalian genomes. Cytogenet. Genome Res. 2012, 137, 97–112. [Google Scholar] [CrossRef]
- Bos, J.; Heyting, C.; Borst, P.; Arnberg, A.; Van Bruggen, E. An insert in the single gene for the large ribosomal RNA in yeast mitochondrial DNA. Nature 1978, 275, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, M.; Coen, D.; Deutsch, J.; Dujon, B.; Netter, P.; Petrochilo, E.; Slonimski, P. Recombination of mitochondria in saccharomyces-cerevisiae. Bull. de l Inst. Pasteur 1971, 69, 215–239. [Google Scholar]
- Jacquier, A.; Dujon, B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 1985, 41, 383–394. [Google Scholar] [PubMed]
- Stoddard, B.L. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 2011, 19, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005, 38, 49–95. [Google Scholar] [CrossRef]
- Taylor, G.K.; Stoddard, B.L. Structural, functional and evolutionary relationships between homing endonucleases and proteins from their host organisms. Nucleic Acids Res. 2012, 40, 5189–5200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hafez, M.; Hausner, G. Homing endonucleases: DNA scissors on a mission. Genome 2012, 55, 553–569. [Google Scholar] [CrossRef]
- Pâques, F.; Duchateau, P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr. Gene Ther. 2007, 7, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pellenz, S.; Ulge, U.; Stoddard, B.L.; Monnat, R.J., Jr. Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009, 37, 1650–1662. [Google Scholar] [CrossRef][Green Version]
- Arnould, S.; Delenda, C.; Grizot, S.; Desseaux, C.; Paques, F.; Silva, G.; Smith, J. The I-CreI meganuclease and its engineered derivatives: Applications from cell modification to gene therapy. Protein Eng. Des. Sel. 2011, 24, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, L.P.; Chandrasegaran, S. Functional domains in Fok I restriction endonuclease. Proc. Natl. Acad. Sci. USA 1992, 89, 4275–4279. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Chandrasegaran, S. Chimeric restriction endonuclease. Proc. Natl. Acad. Sci. USA 1994, 91, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef]
- Persikov, A.V.; Wetzel, J.L.; Rowland, E.F.; Oakes, B.L.; Xu, D.J.; Singh, M.; Noyes, M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic Acids Res. 2015, 43, 1965–1984. [Google Scholar] [CrossRef]
- Chandrasegaran, S.; Smith, J. Chimeric restriction enzymes: What is next? Biol. Chem. 1999, 380, 841–848. [Google Scholar] [CrossRef]
- Smith, J.; Bibikova, M.; Whitby, F.G.; Reddy, A.; Chandrasegaran, S.; Carroll, D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000, 28, 3361–3369. [Google Scholar] [CrossRef]
- Bibikova, M.; Carroll, D.; Segal, D.J.; Trautman, J.K.; Smith, J.; Kim, Y.-G.; Chandrasegaran, S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001, 21, 289–297. [Google Scholar] [CrossRef]
- Şöllü, C.; Pars, K.; Cornu, T.I.; Thibodeau-Beganny, S.; Maeder, M.L.; Joung, J.K.; Heilbronn, R.; Cathomen, T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010, 38, 8269–8276. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Vo, T.D.; Mendel, M.C.; Greenberg, S.G.; Wang, J.; Xia, D.F.; Miller, J.C.; Urnov, F.D.; Gregory, P.D.; Holmes, M.C. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods 2011, 8, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Singh, M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014, 42, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Garton, M.; Najafabadi, H.S.; Schmitges, F.W.; Radovani, E.; Hughes, T.R.; Kim, P.M. A structural approach reveals how neighbouring C2H2 zinc fingers influence DNA binding specificity. Nucleic Acids Res. 2015, 43, 9147–9157. [Google Scholar] [CrossRef] [PubMed]
- Bonas, U.; Stall, R.E.; Staskawicz, B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. MGG 1989, 218, 127–136. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Dey, S.; Chakravarty, A.; Guha Biswas, P.; De Guzman, R.N. The type III secretion system needle, tip, and translocon. Protein Sci. 2019, 28, 1582–1593. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Yan, C.; Pan, X.; Mahfouz, M.; Wang, J.; Zhu, J.-K.; Shi, Y.; Yan, N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 2012, 335, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Bradley, P. Structural modeling of TAL effector–DNA interactions. Protein Sci. 2012, 21, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.; Chandrahas, A.; Bleris, L. Transcription activator-like effectors: A toolkit for synthetic biology. ACS Synth. Biol. 2014, 3, 708–716. [Google Scholar] [CrossRef]
- Streubel, J.; Blücher, C.; Landgraf, A.; Boch, J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 2012, 30, 593–595. [Google Scholar] [CrossRef]
- Richter, A.; Streubel, J.; Boch, J. TAL effector DNA-binding principles and specificity. In TALENs; Springer: Berlin/Heidelberg, Germany, 2016; pp. 9–25. [Google Scholar]
- Carlson, D.F.; Tan, W.; Lillico, S.G.; Stverakova, D.; Proudfoot, C.; Christian, M.; Voytas, D.F.; Long, C.R.; Whitelaw, C.B.A.; Fahrenkrug, S.C. Efficient TALEN-mediated gene knockout in livestock. Proc. Natl. Acad. Sci. USA 2012, 109, 17382–17387. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Mussolino, C.; Morbitzer, R.; Lütge, F.; Dannemann, N.; Lahaye, T.; Cathomen, T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011, 39, 9283–9293. [Google Scholar] [CrossRef]
- Schreiber, T.; Sorgatz, A.; List, F.; Blüher, D.; Thieme, S.; Wilmanns, M.; Bonas, U. Refined requirements for protein regions important for activity of the TALE AvrBs3. PLoS ONE 2015, 10, e0120214. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wu, X.; Chai, J.; Han, Z. Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res. 2012, 22, 1716–1720. [Google Scholar] [CrossRef]
- Doyle, E.L.; Stoddard, B.L.; Voytas, D.F.; Bogdanove, A.J. TAL effectors: Highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 2013, 23, 390–398. [Google Scholar] [CrossRef][Green Version]
- Cong, L.; Zhou, R.; Kuo, Y.-c.; Cunniff, M.; Zhang, F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat. Commun. 2012, 3, 1–6. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Starker, C.G.; Voytas, D.F. Efficient design and assembly of custom TALENs using the Golden Gate platform. In Chromosomal Mutagenesis; Springer: Berlin/Heidelberg, Germany, 2015; pp. 133–159. [Google Scholar]
- Morbitzer, R.; Elsaesser, J.; Hausner, J.; Lahaye, T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011, 39, 5790–5799. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.; Gruetzner, R.; Werner, S.; Engler, C.; Marillonnet, S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 2011, 6, e19722. [Google Scholar] [CrossRef]
- Zhang, F.; Cong, L.; Lodato, S.; Kosuri, S.; Church, G.M.; Arlotta, P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011, 29, 149–153. [Google Scholar] [CrossRef]
- Deng, P.; Carter, S.; Fink, K. Design, Construction, and Application of Transcription Activation-Like Effectors. In Viral Vectors for Gene Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 47–58. [Google Scholar]
- Nitsch, S.; Mussolino, C. Generation of TALE-based designer epigenome modifiers. In Epigenome Editing; Springer: Berlin/Heidelberg, Germany, 2018; pp. 89–109. [Google Scholar]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Charpentier, E. CRISPR-Cas: Biology, mechanisms and relevance. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150496. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The biology of CRISPR-Cas: Backward and forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.A.; Kocak, D.D.; Fitzgibbon, C.J.; McMenemy, J.; Gersbach, C.A.; Marszalek, P.E. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2015, 43, 8924–8941. [Google Scholar] [CrossRef]
- Sternberg, S.H.; LaFrance, B.; Kaplan, M.; Doudna, J.A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 2015, 527, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Le Rhun, A.; Escalera-Maurer, A.; Bratovič, M.; Charpentier, E. CRISPR-Cas in Streptococcus pyogenes. RNA Biol. 2019, 16, 380–389. [Google Scholar] [CrossRef]
- Furuhata, Y.; Kato, Y. Asymmetric Roles of Two Histidine Residues in Streptococcus pyogenes Cas9 Catalytic Domains upon Chemical Rescue. Biochemistry 2021, 60, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.; Zolekar, A.; Babu, K.; Lin, V.J.; Hayatshahi, H.S.; Rajan, R.; Wang, Y.-C.; Liu, J. Structural and functional insights into the bona fide catalytic state of Streptococcus pyogenes Cas9 HNH nuclease domain. Elife 2019, 8, e46500. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; Van Der Oost, J.; Regev, A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Zhong, G.; Wang, H.; Li, Y.; Tran, M.H.; Farzan, M. Cpf1 proteins excise CRISPR RNAs from mRNA transcripts in mammalian cells. Nat. Chem. Biol. 2017, 13, 839–841. [Google Scholar] [CrossRef]
- Yamano, T.; Nishimasu, H.; Zetsche, B.; Hirano, H.; Slaymaker, I.M.; Li, Y.; Fedorova, I.; Nakane, T.; Makarova, K.S.; Koonin, E.V. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 2016, 165, 949–962. [Google Scholar] [CrossRef]
- Tran, M.H.; Park, H.; Nobles, C.L.; Karunadharma, P.; Pan, L.; Zhong, G.; Wang, H.; He, W.; Ou, T.; Crynen, G. A more efficient CRISPR-Cas12a variant derived from Lachnospiraceae bacterium MA2020. Mol. Ther. -Nucleic Acids 2021, 24, 40–53. [Google Scholar] [CrossRef]
- Fu, Y.; Sander, J.D.; Reyon, D.; Cascio, V.M.; Joung, J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014, 32, 279–284. [Google Scholar] [CrossRef]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Janik, E.; Niemcewicz, M.; Ceremuga, M.; Krzowski, L.; Saluk-Bijak, J.; Bijak, M. Various Aspects of a Gene Editing System—CRISPR–Cas9. Int. J. Mol. Sci. 2020, 21, 9604. [Google Scholar] [CrossRef] [PubMed]
- Grundy, G.J.; Moulding, H.A.; Caldecott, K.W.; Rulten, S.L. One ring to bring them all—the role of Ku in mammalian non-homologous end joining. DNA Repair 2014, 17, 30–38. [Google Scholar] [CrossRef]
- Fell, V.L.; Schild-Poulter, C. The Ku heterodimer: Function in DNA repair and beyond. Mutat. Res./Rev. Mutat. Res. 2015, 763, 15–29. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef] [PubMed]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Trevino, A.E.; Zhang, F. Genome editing using Cas9 nickases. Methods Enzymol. 2014, 546, 161–174. [Google Scholar] [PubMed]
- Nakade, S.; Tsubota, T.; Sakane, Y.; Kume, S.; Sakamoto, N.; Obara, M.; Daimon, T.; Sezutsu, H.; Yamamoto, T.; Sakuma, T. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014, 5, 1–8. [Google Scholar] [CrossRef]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.-I.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tsunekawa, Y.; Hernandez-Benitez, R.; Wu, J.; Zhu, J.; Kim, E.J.; Hatanaka, F.; Yamamoto, M.; Araoka, T.; Li, Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016, 540, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Bloomer, H.; Smith, R.H.; Hakami, W.; Larochelle, A. Genome editing in human hematopoietic stem and progenitor cells via CRISPR-Cas9-mediated homology-independent targeted integration. Mol. Ther. 2021, 29, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Schellenberger, V.; Wang, C.-w.; Geething, N.C.; Spink, B.J.; Campbell, A.; To, W.; Scholle, M.D.; Yin, Y.; Yao, Y.; Bogin, O. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat. Biotechnol. 2009, 27, 1186–1190. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C: G-to-T: A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Liu, Y.; Yang, B.; Wang, X.; Wei, J.; Lu, Z.; Zhang, Y.; Wu, J.; Huang, X. Base editing with a Cpf1–cytidine deaminase fusion. Nat. Biotechnol. 2018, 36, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.F.; Zhao, K.T.; Eton, E.; Lapinaite, A.; Newby, G.A.; Thuronyi, B.W.; Wilson, C.; Koblan, L.W.; Zeng, J.; Bauer, D.E. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 2020, 38, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Lapinaite, A.; Knott, G.J.; Palumbo, C.M.; Lin-Shiao, E.; Richter, M.F.; Zhao, K.T.; Beal, P.A.; Liu, D.R.; Doudna, J.A. DNA capture by a CRISPR-Cas9–guided adenine base editor. Science 2020, 369, 566–571. [Google Scholar] [CrossRef]
- Grünewald, J.; Zhou, R.; Lareau, C.A.; Garcia, S.P.; Iyer, S.; Miller, B.R.; Langner, L.M.; Hsu, J.Y.; Aryee, M.J.; Joung, J.K. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020, 38, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Sakata, R.C.; Ishiguro, S.; Mori, H.; Tanaka, M.; Tatsuno, K.; Ueda, H.; Yamamoto, S.; Seki, M.; Masuyama, N.; Nishida, K. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020, 38, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, B.; Chen, L.; Xie, L.; Yu, W.; Wang, Y.; Li, L.; Yin, S.; Yang, L.; Hu, H. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020, 38, 856–860. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Marzec, M.; Brąszewska-Zalewska, A.; Hensel, G. Prime editing: A new way for genome editing. Trends Cell Biol. 2020, 30, 257–259. [Google Scholar] [CrossRef]
- Hsu, J.Y.; Grünewald, J.; Szalay, R.; Shih, J.; Anzalone, A.V.; Lam, K.C.; Shen, M.W.; Petri, K.; Liu, D.R.; Joung, J.K. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021, 12, 1–6. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, M.A.; Corn, J.E.; Carroll, D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods 2017, 121, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Chau, C.; Actis, P.; Hewitt, E. Methods for protein delivery into cells: From current approaches to future perspectives. Biochem. Soc. Trans. 2020, 48, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Loh, O.Y.; Ho, A.M.; Rim, J.E.; Kohli, P.; Patankar, N.A.; Espinosa, H.D. Electric field-induced direct delivery of proteins by a nanofountain probe. Proc. Natl. Acad. Sci. USA 2008, 105, 16438–16443. [Google Scholar] [CrossRef]
- Yang, R.; Lemaître, V.; Huang, C.; Haddadi, A.; McNaughton, R.; Espinosa, H.D. Monoclonal cell line generation and CRISPR/Cas9 manipulation via single-cell electroporation. Small 2018, 14, 1702495. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, H.D.; Mukherjee, P.; Patino, C. Nanofountain Probe Electroporation for Monoclonal Cell Line Generation. In Electroporation Protocols; Springer: Berlin/Heidelberg, Germany, 2020; pp. 59–68. [Google Scholar]
- Xie, X.; Xu, A.M.; Leal-Ortiz, S.; Cao, Y.; Garner, C.C.; Melosh, N.A. Nanostraw–electroporation system for highly efficient intracellular delivery and transfection. ACS Nano 2013, 7, 4351–4358. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Chen, H.; Qiu, R.; Hanna, M.; Ma, E.; Hjort, M.; Zhang, A.; Lewis, R.; Wu, J.; Melosh, N. Universal intracellular biomolecule delivery with precise dosage control. Sci. Adv. 2018, 4, eaat8131. [Google Scholar] [CrossRef]
- Cao, Y.; Ma, E.; Cestellos-Blanco, S.; Zhang, B.; Qiu, R.; Su, Y.; Doudna, J.A.; Yang, P. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc. Natl. Acad. Sci. USA 2019, 116, 7899–7904. [Google Scholar] [CrossRef]
- Briggs, J.A.; Simon, M.N.; Gross, I.; Kräusslich, H.-G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef]
- Lingappa, J.R.; Reed, J.C.; Tanaka, M.; Chutiraka, K.; Robinson, B.A. How HIV-1 Gag assembles in cells: Putting together pieces of the puzzle. Virus Res. 2014, 193, 89–107. [Google Scholar] [CrossRef]
- Yip, B.H. Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules 2020, 10, 839. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Dang, Y.; Abraham, S.; Ma, H.; Zhang, J.; Guo, H.; Cai, Y.; Mikkelsen, J.; Wu, H.; Shankar, P. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016, 23, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massouridès, E.; Sohier, T.J.; Corbin, A. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Montagna, C.; Petris, G.; Casini, A.; Maule, G.; Franceschini, G.M.; Zanella, I.; Conti, L.; Arnoldi, F.; Burrone, O.R.; Zentilin, L. VSV-G-enveloped vesicles for traceless delivery of CRISPR-Cas9. Mol. Ther. Nucleic Acids 2018, 12, 453–462. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Bölker, M.; Kahmann, R. Translation of the bacteriophage Mu mom gene is positively regulated by the phage com gene product. Cell 1989, 57, 1201–1210. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Kahmann, R. Translational stimulation: RNA sequence and structure requirements for binding of Com protein. Cell 1991, 65, 259–269. [Google Scholar] [CrossRef]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’genome editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef]
- Lyu, P.; Wang, L.; Lu, B. Virus-like particle mediated CRISPR/Cas9 delivery for efficient and safe genome editing. Life 2020, 10, 366. [Google Scholar] [CrossRef]
- Lu, Z.; Yao, X.; Lyu, P.; Yadav, M.; Yoo, K.; Atala, A.; Lu, B. Lentiviral Capsid-Mediated Streptococcus pyogenes Cas9 Ribonucleoprotein Delivery for Efficient and Safe Multiplex Genome Editing. CRISPR J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Muratori, C.; Bona, R.; Federico, M. Lentivirus-based virus-like particles as a new protein delivery tool. In Lentivirus Gene Engineering Protocols; Springer: Berlin/Heidelberg, Germany, 2010; pp. 111–124. [Google Scholar]
- Peretti, S.; Schiavoni, I.; Pugliese, K.; Federico, M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol. Ther. 2005, 12, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Tessmer, U.; Schubert, U.; Kräusslich, H.-G. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J. Virol. 2000, 74, 9727–9731. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Xiao, H.; Kim, J.; Seshaiah, P.; Natsoulis, G.; Boeke, J.D.; Hahn, B.H.; Kappes, J.C. Targeting foreign proteins to human immunodeficiency virus particles via fusion with Vpr and Vpx. J. Virol. 1995, 69, 3389–3398. [Google Scholar] [CrossRef]
- Lu, B.; Javidi-Parsijani, P.; Makani, V.; Mehraein-Ghomi, F.; Sarhan, W.M.; Sun, D.; Yoo, K.W.; Atala, Z.P.; Lyu, P.; Atala, A. Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing. Nucleic Acids Res. 2019, 47, e44. [Google Scholar] [CrossRef]
- Banaszynski, L.A.; Liu, C.W.; Wandless, T.J. Characterization of the FKBP.Rapamycin.FRB ternary complex. J. Am. Chem. Soc. 2005, 127, 4715–4721. [Google Scholar] [PubMed]
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with binding domain of human FRAP. Science 1996, 273, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, X.-F.; Brown, E.J.; Schreiber, S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 1995, 92, 4947–4951. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Choi, J.; Clardy, J. Refined structure of the FKBP12–rapamycin–FRB ternary complex at 2.2 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1–18. [Google Scholar]
- Van Niel, G.; d’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular microvesicles/exosomes: Discovery, disbelief, acceptance, and the future? Leukemia 2020, 34, 3126–3135. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed]
- Pols, M.S.; Klumperman, J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009, 315, 1584–1592. [Google Scholar] [CrossRef]
- Yao, X.; Lyu, P.; Yoo, K.; Yadav, M.K.; Singh, R.; Atala, A.; Lu, B. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J. Extracell. Vesicles 2021, 10, e12076. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, X.; Xie, F.; Xu, B.; Xie, P.; Yang, T.; Shi, Q.; Zhang, C.-Y.; Zhang, Y.; Chen, J. An engineered exosome for delivering sgRNA: Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020, 8, 2966–2976. [Google Scholar] [CrossRef]
- Tricarico, C.; Clancy, J.; D'Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Kanada, M.; Bachmann, M.H.; Hardy, J.W.; Frimannson, D.O.; Bronsart, L.; Wang, A.; Sylvester, M.D.; Schmidt, T.L.; Kaspar, R.L.; Butte, M.J. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 2015, 112, E1433–E1442. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef] [PubMed]
| Domain Where Mutations Engineered | Functional Changes | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mutants | RuvC-I (1–55) | BH (56–93) | REC Lobe (94–717) | RuvC-II (718–774) | HNH (775–908) | RuvC-III (909–1098) | PI (1099–1368) | ||
| SpG | D1135L, S1136W, G1218Q, E1219Q, R1335Q, T1337R | The recognition sequence of PAM changed from NGG to NGN. | [69] | ||||||
| SpRY | A61R | L1111R, D1135L S1136W, G1218Q E1219Q, N1317R, A1322R, R1333P, R1335Q, T1337R | The recognition sequence of PAM changed from NGG to PAMless (NRN > NYN). | ||||||
| eSpCas9(1.1) | K848A | K1003A R1060A | eSpCas9(1.1) displayed efficient and precise genome editing in human cells. | [70] | |||||
| SpCas9-HF1 | N497A R661A Q695A | Q926A | SpCas9-HF1 performed with high on-target activity and reduced off-target editing. | [71,72] | |||||
| nSpCas9 | D10A | RuvC nuclease activity was eliminated. (nickase) | [58] | ||||||
| SpCas9(H840A) | H840A | HNH nuclease activity was diminished. | |||||||
| dSpCas9 | D10A | H840A | Dead SpCas9 lost both of the RuvC and HNH nuclease activities. | ||||||
| SpCas9(N863A) | N863A | HNH nuclease activity was eliminated. | [67] | ||||||
| SpCas9(D839A) | D839A | HNH nuclease activity was diminished. | [66] | ||||||
| SpCas9(H983A) | H983A | RuvC nuclease activity was eliminated. | [65] | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chuang, C.-K.; Lin, W.-M. Points of View on the Tools for Genome/Gene Editing. Int. J. Mol. Sci. 2021, 22, 9872. https://doi.org/10.3390/ijms22189872
Chuang C-K, Lin W-M. Points of View on the Tools for Genome/Gene Editing. International Journal of Molecular Sciences. 2021; 22(18):9872. https://doi.org/10.3390/ijms22189872
Chicago/Turabian StyleChuang, Chin-Kai, and Wei-Ming Lin. 2021. "Points of View on the Tools for Genome/Gene Editing" International Journal of Molecular Sciences 22, no. 18: 9872. https://doi.org/10.3390/ijms22189872
APA StyleChuang, C.-K., & Lin, W.-M. (2021). Points of View on the Tools for Genome/Gene Editing. International Journal of Molecular Sciences, 22(18), 9872. https://doi.org/10.3390/ijms22189872






