Molecular Proteomics and Signalling of Human Platelets in Health and Disease
Abstract
1. Introduction
2. Overview of Platelet Proteomic Literature
3. Basic Platelet Proteome
4. Proteome Changes in Ageing Platelets
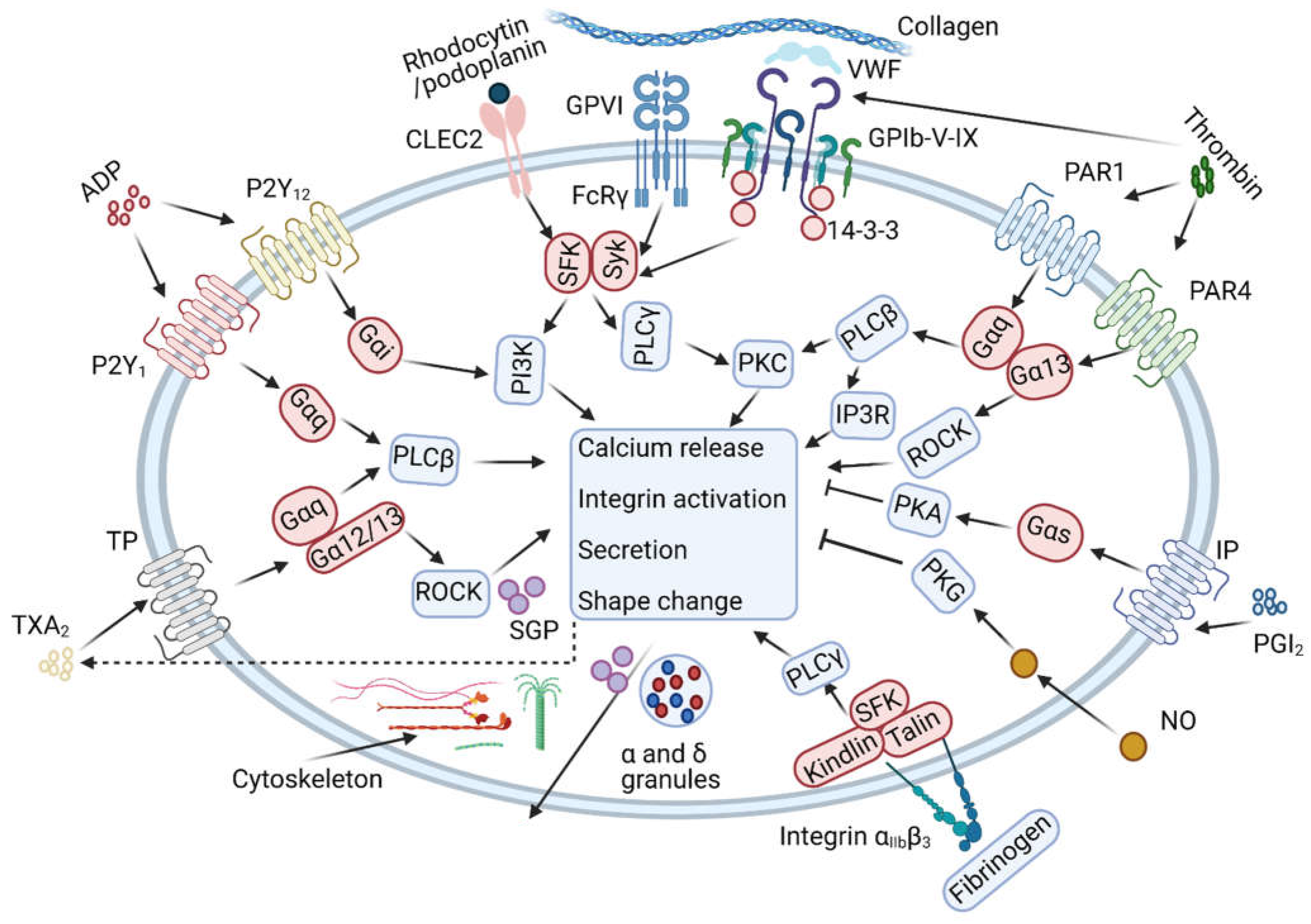
5. Collagen Receptor Glycoprotein VI (GPVI)
6. Signalling C-Type Lectin Receptor 2 (CLEC-2)
7. Thrombin and Protease-Activated Receptors (PAR1, PAR4)
8. Aspirin and Thromboxane A2
9. ADP Receptors and Platelet Inhibitors
10. Platelet Proteomics of Patients with Platelet Defects or Cardiovascular Disease
11. Practical and Technical Considerations
12. Future Perspectives and Challenges Ahead
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yau, J.W.; Teoh, H.; Verma, S. Endothelial Cell Control of Thrombosis. BMC Cardiovasc. Disord. 2015, 15, 130. [Google Scholar] [CrossRef]
- Van der Meijden, P.E.; Heemskerk, J.W. Platelet Biology and Functions: New Concepts and Clinical Perspectives. Nat. Rev. Cardiol. 2019, 16, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Vilahur, G. Thrombosis Formation on Atherosclerotic Lesions and Plaque Rupture. J. Int. Med. 2014, 276, 618–632. [Google Scholar] [CrossRef]
- Boyanova, D.; Nilla, S.; Birschmann, I.; Dandekar, T.; Dittrich, M. PlateletWeb: A Systems Biologic Analysis of Signaling Networks in Human Platelets. Blood 2012, 119, e22–e34. [Google Scholar] [CrossRef] [PubMed]
- Versteeg, H.H.; Heemskerk, J.W.; Levi, M.; Reitsma, P.S. New Fundamentals in Hemostasis. Physiol. Rev. 2013, 93, 327–358. [Google Scholar] [CrossRef] [PubMed]
- Weyrich, A.S.; Schwertz, H.; Kraiss, L.W.; Zimmerman, G.A. Protein Synthesis by Platelets: Historical and New Perspectives. J. Thromb. Haemost. 2009, 7, 241–246. [Google Scholar] [CrossRef]
- Bray, P.F.; McKenzie, S.E.; Edelstein, L.C.; Nagalla, S.; Delgrosso, K.; Ertel, A.; Kupper, J.; Jing, Y.; Londin, E.; Loher, P.; et al. The Complex Transcriptional Landscape of the Anucleate Human Platelet. BMC Genom. 2013, 14, 1. [Google Scholar] [CrossRef]
- Loosse, C.; Swieringa, F.; Heemskerk, J.W.; Sickmann, A.; Lorenz, C. Platelet Proteomics: From Discovery to Diagnosis. Exp. Rev. Proteom. 2018, 15, 467–476. [Google Scholar] [CrossRef]
- Mann, M.; Jensen, O.N. Proteomic Analysis of Post-Translational Modifications. Nat. Biotech. 2003, 21, 255–261. [Google Scholar] [CrossRef]
- Rosenberger, G.; Koh, C.C.; Guo, T.N.; Rost, H.L.; Kouvonen, P.; Collins, B.; Heusel, M.; Liu, Y.S.; Caron, E.; Vichalkovski, A.; et al. A Repository of Assays to Quantify 10,000 Human Proteins by SWATH-MS. Sci. Data 2014, 1, 140031. [Google Scholar] [CrossRef]
- Huang, J.; Swieringa, F.; Solari, F.A.; Provenzale, I.; Grassi, L.; De Simone, I.; Baaten, C.; Cavill, R.; Sickmann, A.; Frontini, M.; et al. Assessment of a Complete and Classified Platelet Proteome from Genome-Wide Transcripts of Human Platelets and Megakaryo-Cytes Covering Platelet Functions. Sci. Rep. 2021, 11, 12358. [Google Scholar] [CrossRef]
- Flores-Villalva, S.; Rogriguez-Hernandez, E.; Rubio-Venegas, Y.; Canto-Alarcon, J.G.; Milian-Suazo, F. What Can Proteomics Tell Us About Tuberculosis? J. Microbiol. Biotechnol. 2015, 25, 1181–1194. [Google Scholar] [CrossRef]
- Rijkers, M.; van den Eshof, B.L.; van der Meer, P.F.; van Alphen, F.P.; de Korte, D.; Leebeek, F.W.; Meijer, A.B.; Voorberg, J.; Jansen, A.J. Monitoring Storage Induced Changes in The Platelet Proteome Employing Label Free Quantitative Mass Spectrometry. Sci. Rep. 2017, 7, 11045. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.N.; Wang, J.; Li, Q.Q.; Zhang, Y.; Zhang, X.M. Enzyme and Chemical Assisted N-Terminal Blocked Peptides Analysis, Enchant, as A Selective Proteomics Approach Complementary to Conventional Shotgun Approach. J. Proteome Res. 2018, 17, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.J.; Reumiller, C.M.; Ercan, H.; Resch, U.; Butt, E.; Heber, S.; Liutkeviciute, Z.; Basilio, J.; Schmid, J.A.; Assinger, A.; et al. Comparative Proteomics Reveals Unexpected Quantitative Phosphorylation Differences Linked to Platelet Activation State. Sci. Rep. 2019, 9, 19009. [Google Scholar] [CrossRef] [PubMed]
- Tabb, D.L.; Vega-Montoto, L.; Rudnick, P.A.; Variyath, A.M.; Ham, A.J.L.; Bunk, D.M.; Kilpatrick, L.E.; Billheimer, D.D.; Blackman, R.K.; Cardasis, H.L.; et al. Repeatability and Reproducibility in Proteomic Identifications by Liquid Chromatography-Tandem Mass Spectrometry. J. Proteome Res. 2010, 9, 761–776. [Google Scholar] [CrossRef]
- Cremer, S.E.; Catalfamo, J.L.; Goggs, R.; Seemann, S.E.; Kristensen, A.T.; Brooks, M.B. Proteomic Profiling of The Thrombin-Activated Canine Platelet Secretome. PLoS ONE 2019, 14, e0224891. [Google Scholar] [CrossRef]
- Cimmino, G.; Tarallo, R.; Nassa, G.; De Filippo, M.R.; Giurato, G.; Ravo, M.; Rizzo, F.; Conte, S.; Pellegrino, G.; Cirillo, P.; et al. Activating Stimuli Induce Platelet Microrna Modulation and Proteome Reorganisation. Thromb. Haemost. 2015, 114, 96–108. [Google Scholar]
- Sabrkhany, S.; Kuijpers, M.J.; Knol, J.C.; Damink, S.; Dingemans, A.M.; Verheul, H.M.; Piersma, S.R.; Pham, T.V.; Griffioen, A.W.; Egbrink, M.; et al. Exploration of The Platelet Proteome in Patients with Early-Stage Cancer. J. Proteom. 2018, 177, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, I.; Barrachina, M.N.; Hermida-Nogueira, L.; Casas, V.; Moran, L.A.; Lacerenza, S.; Pinto-Llorente, R.; Eble, J.A.; de los Rios, V.; Dominguez, E.; et al. A Comprehensive Tyrosine Phosphoproteomic Analysis Reveals Novel Components of The Platelet Clec-2 Signaling Cascade. Thromb. Haemost. 2020, 120, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.P.; Unsworth, A.J.; Gibbins, J.M. Platelet Signaling: A Complex Interplay between Inhibitory and Activatory Networks. J. Thromb. Haemost. 2016, 14, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Dowal, L.; Yang, W.; Freeman, M.R.; Steen, H.; Flaumenhaft, R. Proteomic Analysis of Palmitoylated Platelet Proteins. Blood 2011, 118, e62–e73. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chae, S.; Park, J.; Bae, J.; Go, E.; Kim, S.J.; Kim, H.; Hwang, D.; Lee, S.W.; Lee, S.Y. Comprehensive Proteome Profiling of Platelet Identified A Protein Profile Predictive of Responses to An Antiplatelet Agent Sarpogrelate. Mol. Cell. Proteom. 2016, 15, 3461–3472. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, J.M.; Vaudel, M.; Gambaryan, S.; Radau, S.; Walter, U.; Martens, L.; Geiger, J.; Sickmann, A.; Zahedi, R.P. The First Comprehensive and Quantitative Analysis of Human Platelet Protein Composition Allows the Comparative Analysis of Structural and Functional Pathways. Blood 2012, 120, e73–e82. [Google Scholar] [CrossRef]
- Burkhart, J.M.; Gambaryan, S.; Watson, S.P.; Jurk, K.; Walter, U.; Sickmann, A.; Heemskerk, J.W.; Zahedi, R.P. What Can Proteomics Tell Us About Platelets? Circ. Res. 2014, 114, 1204–1219. [Google Scholar] [CrossRef]
- Klockenbusch, C.; Walsh, G.M.; Brown, L.M.; Hoffman, M.D.; Ignatchenko, V.; Kislinger, T.; Kast, J. Global Proteome Analysis Identifies Active Immunoproteasome Subunits in Human Platelets. Mol. Cell. Proteom. 2014, 13, 3308–3319. [Google Scholar] [CrossRef]
- Handtke, S.; Steil, L.; Palankar, R.; Conrad, J.; Cauhan, S.; Kraus, K.; Ferrara, M.; Dhople, V.; Wesche, J.; Völker, U.; et al. Role of Platelet Size Revisited: Function and Protein Composition of Large and Small Platelets. Thromb. Haemost. 2019, 119, 407–420. [Google Scholar] [CrossRef]
- Solari, F.A.; Mattheij, N.J.; Burkhart, J.M.; Swieringa, F.; Collins, P.W.; Cosemans, J.; Sickmann, A.; Heemskerk, J.W.; Zahedi, R.P. Combined Quantification of the Global Proteome, Phosphoproteome, and Proteolytic Cleavage to Characterize Altered Platelet Functions in the Human Scott Syndrome. Mol. Cell. Proteom. 2016, 15, 3154–3169. [Google Scholar] [CrossRef]
- Swieringa, F.; Solari, F.A.; Pagel, O.; Beck, F.; Huang, J.; Feijge, M.A.H.; Jurk, K.; Körver-Keularts, I.M.L.W.; Mattheij, N.J.A.; Faber, J.; et al. Impaired Iloprost-Induced Platelet Inhibition and Phosphoproteome Changes in Patients with Confirmed Pseudohypoparathyroidism Type Ia, Linked to Genetic Mutations in GNAS. Sci. Rep. 2020, 10, 11389. [Google Scholar] [CrossRef]
- Bergemalm, D.; Ramström, S.; Kardeby, C.; Hultenby, K.; Eremo, A.G.; Sihlbom, C.; Bergström, J.; Palmblad, J.; Aström, M. Platelet Proteome and Function in X-Linked Thrombocytopenia with Thalassemia and In Silico Comparisons with Gray Platelet Syndrome. Haematologica 2020. [Google Scholar] [CrossRef]
- Sims, M.C.; Mayer, L.; Collins, J.H.; Bariana, T.K.; Megy, K.; Lavenu-Bombled, C.; Seyres, D.; Kollipara, L.; Burden, F.S.; Greene, D.; et al. Novel Manifestations of Immune Dysregulation and Granule Defects in Gray Platelet Syndrome. Blood 2020, 136, 1956–1967. [Google Scholar] [CrossRef] [PubMed]
- Rocheleau, A.D.; Melrose, A.R.; Cunliffe, J.M.; Klimek, J.; Babur, O.; Yunga, S.T.; Ngo, A.T.P.; Pang, J.Q.; David, L.L.; McCarty, O.J.; et al. Identification, Quantification, and System Analysis of Protein N-Epsilon Lysine Methylation in Anucleate Blood Platelets. Proteomics 2019, 19, e1900001. [Google Scholar] [CrossRef] [PubMed]
- Prudova, A.; Serrano, K.; Eckhard, U.; Fortelny, N.; Devine, D.V.; Overall, C.M. TAILS N-Terminomics of Human Platelets Reveals Pervasive Metalloproteinase-Dependent Proteolytic Processing in Storage. Blood 2014, 124, e49–e60. [Google Scholar] [CrossRef] [PubMed]
- Schoenichen, C.; Bode, C.; Duerschmied, D. Role of Platelet Serotonin in Innate Immune Cell Recruitment. Front. Biosci. 2019, 24, 514–526. [Google Scholar]
- Pang, A.; Cui, Y.; Chen, Y.; Cheng, N.; Delaney, M.K.; Gu, M.; Stojanovic-Terpo, A.; Zhu, C.; Du, X. Shear-Induced Integrin Signaling in Platelet Phosphatidylserine Exposure, Microvesicle Release, and Coagulation. Blood 2018, 132, 533–543. [Google Scholar] [CrossRef]
- Zimmerman, G.A.; Weyrich, A.S. Signal-Dependent Protein Synthesis by Activated Platelets: New Pathways to Altered Phenotype and Function. Arterioscler. Thromb. Vasc. Biol. 2008, 28, s17–s24. [Google Scholar] [CrossRef]
- Ng, M.S.; Tung, J.P.; Fraser, J.F. Platelet Storage Lesions: What More Do We Know Now? Transfus. Med. Rev. 2018, 32, 144–154. [Google Scholar] [CrossRef]
- Thiele, T.; Braune, J.; Dhople, V.; Hammer, E.; Scharf, C.; Greinacher, A.; Volker, U.; Steil, L. Proteomic Profile of Platelets during Reconstitution of Platelet Counts after Apheresis. Proteom. Clin. Appl. 2016, 10, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Prudent, M.; Crettaz, D.; Delobel, J.; Tissot, J.D.; Lion, N. Proteomic Analysis of Intercept-Treated Platelets. J. Proteom. 2012, 76, 316–328. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, T.; Fan, Y.; Zhao, S. A Proteomic Approach Reveals the Variation in Human Platelet Protein Composition after Storage at Different Temperatures. Platelets 2019, 30, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.; Padula, M.P.; Marks, D.C.; Johnson, L. Refrigerated Storage of Platelets Initiates Changes in Platelet Surface Marker Expression and Localization of Intracellular Proteins. Transfusion 2016, 56, 2548–2559. [Google Scholar] [CrossRef]
- Salunkhe, V.; De Cuyper, I.M.; Papadopoulos, P.; van der Meer, P.F.; Daal, B.D.; Villa-Fajardo, M.; de Korte, D.; van den Berg, T.K.; Gutiérrez, L. A Comprehensive Proteomics Study on Platelet Concentrates: Platelet Proteome, Storage Time and Mirasol Pathogen Reduction Technology. Platelets 2019, 30, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Schubert, P.; Culibrk, B.; Karwal, S.; Goodrich, R.P.; Devine, D.V. Protein Translation Occurs in Platelet Concentrates Despite Riboflavin/UV Light Pathogen Inactivation Treatment. Proteom. Clin. Appl. 2016, 10, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Sonego, G.; Abonnenc, M.; Crettaz, D.; Lion, N.; Tissot, J.D.; Prudent, M. Irreversible Oxidations of Platelet Proteins after Riboflavin-UVB Pathogen Inactivation. Transfus. Clin. Biol. 2020, 27, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Hermida-Nogueira, L.; Barrachina, M.N.; Izquierdo, I.; García-Vence, M.; Lacerenza, S.; Bravo, S.; Castrillo, A.; García, Á. Proteomic Analysis of Extracellular Vesicles Derived From Platelet Concentrates Treated with Mirasol Identifies Biomarkers of Platelet Storage Lesion. J. Proteom. 2020, 210, 103529. [Google Scholar] [CrossRef] [PubMed]
- Aloui, C.; Barlier, C.; Claverol, S.; Fagan, J.; Awounou, D.; Tavernier, E.; Guyotat, D.; Hamzeh-Cognasse, H.; Cognasse, F.; Garraud, O.; et al. Differential Protein Expression of Blood Platelet Components Associated with Adverse Transfusion Reactions. J. Proteom. 2019, 194, 25–36. [Google Scholar] [CrossRef]
- Aloui, C.; Barlier, C.; Awounou, D.; Thiam, S.; Fagan, J.; Claverol, S.; Tavernier, E.; Mounier, C.; Hamzeh-Cognasse, H.; Cognasse, F.; et al. Dysregulated Pathways and Differentially Expressed Proteins Associated with Adverse Transfusion Reactions in Different Types of Platelet Components. J. Proteom. 2020, 218, 103717. [Google Scholar] [CrossRef]
- Nieswandt, B.; Watson, S.P. Platelet-Collagen Interaction: Is GPVI the Central Receptor? Blood 2003, 102, 449–461. [Google Scholar] [CrossRef]
- Perrella, G.; Huang, J.; Provenzale, I.; Swieringa, F.; Heubel-Moenen, F.C.; Farndale, R.W.; Roest, M.; van der Meijden, P.E.; Thomas, M.; Ariëns, R.A.; et al. Non-Redundant Roles of Platelet Glycoprotein VI and Integrin aIIbb3 in Fibrin-Mediated Microthrombus Formation. Arterioscler. Thromb. Vasc. Biol. 2021, 41, e97–e111. [Google Scholar] [CrossRef]
- Nagy, M.; van Geffen, J.P.; Stegner, D.; Adams, D.; Braun, A.; de Witt, S.M.; Elvers, M.; Kuijpers, M.J.; Kunzelmann, K.; Oury, C.; et al. Comparative Analysis of Microfluidics Thrombus Formation in Multiple Genetically Modified Mice: Link to Thrombosis and Hemostasis. Front. Cardiovasc. Med. 2019, 6, 99. [Google Scholar] [CrossRef]
- Baaten, C.C.; Meacham, S.; de Witt, S.M.; Feijge, M.A.; Adams, D.J.; Akkerman, J.W.; Cosemans, J.M.; Grassi, L.; Jupe, S.; Kostadima, M.; et al. A Synthesis Approach of Mouse Studies to Identify Genes and Proteins in Arterial Thrombosis and Bleeding. Blood 2018, 132, e35–e46. [Google Scholar] [CrossRef]
- Konstantinides, S.; Ware, J.; Marchese, P.; Almus-Jacobs, F.; Loskutoff, D.J.; Ruggeri, Z.M. Distinct Antithrombotic Consequences of Platelet Glycoprotein Iba and VI Deficiency in a Mouse Model of Arterial Thrombosis. J. Thromb. Haemost. 2006, 4, 2014–2021. [Google Scholar] [CrossRef]
- Watson, S.P.; Herbert, J.M.; Pollitt, A.Y. GPVI and CLEC-2 in Hemostasis and Vascular Integrity. J. Thromb. Haemost. 2010, 8, 1457–1467. [Google Scholar] [CrossRef]
- Hughes, C.E.; Finney, B.A.; Koentgen, F.; Lowe, K.L.; Watson, S.P. The N-Terminal SH2 Domain of Syk Is Required for (hem)ITAM, but Not Integrin, Signaling in Mouse Platelets. Blood 2015, 125, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Matus, V.; Valenzuela, G.; Saez, C.G.; Hidalgo, P.; Lagos, M.; Aranda, E.; Panes, O.; Pereira, J.; Pillois, X.; Nurden, A.T.; et al. An Adenine Insertion in Exon 6 of Human GP6 Generates a Truncated Protein Associated with a Bleeding Disorder in Four Chilean Families. J. Thromb. Haemost. 2013, 11, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Jandrot-Perrus, M.; Hermans, C.; Mezzano, D. Platelet Glycoprotein VI Genetic Quantitative and Qualitative Defects. Platelets 2019, 30, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A. Proteome Analysis of Signaling Cascades in Human Platelets. Blood Cells Mol. Dis. 2006, 36, 152–156. [Google Scholar] [CrossRef]
- Schulz, C.; Leuschen, N.V.; Frohlich, T.; Lorenz, M.; Pfeiler, S.; Gleissner, C.A.; Kremmer, E.; Kessler, M.; Khandoga, A.G.; Engelmann, B.; et al. Identification of Novel Downstream Targets of Platelet Glycoprotein VI Activation by Differential Proteome Analysis: Implications for Thrombus Formation. Blood 2010, 115, 4102–4110. [Google Scholar] [CrossRef] [PubMed]
- Babur, O.; Melrose, A.R.; Cunliffe, J.M.; Klimek, J.; Pang, J.; Sepp, A.I.; Zilberman-Rudenko, J.; Tassi Yunga, S.; Zheng, T.; Parra-Izquierdo, I.; et al. Phosphoproteomic Quantitation and Causal Analysis Reveal Pathways in GPVI/ITAM-Mediated Platelet Activation Programs. Blood 2020, 136, 2346–2358. [Google Scholar] [CrossRef]
- Velez, P.; Ocaranza-Sanchez, R.; Lopez-Otero, D.; Grigorian-Shamagian, L.; Rosa, I.; Guitian, E.; Garcia-Acuna, J.M.; Gonzalez-Juanatey, J.R.; Garcia, A. Alteration of Platelet GPVI Signaling in ST-Elevation Myocardial Infarction Patients Demonstrated by a Combination of Pro-Teomic, Biochemical, and Functional Approaches. Sci. Rep. 2016, 6, 39603. [Google Scholar] [CrossRef]
- Barrachina, M.N.; Hermida-Nogueira, L.; Moran, L.A.; Casas, V.; Hicks, S.M.; Sueiro, A.M.; Di, Y.; Andrews, R.K.; Watson, S.P.; Gardiner, E.E.; et al. Phosphoproteomic Analysis of Platelets in Severe Obesity Uncovers Platelet Reactivity and Signaling Pathways Alterations. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 478–490. [Google Scholar] [PubMed]
- Aslan, J.E.; Rigg, R.A.; Nowak, M.S.; Loren, C.P.; Baker-Groberg, S.M.; Pang, J.; David, L.L.; McCarty, O.J. Lysine Acetyltransfer Supports Platelet Function. J. Thromb. Haemost. 2015, 13, 1908–1917. [Google Scholar] [CrossRef]
- Shah, P.; Yang, W.; Sun, S.; Pasay, J.; Faraday, N.; Zhang, H. Platelet Glycoproteins Associated with Aspirin-Treatment upon Platelet Activation. Proteomics 2017, 17, 1600199. [Google Scholar] [CrossRef] [PubMed]
- Toonstra, C.; Hu, Y.; Zhang, H. Deciphering the Roles of N-Glycans on Collagen-Platelet Interactions. J. Proteome Res. 2019, 18, 2467–2477. [Google Scholar] [CrossRef]
- Unsworth, A.J.; Bombik, I.; Pinto-Fernandez, A.; McGouran, J.F.; Konietzny, R.; Zahedi, R.P.; Watson, S.P.; Kessler, B.M.; Pears, C.J. Human Platelet Protein Ubiquitylation and Changes Following GPVI Activation. Thromb. Haemost. 2019, 119, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Durrant, T.N.; Hutchinson, J.L.; Heesom, K.J.; Anderson, K.E.; Stephens, L.R.; Hawkins, P.T.; Marshall, A.J.; Moore, S.F.; Hers, I. In-Depth PtdIns(3,4, 5)P3 Signalosome Analysis Identifies DAPP1 as a Negative Regulator of GPVI-Driven Platelet Function. Blood Adv. 2017, 1, 918–932. [Google Scholar] [CrossRef]
- Izquierdo, I.; Barrachina, M.N.; Hermida-Nogueira, L.; Casas, V.; Eble, J.E.; Carrascal, M.; Abian, J.; García, A. Platelet Membrane Lipid Rafts Protein Composition Varies Following GPVI and CLEC-2 Receptors Activation. J. Proteom. 2019, 195, 88–97. [Google Scholar] [CrossRef]
- Bleijerveld, O.B.; van Holten, T.C.; Preisinger, C.; van der Smagt, J.J.; Farndale, R.W.; Kleefstra, T.; Willemsen, M.H.; Urbanus, R.T.; de Groot, P.G.; Heck, A.J.; et al. Targeted Phosphotyrosine Profiling of Glycoprotein VI Signaling Implicates Oligophrenin-1 in Platelet Filopodia Formation. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1538–1543. [Google Scholar] [CrossRef][Green Version]
- De Witt, S.; Verdoold, R.; Cosemans, J.M.E.M.; Heemskerk, J.W.M. Insights into Platelet-Based Control of Coagulation. Thromb. Res. 2014, 133, 139–148. [Google Scholar] [CrossRef]
- Heemskerk, J.W.; Cosemans, J.M.; van der Meijden, P.E. Platelets and Coagulation. In Platelets in Thrombotic and Non-Thrombotic Disorders; Gresele, P., Kleiman, N.S., Lopez, J.A., Page, C.P., Eds.; Springer: Cham, Switzerland, 2017; pp. 447–462. ISBN 978-3-319-47460-1. [Google Scholar]
- Agbani, E.O.; van den Bosch, M.T.; Brown, E.; Williams, C.M.; Mattheij, N.J.; Cosemans, J.M.; Collins, P.W.; Heemskerk, J.W.; Hers, I.; Poole, A.W. Coordinated Membrane Ballooning and Procoagulant Spreading in Human Platelets. Circulation 2015, 132, 1414–1424. [Google Scholar] [CrossRef]
- Rayes, J.; Watson, S.P.; Nieswandt, B. Functional Significance of the Platelet Immune Receptors GPVI and CLEC-2. J. Clin. Investig. 2019, 129, 12–23. [Google Scholar] [CrossRef]
- Suzuki-Inoue, K.; Fuller, G.L.; Garcia, A.; Eble, J.A.; Pohlmann, S.; Inoue, O.; Gartner, T.K.; Hughan, S.C.; Pearce, A.C.; Laing, G.D.; et al. A Novel Syk-Dependent Mechanism of Platelet Activation by the C-Type Lectin Receptor CLEC-2. Blood 2006, 107, 542–549. [Google Scholar] [CrossRef]
- Manne, B.K.; Badolia, R.; Dangelmaier, C.; Eble, J.A.; Ellmeier, W.; Kahn, M.; Kunapuli, S.P. Distinct Pathways Regulate Syk Protein Activation Downstream of Immune Tyrosine Activation Motif (ITAM) and hemITAM Receptors in Platelets. J. Biol. Chem. 2015, 290, 11557–11568. [Google Scholar] [CrossRef]
- Parguina, A.F.; Alonso, J.; Rosa, I.; Velez, P.; Gonzalez-Lopez, M.J.; Guitian, E.; Ebble, J.A.; Loza, M.I.; Garcia, A. A Detailed Proteomic Analysis of Rhodocytin-Activated Platelets Reveals Novel Clues on the CLEC-2 Signalosome: Implications for CLEC-2 Signaling Regulation. Blood 2012, 120, e117–e126. [Google Scholar] [CrossRef]
- Coughlin, S.R. Thrombin Signalling and Protease-Activated Receptors. Nature 2000, 407, 258–264. [Google Scholar] [CrossRef]
- Wu, J.; Heemskerk, J.W.; Baaten, C.C. Platelet Membrane Receptor Proteolysis: Implications for Platelet Function. Front. Cardiovasc. Med. 2020, 7, 608391. [Google Scholar] [CrossRef]
- Estevez, B.; Kim, K.; Delaney, M.K.; Stojanovic-Terpo, A.; Shen, B.; Ruan, C.G.; Cho, J.Y.; Ruggeri, Z.M.; Du, X.P. Signaling-Mediated Cooperativity Between Glycoprotein Ib-IX and Protease-Activated Receptors in Thrombin-Induced Platelet Activation. Blood 2016, 127, 626–636. [Google Scholar] [CrossRef]
- Mammadova-Bach, E.; Nagy, M.; Heemskerk, J.W.; Nieswandt, N.; Braun, A. Store-Operated Calcium Entry in Blood Cells in Thrombo-Inflammation. Cell Calcium 2019, 77, 39–48. [Google Scholar] [CrossRef]
- Fernandez, D.I.; Kuijpers, M.J.; Heemskerk, J.W. Platelet Calcium Signalling by G-Protein Coupled and ITAM-Linked Receptors Regulating Anoctamin-6 and Procoagulant Activity. Platelets 2020, 32, 863–871. [Google Scholar] [CrossRef]
- Kim, S.; Foster, C.; Lecchi, A.; Quinton, T.M.; Prosser, D.M.; Jin, J.G.; Cattaneo, M.; Kunapuli, S.P. Protease-Activated Receptors 1 and 4 Do Not Stimulate Gi Signaling Pathways in the Absence of Secreted ADP and Cause Human Platelet Aggregation Independently of Gi Signaling. Blood 2002, 99, 3629–3636. [Google Scholar] [CrossRef]
- Moers, A.; Nieswandt, B.; Massberg, S.; Wettschureck, N.; Gruner, S.; Konrad, I.; Schulte, V.; Aktas, B.; Gratacap, M.P.; Simon, M.I.; et al. G13 Is an Essential Mediator of Platelet Activation in Hemostasis and Thrombosis. Nat. Med. 2003, 9, 1418–1422. [Google Scholar] [CrossRef]
- Pagel, O.; Walter, E.; Jurk, K.; Zahedi, R.P. Taking the Stock of Granule Cargo: Platelet Releasate Proteomics. Platelets 2017, 28, 119–128. [Google Scholar] [CrossRef]
- Velez, P.; Izquierdo, I.; Rosa, I.; Garcia, A. A 2D-DIGE-Based Proteomic Analysis Reveals Differences in the Platelet Releasate Composition When Comparing Thrombin and Collagen Stimulations. Sci. Rep. 2015, 5, 8198. [Google Scholar] [CrossRef]
- Szklanna, P.B.; Parsons, M.E.; Wynne, K.; O’Connor, H.; Egan, K.; Allen, S.; Ní Áinle, F.; Maguire, P.B. The Platelet Releasate Is Altered in Human Pregnancy. Proteom. Clin. Appl. 2019, 13, e1800162. [Google Scholar] [CrossRef]
- Parsons, M.E.; Szklanna, P.B.; Guerrero, J.A.; Wynne, K.; Dervin, F.; O’Connell, K.; Allen, S.; Egan, K.; Bennett, C.; McGuigan, C.; et al. Platelet Releasate Proteome Profiling Reveals a Core Set of Proteins with Low Variance between Healthy Adults. Proteomics 2018, 18, e1800219. [Google Scholar] [CrossRef]
- Milioli, M.; Ibanez-Vea, M.; Sidoli, S.; Palmisano, G.; Careri, M.; Larsen, M.R. Quantitative Proteomics Analysis of Platelet-Derived Microparticles Reveals Distinct Protein Signatures When Stimulated by Different Physiological Agonists. J. Proteom. 2015, 121, 55–66. [Google Scholar] [CrossRef]
- Grande, R.; Dovizio, M.; Marcone, S.; Szklanna, P.B.; Bruno, A.; Ebhardt, H.A.; Cassidy, H.; Ainle, F.N.; Caprodossi, A.; Lanuti, P.; et al. Platelet-Derived Microparticles from Obese Individuals: Characterization of Number, Size, Proteomics, and Crosstalk with Cancer and Endothelial Cells. Front. Pharmacol. 2019, 10, 7. [Google Scholar] [CrossRef]
- Garcia, A.; Prabhakar, S.; Hughan, S.; Anderson, T.W.; Brock, C.J.; Pearce, A.C.; Dwek, R.A.; Watson, S.P.; Hebestreit, H.F.; Zitzmann, N. Differential Proteome Analysis of TRAP-Activated Platelets: Involvement of DOK-2 and Phosphorylation of RGS Proteins. Blood 2004, 103, 2088–2095. [Google Scholar] [CrossRef]
- Tsai, H.J.; Chien, K.Y.; Liao, H.R.; Shih, M.S.; Lin, Y.C.; Chang, Y.W.; Cheng, J.C.; Tseng, C.P. Functional Links between Disabled-2 Ser723 Phosphorylation and Thrombin Signaling in Human Platelets. J. Thromb. Haemost. 2017, 15, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Zufferey, A.; Ibberson, M.; Reny, J.L.; Nolli, S.; Schvartz, D.; Docquier, M.; Xenarios, I.; Sanchez, J.C.; Fontana, P. New Molecular Insights into Modulation of Platelet Reactivity in Aspirin-Treated Patients Using a Network-Based Approach. Hum. Genet. 2016, 135, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Fontana, P.; Zufferey, A.; Daali, Y.; Reny, J.L. Antiplatelet Therapy: Targeting the TxA2 Pathway. J. Cardiovasc. Translat. Res. 2014, 7, 29–38. [Google Scholar] [CrossRef]
- Offermanns, S. Activation of Platelet Function through G Protein-Coupled Receptors. Circ. Res. 2006, 99, 1293–1304. [Google Scholar] [CrossRef]
- Stokhuijzen, E.; Koornneef, J.M.; Nota, B.; van den Eshof, B.L.; van Alphen, F.P.; van den Biggelaar, M.; van der Zwaan, C.; Kuijk, C.; Mertens, K.; Fijnvandraat, K.; et al. Differences between Platelets Derived from Neonatal Cord Blood and Adult Peripheral Blood Assessed by Mass Spectrometry. J. Proteome Res. 2017, 16, 3567–3575. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, M.; Debski, J.; Szahidewicz-Krupska, E.; Turek-Jakubowska, A.; Gawrys, J.; Gawrys, K.; Skomro, R.; Derkacz, A.; Doroszko, A. Platelet Carbonic Anhydrase II, a Forgotten Enzyme, May Be Responsible for Aspirin Resistance. Oxid. Med. Cell Longev. 2017, 2017, 3132063. [Google Scholar] [CrossRef] [PubMed]
- Finamore, F.; Reny, J.L.; Malacarne, S.; Fontana, P.; Sanchez, J.C. Shotgun Proteomics Data on the Impact of Hyperglycaemia on Platelet Protein Acetylation by Aspirin. Data Brief 2018, 21, 2475–2481. [Google Scholar] [CrossRef]
- Finamore, F.; Reny, J.L.; Malacarne, S.; Fontana, P.; Sanchez, J.C. A High Glucose Level Is Associated with Decreased Aspirin-Mediated Acetylation of Platelet Cyclooxygenase (COX)-1 at Serine 529: A Pilot Study. J. Proteom. 2019, 192, 258–266. [Google Scholar] [CrossRef]
- Gachet, C.; Hechler, B.; Leon, C.; Vial, C.; Leray, C.; Ohlmann, P.; Cazenave, J.P. Activation of ADP Receptors and Platelet Function. Thromb. Haemost. 1997, 78, 271–275. [Google Scholar] [CrossRef]
- Monroe, D.M.; Hoffman, M. What Does It Take to Make the Perfect Clot? Arterioscler. Thromb. Vasc. Biol. 2006, 26, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Rukoyatkina, N.; Walter, U.; Friebe, A.; Gambaryan, S. Differentiation of cGMP-Dependent and-Independent Nitric Oxide Effects on Platelet Apoptosis and Reactive Oxygen Species Production Using Platelets Lacking Soluble Guanylyl Cyclase. Thromb. Haemost. 2011, 106, 922–933. [Google Scholar] [CrossRef]
- Beck, F.; Geiger, J.; Gambaryan, S.; Veit, J.; Vaudel, M.; Nollau, P.; Kohlbacher, O.; Martens, L.; Walter, U.; Sickmann, A.; et al. Time-Resolved Characterization of cAMP/PKA-Dependent Signaling Reveals That Platelet Inhibition Is a Concerted Process Involving Multiple Signaling Pathways. Blood 2014, 123, e1–e10. [Google Scholar] [CrossRef]
- Beck, F.; Geiger, J.; Gambaryan, S.; Solari, F.A.; Dell’Aica, M.; Loroch, S.; Mattheij, N.J.; Mindukshev, I.; Potz, O.; Jurk, K.; et al. Temporal Quantitative Phosphoproteomics of ADP Stimulation Reveals Novel Central Nodes in Platelet Activation and Inhibition. Blood 2017, 129, e1–e12. [Google Scholar] [CrossRef]
- Radomski, M.W.; Palmer, R.M.; Moncada, S. Comparative Pharmacology of Endothelium-Derived Relaxing Factor, Nitric Oxide and Prostacyclin in Platelets. Br. J. Pharmacol. 1987, 92, 181–187. [Google Scholar] [CrossRef]
- Makhoul, S.; Walter, E.; Pagel, O.; Walter, U.; Sickmann, A.; Gambaryan, S.; Smolenski, A.; Zahedi, R.P.; Jurk, K. Effects of the NO/Soluble Guanylate Cyclase/cGMP System on the Functions of Human Platelets. Nitric Oxide 2018, 76, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Dangel, O.; Mergia, E.; Karlisch, K.; Groneberg, D.; Koesling, D.; Friebe, A. Nitric Oxide-Sensitive Guanylyl Cyclase Is the Only Nitric Oxide Receptor Mediating Platelet Inhibition. J. Thromb. Haemost. 2010, 8, 1343–1352. [Google Scholar] [CrossRef]
- Kumm, E.J.; Pagel, O.; Gambaryan, S.; Walter, U.; Zahedi, R.P.; Smolenski, A.; Jurk, K. The Cell Cycle Checkpoint System MAST(L)-ENSA/ARPP19-PP2A Is Targeted by cAMP/PKA and cGMP/PKG in Anucleate Human Platelets. Cells 2020, 9, 472. [Google Scholar] [CrossRef]
- Makhoul, S.; Dorschel, S.; Gambaryan, S.; Walter, U.; Jurk, K. Feedback Regulation of Syk by Protein Kinase C in Human Platelets. Int. J. Mol. Sci. 2019, 21, 176. [Google Scholar] [CrossRef] [PubMed]
- Loroch, S.; Trabold, K.; Gambaryan, S.; Reiss, C.; Schwierczek, K.; Fleming, I.; Sickmann, A.; Behnisch, W.; Zieger, B.; Zahedi, R.P.; et al. Alterations of the Platelet Proteome in Type I Glanzmann Thrombasthenia Caused by Different Homozygous deIG Frameshift Mutations in ITGA2B. Thromb. Haemost. 2017, 117, 556–569. [Google Scholar] [CrossRef]
- Van Kruchten, R.; Mattheij, N.J.; Saunders, C.; Feijge, M.A.; Swieringa, F.; Wolfs, J.L.; Collins, P.W.; Heemskerk, J.W.; Bevers, E.M. Both TMEM16F-Dependent and TMEM16F-Independent Pathways Contribute to Phosphatidylserine Exposure in Platelet Apoptosis and Platelet Activation. Blood 2013, 121, 1850–1857. [Google Scholar] [CrossRef]
- Van Bergen, M.G.; Marneth, A.E.; Hoogendijk, A.J.; van Alphen, F.P.; van den Akker, E.; Laros-Van Gorkom, B.A.; Hoeks, M.; Simons, A.; de Munnik, S.A.; Janssen, J.J.; et al. Specific Proteome Changes in Platelets from Individuals with GATA1-, GFI1B-, and RUNX1-Linked Bleeding Disorders. Blood 2021, 138, 86–90. [Google Scholar] [CrossRef]
- Bijak, M.; Olejnik, A.; Rokita, B.; Morel, A.; Dziedzic, A.; Miller, E.; Saluk-Bijak, J. Increased Level of Fibrinogen Chains in the Proteome of Blood Platelets in Secondary Progressive Multiple Sclerosis Patients. J. Cell Mol. Med. 2019, 23, 3476–3482. [Google Scholar] [CrossRef] [PubMed]
- Trugilho, M.R.; Hottz, E.D.; Brunoro, G.V.; Teixeira-Ferreira, A.; Carvalho, P.C.; Salazar, G.A.; Zimmerman, G.A.; Bozza, F.A.; Bozza, P.T.; Perales, J. Platelet Proteome Reveals Novel Pathways of Platelet Activation and Platelet-Mediated Immunoregulation in Dengue. PLoS Pathog. 2017, 13, e1006385. [Google Scholar] [CrossRef]
- Yu, H.; Liu, Y.; He, B.; He, T.; Chen, C.; He, J.; Yang, X.; Wang, J.Z. Platelet Biomarkers for a Descending Cognitive Function: A Proteomic Approach. Aging Cell 2021, 20, e13358. [Google Scholar] [CrossRef]
- Parguina, A.F.; Grigorian-Shamajian, L.; Agra, R.M.; Teijeira-Fernandez, E.; Rosa, I.; Alonso, J.; Vinuela-Roldan, J.E.; Seoane, A.; Gonzalez-Juanatey, J.R.; Garcia, A. Proteins Involved in Platelet Signaling Are Differentially Regulated in Acute Coronary Syndrome: A Proteomic Study. PLoS ONE 2010, 5, e13404. [Google Scholar]
- Lopez-Farré, A.J.; Zamorano-Leon, J.J.; Azcona, L.; Modrego, J.; Mateos-Caceres, P.J.; Gonzalez-Armengol, J.; Villarroel, P.; Moreno-Herrero, R.; Rodríguez-Sierra, P.; Segura, A.; et al. Proteomic Changes Related to Bewildered Circulating Platelets in the Acute Coronary Syndrome. Proteomics 2011, 11, 3335–3348. [Google Scholar] [CrossRef]
- Velez, P.; Ocaranza-Sanchez, R.; Lopez-Otero, D.; Grigorian-Shamagian, L.; Rosa, I.; Belen Bravo, S.; Gonzalez-Juanatey, J.R.; Garcia, A. 2D-DIGE-Based Proteomic Analysis of Intracoronary vs. Peripheral Arterial Blood Platelets from Acute Myocardial Infarction Patients: Upregulation of Platelet Activation Biomarkers at the Culprit Site. Proteom. Clin. Appl. 2016, 10, 851–858. [Google Scholar] [CrossRef]
- Maguire, P.B.; Parsons, M.E.; Szklanna, P.B.; Zdanyte, M.; Münzer, P.; Chatterjee, M.; Wynne, K.; Rath, D.; Comer, S.P.; Hayden, M.; et al. Comparative Platelet Releasate Proteomic Profiling of Acute Coronary Syndrome vs. Stable Coronary Artery Disease. Front. Cardiovasc. Med. 2020, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Hell, L.; Lurger, K.; Mauracher, L.M.; Grilz, E.; Reumiller, C.M.; Schmidt, G.J.; Ercan, H.; Koder, S.; Assinger, A.; Basilio, J.; et al. Altered Platelet Proteome in Lupus Anticoagulant-Positive Patients-Protein Disulfide Isomerase and NETosis as New Players in LA-Related Thrombosis. Exp. Mol. Med. 2020, 52, 66–78. [Google Scholar] [CrossRef]
- Van Geffen, J.P.; Swieringa, F.; van Kuijk, K.; Tullemans, B.M.; Solari, F.A.; Peng, B.; Clemetson, K.J.; Farndale, R.W.; Dubois, L.J.; Sickmann, A.; et al. Mild Hyperlipidemia in Mice Aggravates Platelet Responsiveness in Thrombus Formation and Exploration of Platelet Proteome and Lipidome. Sci. Rep. 2020, 10, 21407. [Google Scholar] [CrossRef]
- Malchow, S.; Loosse, C.; Sickmann, A.; Lorenz, C. Quantification of Cardiovascular Disease Biomarkers in Human Platelets by Targeted Mass Spectrometry. Proteomes 2017, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension Trapping (STrap) Sample Preparation Method for Bottom-Up Proteomics Analysis. Proteomics 2014, 14, 1006. [Google Scholar] [CrossRef]
- Ludwig, K.R.; Schroll, M.M.; Hummon, A.B. Comparison of Insolution, FASP, and S-Trap Based Digestion Methods for Bottom-Up Proteomic Studies. J. Proteome Res. 2018, 17, 2480–2490. [Google Scholar] [CrossRef]
- Zhu, Y.; Piehowski, P.D.; Zhao, R.; Chen, J.; Shen, Y.; Moore, R.J.; Shukla, A.K.; Petyuk, V.A.; Campbell-Thompson, M.; Mathews, C.E.; et al. Nanodroplet Processing Platform for Deep and Quantitative Proteome Profiling of 10–100 Mammalian Cells. Nat. Commun. 2018, 9, 882. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, B.; Koch, H.; Medard, G.; Mundt, M.; Kuster, B.; Lemeer, S. Comprehensive and Reproducible Phosphopeptide Enrichment Using Iron Immobilized Metal Ion Affinity Chromatography (Fe-IMAC) Columns. Mol. Cell. Proteom. 2015, 14, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hou, J.; Tanner, J.J.; Cheng, J. Bioinformatics Methods for Mass Spectrometry-Based Proteomics Data Analysis. Int. J. Mol. Sci. 2020, 21, 2873. [Google Scholar] [CrossRef] [PubMed]
- Solari, F.A.; Dell’Aica, M.; Sickmann, A.; Zahedi, R.P. Why Phosphoproteomics Is Still a Challenge. Mol. Biosyst. 2015, 11, 1487–1493. [Google Scholar] [CrossRef]
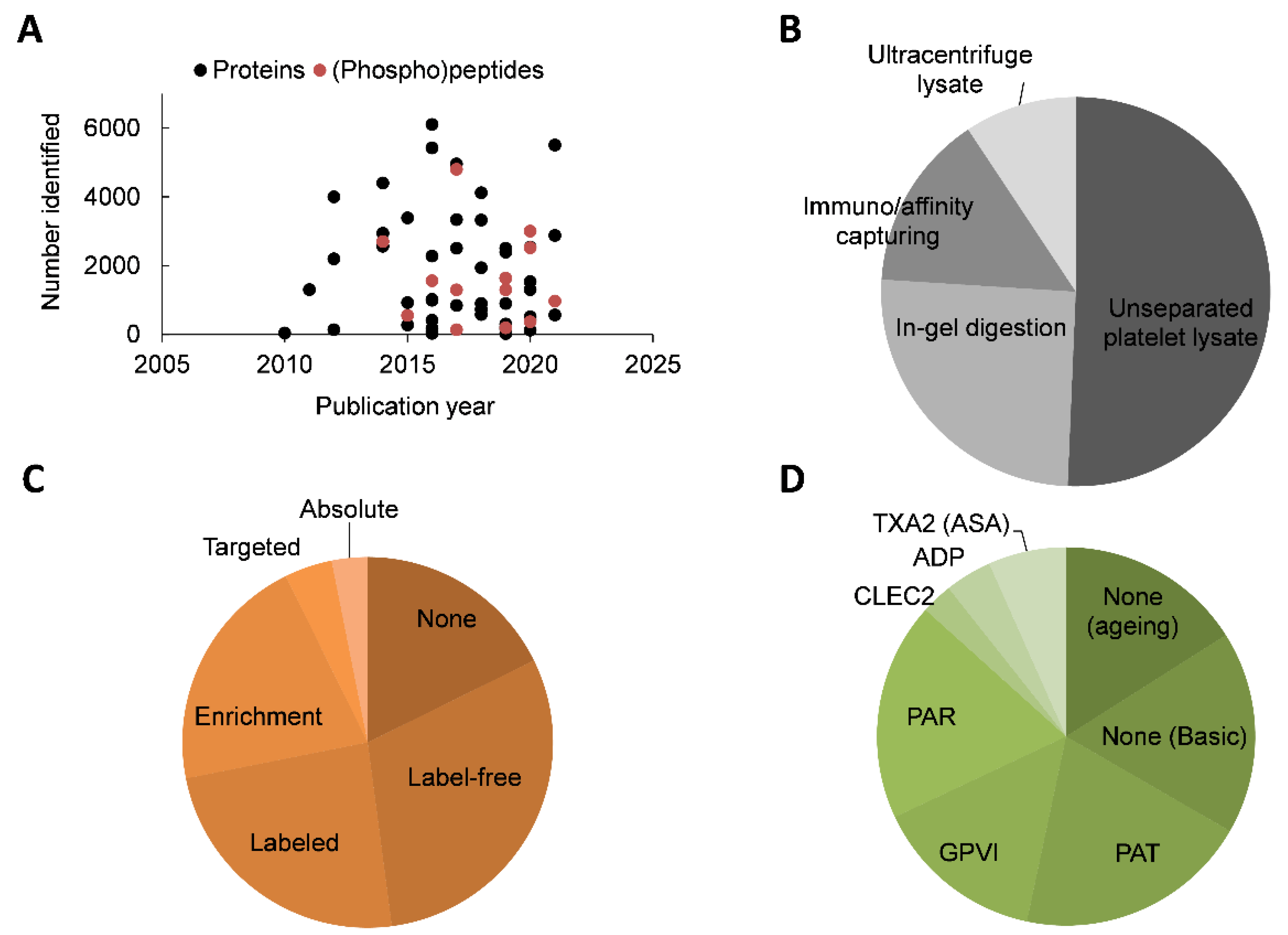

| Section | Year | Purity Checked | Sample Size | Type of (Sub)Proteomes | Reported Limitations | Pathway (GO) |
|---|---|---|---|---|---|---|
| 3-Basic | ≥2011 | 6/12 | 20–250 µg | platelets, granules, palmitoylation, methylation | unclear relation to platelet functions, low sample number | 8/12 |
| 4-Ageing | ≥2012 | 3/12 | 4–500 µg | stored platelets, N-terminome, extracellular vesicles | unclear relation to platelet functions, low sample number | 7/12 |
| 5-GPVI | ≥2010 | 8/11 | 150–2500+ µg | platelets (label-free), phosphorylation (TiO2), acetylation, PRM targeted, ubiquitylation, releasate | limited protein recovery, sample pooling | 7/11 |
| 6-CLEC-2 | ≥2012 | 2/2 | 150 µg | platelets, phosphorylation | limited protein recovery, second mediator interference | 2/2 |
| 7-PARs | ≥2015 | 5/13 | 24–150 µg | platelets (label-free), phosphorylation (TiO2), releasate, extracellular vesicles | sample pooling, low sample number, leukocyte contamination, clinical relevance unclear | 12/13 |
| 8-ASA | ≥2017 | 2/5 | 5–100 µg | platelets, acetylation, glycosylation | unclear clinical relevance | 3/5 |
| 9-ADP/INH | ≥2014 | 3/3 | 100–800 µg | platelets, phosphorylation (TiO2) | low sample number, unclear function of phosphorylation | 2/3 |
| 10-PAT | ≥2010 | 7/15 | 40–600+ µg | platelets (label-free), targeted, phosphorylation, N-terminome | low patient number, inter-patient variation, unclear relation to platelet functions | 8/15 |
| Aim of Study | No. of Regulated Proteins | Selection of Regulated Proteins | Ref. |
|---|---|---|---|
| 3-BASIC. Proteomes of large and small platelets (1 subject) | 80 up or down (9%) | ADP-ribosylation factor 1/3, GTP-binding protein SAR1a, guanylate cyclase soluble subunit α3, voltage-dependent anion channel protein 3, serotransferrin, immunoglobulins, haptoglobin, hemopexin, α1-antitrypsin, vitronectin | [27] |
| 5-GPVI. Platelet proteome in Scott syndrome (1 patient) | 134 up or down (6%) | anoctamin 6, annexin A5, calpain 1, protein S100-A8/9, channel aquaporin-1, pregnancy zone protein, myeloperoxidase, serine-pyruvate aminotransferase, platelet glycoprotein 4, cAMP-dependent protein kinase IIβ regulatory subunit | [28] |
| 10-PAT. Platelet iloprost phosphoproteome of PHP Ia patients (6 patients) | 51 up or down (11%) | phosphorylation of protein kinase A consensus sites: inositol triphosphate receptor associated 1/2, bridging integrator 2, vasodilator-stimulated phosphoprotein, coiled-coil domain-containing protein 9, claudin 5, consortin, Grb2-associated binding protein 2. | [29] |
| 10-PAT. Platelet proteome in X-linked thrombocytopenia (5 patients) | 83 up or down (4%) | prostaglandin G/H synthase 1, solute carrier family 35 member D3, carbonic anhydrase 2, peroxiredoxin 1, tubulin-tyrosine ligase-like protein 12, spectrin α-chain, nexilin, E2 ubiquitin-conjugating enzyme, proteasome subunit α/βtype 4, heat shock 70 kDa protein 1b | [30] |
| 10-PAT. Platelet proteome in Gray platelet syndrome (5 patients) | 123 up or down (% n.d.) | neurobeachin-like protein 2, SPARC, serglycin, latent-transforming growth factor β-binding protein 1, platelet basic protein, thrombospondin-1, platelet factor 4, multimerin-1, von Willebrand factor | [31] |
| 10-PAT. Platelet proteome of patients with early-stage cancer (12 patients) | 85 up or down (3%) | mitochondrial deoxyribonucleotidase and 39 S ribosomal protein, V-type proton ATPase subunit S1, myomegalin, serine/arginine repetitive matrix protein 2, CD34, peptidoglycan recognition protein 1, nuclear ribonucleoprotein A1, histone H2B type 1J, neurochondrin, calmodulin-dependent protein kinase type 1D | [19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhang, P.; Solari, F.A.; Sickmann, A.; Garcia, A.; Jurk, K.; Heemskerk, J.W.M. Molecular Proteomics and Signalling of Human Platelets in Health and Disease. Int. J. Mol. Sci. 2021, 22, 9860. https://doi.org/10.3390/ijms22189860
Huang J, Zhang P, Solari FA, Sickmann A, Garcia A, Jurk K, Heemskerk JWM. Molecular Proteomics and Signalling of Human Platelets in Health and Disease. International Journal of Molecular Sciences. 2021; 22(18):9860. https://doi.org/10.3390/ijms22189860
Chicago/Turabian StyleHuang, Jingnan, Pengyu Zhang, Fiorella A. Solari, Albert Sickmann, Angel Garcia, Kerstin Jurk, and Johan W. M. Heemskerk. 2021. "Molecular Proteomics and Signalling of Human Platelets in Health and Disease" International Journal of Molecular Sciences 22, no. 18: 9860. https://doi.org/10.3390/ijms22189860
APA StyleHuang, J., Zhang, P., Solari, F. A., Sickmann, A., Garcia, A., Jurk, K., & Heemskerk, J. W. M. (2021). Molecular Proteomics and Signalling of Human Platelets in Health and Disease. International Journal of Molecular Sciences, 22(18), 9860. https://doi.org/10.3390/ijms22189860











