Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing
Abstract
1. Introduction
2. Results and Discussion
2.1. Complete Organelle Genomes of N. ‘Joey Tomocik’
2.2. Co-Transcribed Genes in Organelle Genomes of N. ‘Joey Tomocik’
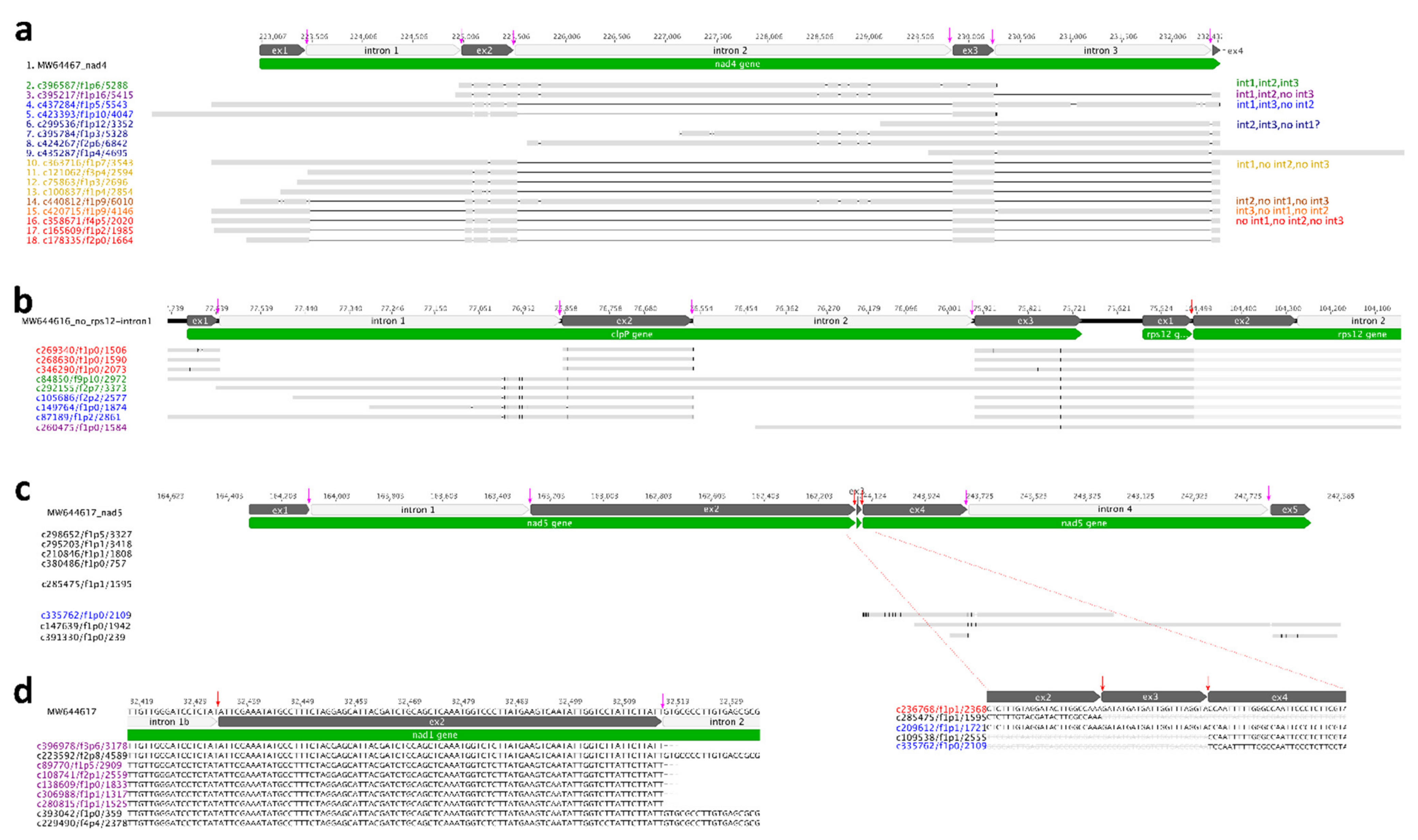
2.3. Diverse Intron Splicing Intermediates in Organelle Genome
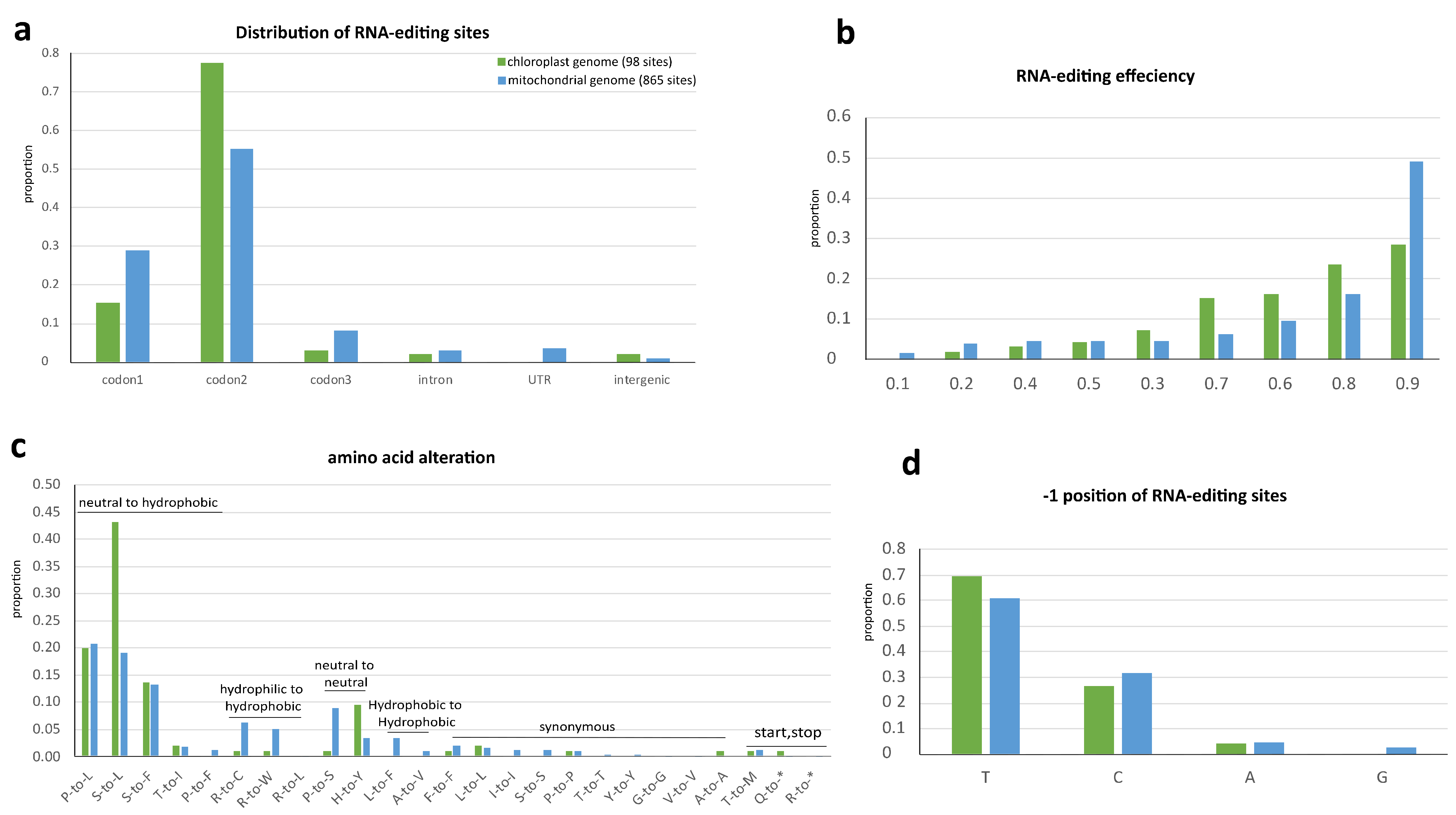
2.4. RNA-Editing in Organelle Genomes of Nymphaea
2.5. Interplay between RNA-Editing and Intron-Splicing
3. Materials and Methods
3.1. Genomic DNA Isolation and Sequencing
3.2. Organelle Genome Assembly and Annotation
3.3. RNA Isolation, Library Construction and Transcriptome Sequencing
3.4. Iso-seq and ssRNA-seq Data Processing and Mapping
3.5. Prediction of Polycistronic Transcript Unit (PTU)
3.6. Identification and Structure Prediction of trans-Spliced Group II Introns
3.7. The Identification of RNA Editing Sites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef] [PubMed]
- Glanz, S.; Kück, U. Trans-splicing of organelle introns—A detour to continuous RNAs. BioEssays 2009, 31, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, B.; Royan, S.; Schallenberg-Rüdinger, M.; Lenz, H.; Castleden, I.R.; McDowell, R.; Vacher, M.A.; Tonti-Filippini, J.; Bond, C.S.; Knoop, V.; et al. The expansion and diversification of pentatricopeptide repeat RNA-editing factors in plants. Mol. Plant 2020, 13, 215–230. [Google Scholar] [CrossRef]
- Zimmerly, S.; Semper, C. Evolution of group II introns. Mob. DNA 2015, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Bonen, L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 2008, 8, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Kazuhiko, U.; Haruo, O. Comparative and functional anatomy of group II catalytic introns—A review. Gene 1989, 82, 5–30. [Google Scholar] [CrossRef]
- Lambowitz, A.M.; Zimmerly, S. Group II Introns: Mobile ribozymes that invade DNA. Cold Spring Harb. Perspect. Biol. 2010, 3, a003616. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef]
- Kim, S.; Yoon, M.-K. Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr. Genet. 2010, 56, 177–188. [Google Scholar] [CrossRef]
- Massel, K.; Silke, J.R.; Bonen, L. Multiple splicing pathways of group II trans-splicing introns in wheat mitochondria. Mitochondrion 2016, 28, 23–32. [Google Scholar] [CrossRef]
- Lukeš, J.; Kaur, B.; Speijer, D. RNA editing in mitochondria and plastids: Weird and widespread. Trends Genet. 2021, 37, 99–102. [Google Scholar] [CrossRef]
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Lamattina, L.; Bonnard, G.; Weil, J.-H.; Grienenberger, J.-M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef] [PubMed]
- Hiesel, R.; Wissinger, B.; Schuster, W.; Brennicke, A. RNA editing in plant mitochondria. Science 1989, 246, 1632–1634. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. RNA editing in plant mitochondria: 20 years later. IUBMB Life 2009, 61, 1101–1104. [Google Scholar] [CrossRef] [PubMed]
- Hoch, B.; Maier, R.M.; Appel, K.; Igloi, G.L.; Kössel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 1991, 353, 178–180. [Google Scholar] [CrossRef]
- Rüdinger, M.; Polsakiewicz, M.; Knoop, V. Organellar RNA editing and plant-Specific extensions of pentatricopeptide repeat proteins in Jungermanniid but not in Marchantiid Liverworts. Mol. Biol. Evol. 2008, 25, 1405–1414. [Google Scholar] [CrossRef]
- Fan, W.; Guo, W.; Funk, L.; Mower, J.P.; Zhu, A. Complete loss of RNA editing from the plastid genome and most highly expressed mitochondrial genes of Welwitschia mirabilis. Sci. China Life Sci. 2019, 62, 498–506. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef]
- Freyer, R.; Kiefer-Meyer, M.C.; Kössel, H. Occurrence of plastid RNA editing in all major lineages of land plants. Proc. Natl. Acad. Sci. USA 1997, 94, 6285–6290. [Google Scholar] [CrossRef]
- Zhang, A.; Fang, J.; Jiang, X.; Wang, T.; Zhang, X. A comprehensive study on chloroplast RNA editing by performing a broad-spectrum RNA-seq analysis. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Shields, D.C.; Wolfe, K.H. Accelerated evolution of sites undergoing mRNA editing in plant mitochondria and chloroplasts. Mol. Biol. Evol. 1997, 14, 344–349. [Google Scholar] [CrossRef][Green Version]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; de Pamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar] [CrossRef]
- Ishibashi, K.; Small, I.; Shikanai, T. Evolutionary model of plastidial rNA editing in angiosperms presumed from genome-wide analysis of Amborella trichopoda. Plant Cell Physiol. 2019, 60, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. Modeling sites of RNA editing as a fifth nucleotide state reveals progressive loss of edited sites from angiosperm mitochondria. Mol. Biol. Evol. 2008, 25, 52–61. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef]
- Gerke, P.; Szövényi, P.; Neubauer, A.; Lenz, H.; Gutmann, B.; McDowell, R.; Small, I.; Schallenberg-Rüdinger, M.; Knoop, V. Towards a plant model for enigmatic U-to-C RNA editing: The organelle genomes, transcriptomes, editomes and candidate RNA editing factors in the hornwort Anthoceros agrestis. New Phytol. 2020, 225, 1974–1992. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef]
- Richardson, A.O.; Rice, D.W.; Young, G.J.; Alverson, A.J.; Palmer, J.D. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Best, C.; Mizrahi, R.; Ostersetzer-Biran, O. Why so complex? The intricacy of genome structure and gene expression, associated with angiosperm mitochondria, may relate to the regulation of embryo quiescence or dormancy-intrinsic blocks to early plant life. Plants 2020, 9, 598. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhao, C.; Chen, F.; Liu, Y.; Zhang, S.; Wu, H.; Zhang, L.; Liu, Y. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 2018, 19, 614. [Google Scholar] [CrossRef]
- Mower, J.P. Variation in protein gene and intron content among land plant mitogenomes. Mitochondrion 2020, 53, 203–213. [Google Scholar] [CrossRef]
- Zhelyazkova, P.; Sharma, C.M.; Förstner, K.U.; Liere, K.; Vogel, J.; Börner, T. The Primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 2012, 24, 123–136. [Google Scholar] [CrossRef]
- Pereira de Souza, A.; Jubier, M.F.; Delcher, E.; Lancelin, D.; Lejeune, B. A trans-splicing model for the expression of the tripartite nad5 gene in wheat and maize mitochondria. Plant Cell 1991, 3, 1363–1378. [Google Scholar] [CrossRef][Green Version]
- Cheng, S.; Lim, B.L. Organelle transcriptomes in plants. Transcriptomics 2013, 2, 1000e106. [Google Scholar]
- Chen, T.-C.; Liu, Y.-C.; Wang, X.; Wu, C.-H.; Huang, C.-H.; Chang, C.-C. Whole plastid transcriptomes reveal abundant RNA editing sites and differential editing status in Phalaenopsis aphrodite subsp. formosana. Bot. Stud. 2017, 58, 38. [Google Scholar] [CrossRef] [PubMed]
- Pommié, C.; Levadoux, S.; Sabatier, R.; Lefranc, G.; Lefranc, M.-P. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004, 17, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Giegé, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324. [Google Scholar] [CrossRef]
- Maier, R.M.; Neckermann, K.; Hoch, B.; Akhmedov, N.B.; Kössel, H. Identification of editing positions in the ndhB transcript from maize chloroplasts reveals sequence similarities between editing sites of chloroplasts and plant mitochondria. Nucleic Acids Res. 1992, 20, 6189–6194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schmitz-Linneweber, C.; Tillich, M.; Herrmann, R.G.; Maier, R.M. Heterologous, splicing-dependent RNA editing in chloroplasts: Allotetraploidy provides trans-factors. EMBO J. 2001, 20, 4874–4883. [Google Scholar] [CrossRef]
- Li-Pook-Than, J.; Carrillo, C.; Niknejad, N.; Calixte, S.; Crosthwait, J.; Bonen, L. Relationship between RNA splicing and exon editing near intron junctions in wheat mitochondria. Physiol. Plant. 2007, 129, 23–33. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, C.; Yagi, Y.; Nakamura, T.; Sugita, M. Two DYW subclass PPR proteins are involved in RNA editing of ccmFc and atp9 transcripts in the moss Physcomitrella patens: First complete set of PPR editing factors in plant mitochondria. Plant Cell Physiol. 2013, 54, 1907–1916. [Google Scholar] [CrossRef]
- Sutton, C.A.; Conklin, P.L.; Pruitt, K.D.; Hanson, M.R. Editing of pre-mRNAs can occur before cis- and trans-splicing in Petunia mitochondria. Mol. Cell. Biol. 1991, 11, 4274–4277. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zanlungo, S.; Quiñones, V.; Moenne, A.; Holuigue, L.; Jordana, X. Splicing and editing of rps10 transcripts in potato mitochondria. Curr. Genet. 1995, 27, 565–571. [Google Scholar] [CrossRef]
- Binder, S.; Marchfelder, A.; Brennicke, A.; Wissinger, B. RNA editing in trans-splicing intron sequences of nad2 mRNAs in Oenothera mitochondria. J. Biol. Chem. 1992, 267, 7615–7623. [Google Scholar] [CrossRef]
- Wissinger, B.; Schuster, W.; Brennicke, A. Trans splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell 1991, 65, 473–482. [Google Scholar] [CrossRef]
- Börner, G.V.; Mörl, M.; Wissinger, B.; Brennicke, A.; Schmelzer, C. RNA editing of a group II intron in Oenothera as a prerequisite for splicing. Mol. Gen. Genet. 1995, 246, 739–744. [Google Scholar] [CrossRef]
- Farré, J.-C.; Aknin, C.; Araya, A.; Castandet, B. RNA editing in mitochondrial trans-Introns is required for splicing. PLoS ONE 2012, 7, e52644. [Google Scholar] [CrossRef] [PubMed]
- Morawala-Patell, V.; Gualberto, J.M.; Lamattina, L.; Grienenberger, J.M.; Bonnard, G. Cis- and trans-splicing and RNA editing are required for the expression of nad2 in wheat mitochondria. Mol. Gen. Genet. 1998, 258, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Song, S.; Yang, Y.-Z.; Lu, F.; Zhang, M.-D.; Sun, F.; Jia, R.; Song, R.; Tan, B.-C. DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. Plant J. 2020, 103, 1767–1782. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Soorni, A.; Haak, D.; Zaitlin, D.; Bombarely, A. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genom. 2017, 18, 49. [Google Scholar] [CrossRef]
- Yang, J.-B.; Li, D.-Z.; Li, H.-T. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol. Ecol. Resour. 2014, 14, 1024–1031. [Google Scholar] [CrossRef]
- Kovar, L.; Nageswara-Rao, M.; Ortega-Rodriguez, S.; Dugas, D.V.; Straub, S.; Cronn, R.; Strickler, S.R.; Hughes, C.E.; Hanley, K.A.; Rodriguez, D.N.; et al. PacBio-Based mitochondrial genome assembly of Leucaena trichandra (Leguminosae) and an intrageneric assessment of mitochondrial RNA editing. Genome Biol. Evol. 2018, 10, 2501–2517. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Biosciences, P. SMRT® Tools Reference Guide; SMRT Corporation LTD: Singapore, 2018. [Google Scholar]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; de Pamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar]
- Qu, X.-J.; Moore, M.J.; Li, D.-Z.; Yi, T.-S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 2019, 15, 50. [Google Scholar] [CrossRef]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Salmela, L.; Rivals, E. LoRDEC: Accurate and efficient long read error correction. Bioinformatics 2014, 30, 3506–3514. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Lang, B.F.; Laforest, M.-J.; Burger, G. Mitochondrial introns: A critical view. Trends Genet. 2007, 23, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Wu, S.; Liu, W.; Aljohi, H.A.; Alromaih, S.A.; Alanazi, I.O.; Lin, Q.; Yu, J.; Hu, S. REDO: RNA editing detection in plant organelles based on variant calling results. J. Comput. Biol. 2018, 25, 509–516. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Lenz, H.; Hein, A.; Knoop, V. Plant organelle RNA editing and its specificity factors: Enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinform. 2018, 19, 255. [Google Scholar] [CrossRef] [PubMed]




| Chloroplast Genome | Mitochondrial Genome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Atri | NJoey | Ncol | Ssph | Ltul | Atri | NJoey | Ncol | Ssph | Ltul | |
| Accession number | NC_005086 | MW644616 | MT107631 | NC_037145 | NC_008326 | KF754799–KF754803 | MW644617 | NC_037468 | NC_042758 | NC_021152 |
| Genome size (bp) | 162,686 | 159,968 | 159,842 | 146,843 | 159,886 | 3,866,039 | 335,042 | 617,195 | 1,101,768 | 553,721 |
| No. of genes (unique) | 132 (114) | 132 (114) | 132 (114) | 125 (113) | 131 (113) | 63 | 73 (65) | 66 (60) | 70 (58) | 65 (62) |
| No. of protein genes (unique) | 87 (80) | 87 (80) | 87 (80) | 82 (79) | 86 (79) | 40 (40) | 43 (41) | 43 (41) | 43 (41) | 41 (41) |
| No. of tRNA genes (unique) | 37 (30) | 37 (30) | 37 (30) | 35 (30) | 37 (30) | 10 (9) | 24 (21) | 20 (16) | 22 (14) | 21 (18) |
| No. of rRNA genes (unique) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 8 (4) | 3 (3) | 6 (3) | 3 (3) | 5 (3) | 3 (3) |
| Genome GC content | 38.30% | 39.10% | 39.14% | 39.60% | 39.20% | 45.90% | 48.70% | 45.10% | 46.40% | 47.70% |
| cis-splicing intron (unique) | 24 (20) | 24 (20) | 24 (20) | 24 (20) | 24 (20) | 19 (19) 1 | 19 (19) | 19 (19) | 18 (18) 1 | 19 (19) |
| trans-splicing intron | 19 | 1 | 1 | 1 | 1 | 6 | 6 | 6 | 6 | 6 |
| PTU | Genes Covered by PTU |
|---|---|
| PTU1_chloroplast | trnQ-UUG, rps16, trnK-UUU(matK), psbA, trnH-GUG |
| PTU2_chloroplast | psbK, psbI, trnG-UCC |
| PTU3_chloroplast | trnE-UUC, trnY-GUA, trnD-GUC, psbM, rpoB, rpoC1, rpoC2, rps2, atpI, atpH, atpF, atpA |
| PTU4_chloroplast | trnC-GCA, petN |
| PTU5_chloroplast | trnT-GGU, psbD, psbC, psbZ, trnG-GCC |
| PTU6_chloroplast | rps4, ycf3, psaA, psaB, rps14, trnfM-CAU |
| PTU7_chloroplast | atpB, atpE, trnV-UAC, ndhC, ndhK, ndhJ |
| PTU8_chloroplast | rbcL, accD, psaI, ycf4, cemA, petA |
| PTU9_chloroplast | psbE, psbF, psbL, psbJ |
| PTU10_chloroplast | petL, petG, psaJ, rpl33, rps18 |
| PTU11_chloroplast | clpP, rps12_5′(exon 1), rpl20 |
| PTU12_chloroplast | psbB, psbT, psbH, petB, petD |
| PTU13_chloroplast | trnI-CAU, rpl23, rpl2, rps19, rpl22, rps3, rpl16, rpl14, rps8, infA, rpl36, rps11, rpoA |
| PTU14_chloroplast | ycf2(partial), ycf15 |
| PTU15_chloroplast | rps12_3′ (exon 2-exon 3), rps7, ndhB |
| PTU16_chloroplast | rrn16, trnI-GAU, trnA-UGC, rrn23, rrn4.5, rrn5, trnR-ACG |
| PTU17_chloroplast | ndhF, trnN-GUU |
| PTU18_chloroplast | ycf1(partial), rps15, ndhH, ndhA, ndhI, ndhG, ndhE, psaC, ndHD |
| PTU1_mitochondria | rps2, nad1(partial, exon 2-exon 3) |
| PTU2_mitochondria | rps10, cox1 |
| PTU3_mitochondria | nad3, rps12, nad5_5′(exon 1) |
| PTU4_mitochondria | rpl2, rps19, rps3 |
| PTU5_mitochondria | nad1(partial, exon 4), atp6 |
| Group II Introns | Plastome Introns | Iso-seq 1 | Trinity 1 | Mitogenome Introns | Iso-seq | Trinity |
|---|---|---|---|---|---|---|
| trans-splicing | rps12-i1 | ■ | ■ | nad1-i1 | □ | ■ |
| nad1-i3 | ■ | ■ | ||||
| nad1-i4 | □ | ■ | ||||
| nad2-i2 | □ | ■ | ||||
| nad5-i2 | ■ | ■ | ||||
| nad5-i3 | ■ | ■ | ||||
| cis-splicing | rps12-i2 | □ | ■ | nad1-i2 | ■ | ■ |
| ycf3-i1 | □ | ■ | nad2-i1 | □ | ■ | |
| ycf3-i2 | □ | ■ | nad2-i3 | □ | ■ | |
| clpP-i1 | ■ | ■ | nad2-i4 | □ | ■ | |
| clpP-i2 | ■ | ■ | nad5-i1 | ■ | ■ | |
| rps16-i | ■ | ■ | nad5-i4 | ■ | ■ | |
| atpF-i | □ | ■ | nad7-i1 | ■ | ■ | |
| ndhA-i | □ | ■ | nad7-i2 | □ | ■ | |
| petB-i | □ | ■ | nad7-i3 | ■ | ■ | |
| petD-i | □ | ■ | nad7-i4 | ■ | ■ | |
| rpl16-i | □ | ■ | nad4-i1 | ■ | ■ | |
| rpoC1-i | □ | ■ | nad4-i2 | ■ | ■ | |
| trnG-UCC-i | □ | □ | nad4-i3 | ■ | ■ | |
| trnK-UUU-i | □ | □ | cox2-i1 | ■ | ■ | |
| trnL-UAA-i | □ | □ | cox2-i2 | ■ | ■ | |
| trnV-UAC-i | □ | □ | ccmFC-i | □ | ■ | |
| 2ndhB-i | □ | ■ | rpl2-i | ■ | ■ | |
| 2trnA-UGC-i | □ | □ | rps3-i | ■ | ■ | |
| 2trnI-GAU-i | □ | □ | rps10-i | □ | ■ | |
| 2rpl2-i | ■ | ■ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.-S.; Zhu, A.; Yang, J.-B.; Fan, W.; Li, D.-Z. Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing. Int. J. Mol. Sci. 2021, 22, 9842. https://doi.org/10.3390/ijms22189842
He Z-S, Zhu A, Yang J-B, Fan W, Li D-Z. Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing. International Journal of Molecular Sciences. 2021; 22(18):9842. https://doi.org/10.3390/ijms22189842
Chicago/Turabian StyleHe, Zheng-Shan, Andan Zhu, Jun-Bo Yang, Weishu Fan, and De-Zhu Li. 2021. "Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing" International Journal of Molecular Sciences 22, no. 18: 9842. https://doi.org/10.3390/ijms22189842
APA StyleHe, Z.-S., Zhu, A., Yang, J.-B., Fan, W., & Li, D.-Z. (2021). Organelle Genomes and Transcriptomes of Nymphaea Reveal the Interplay between Intron Splicing and RNA Editing. International Journal of Molecular Sciences, 22(18), 9842. https://doi.org/10.3390/ijms22189842






