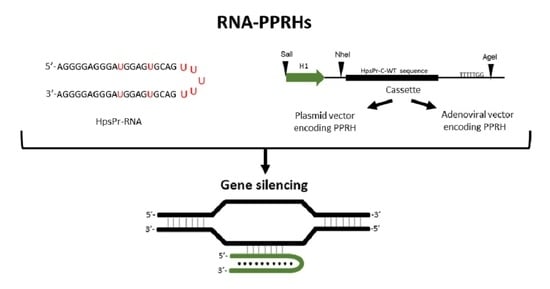
PolyPurine Reverse Hoogsteen Hairpins Work as RNA Species for Gene Silencing
Abstract
:1. Introduction
2. Results
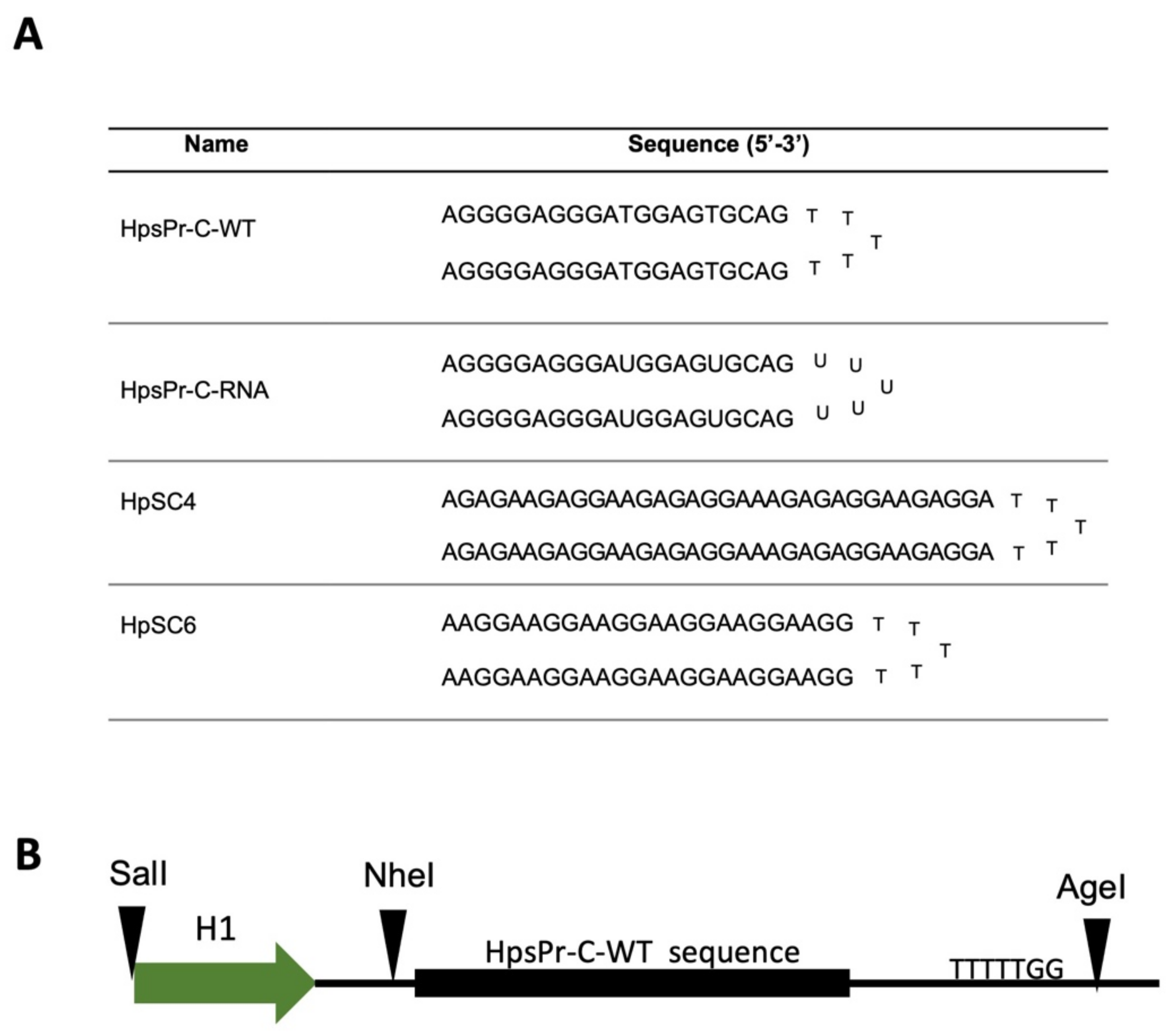
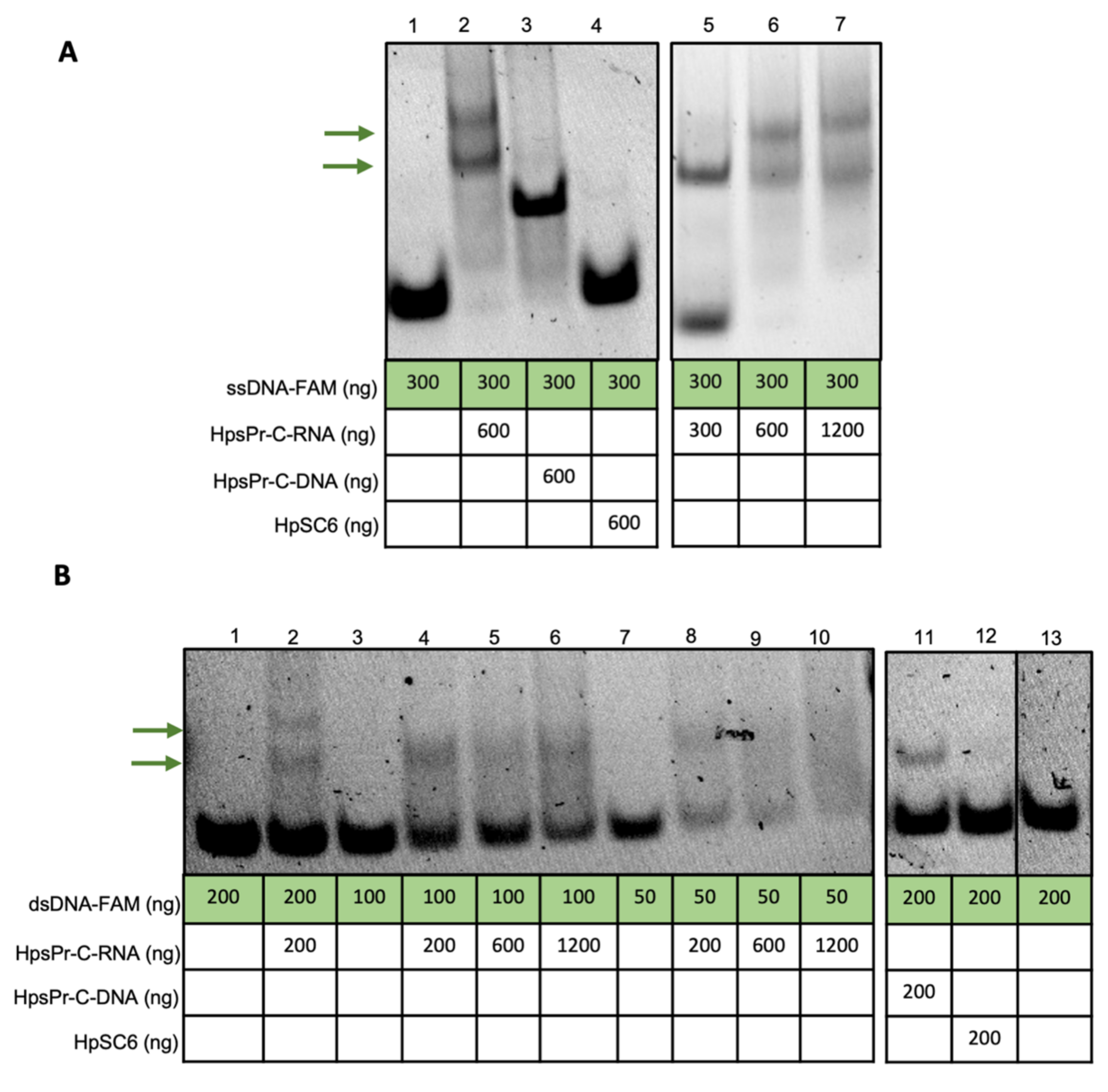
2.1. RNA-PPRH Binding Analyses
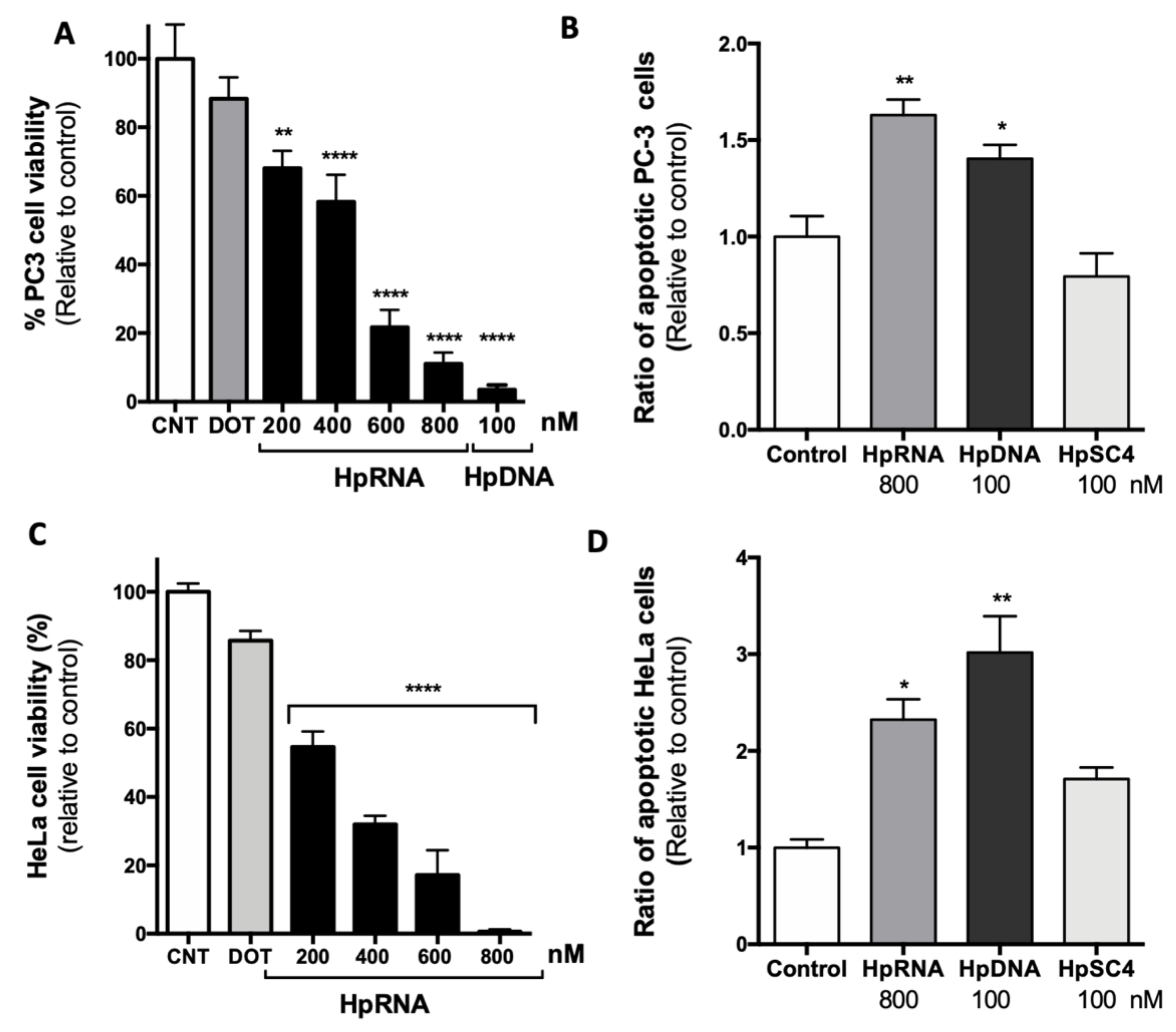
2.2. Effect of a Synthetic RNA-PPRH Targeting Survivin on Viability and Apoptosis
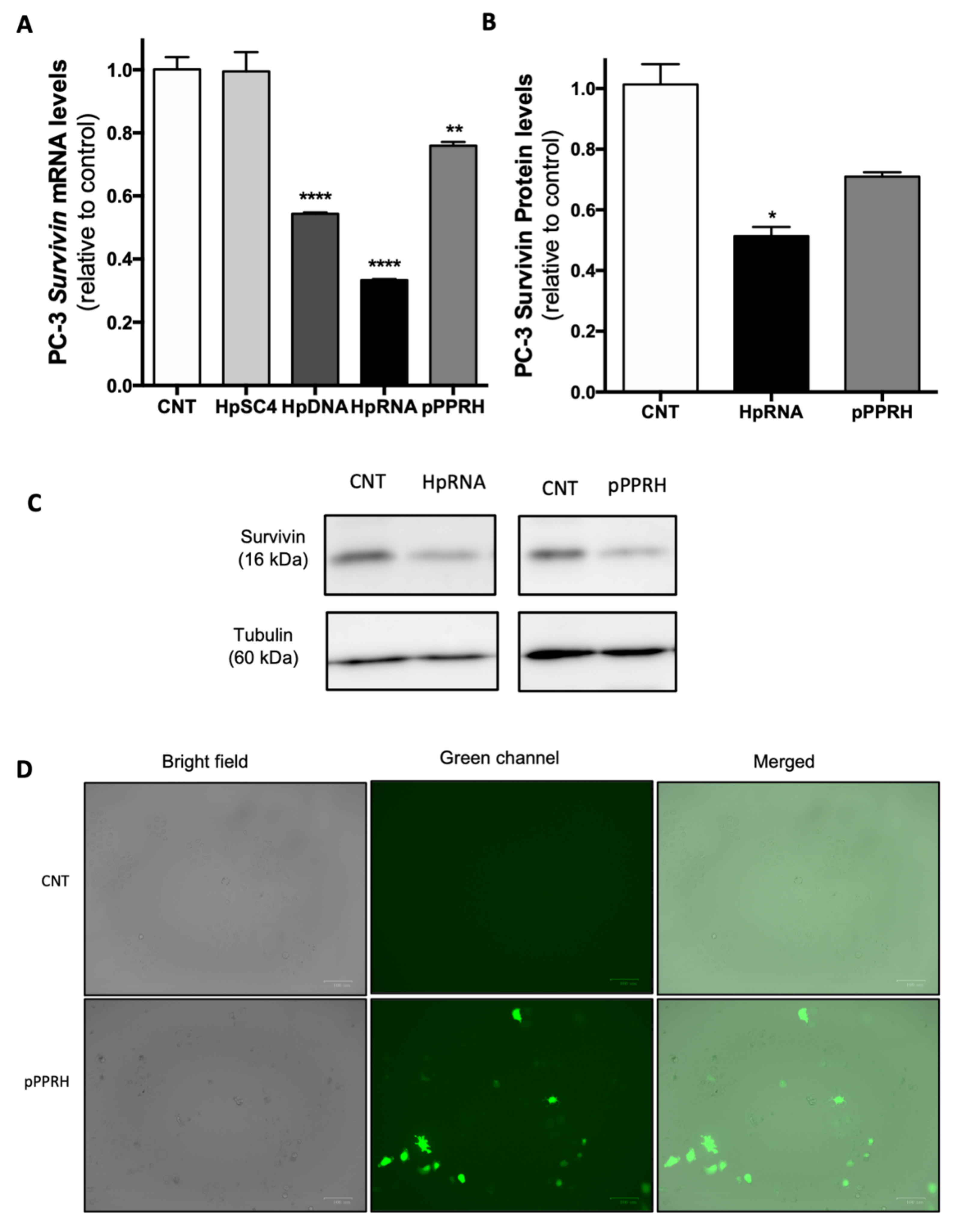
2.3. Effect on the Levels of Survivin mRNA and Protein upon RNA-PPRH Transfection
2.4. Cell Viability Assays with AdV Vectors
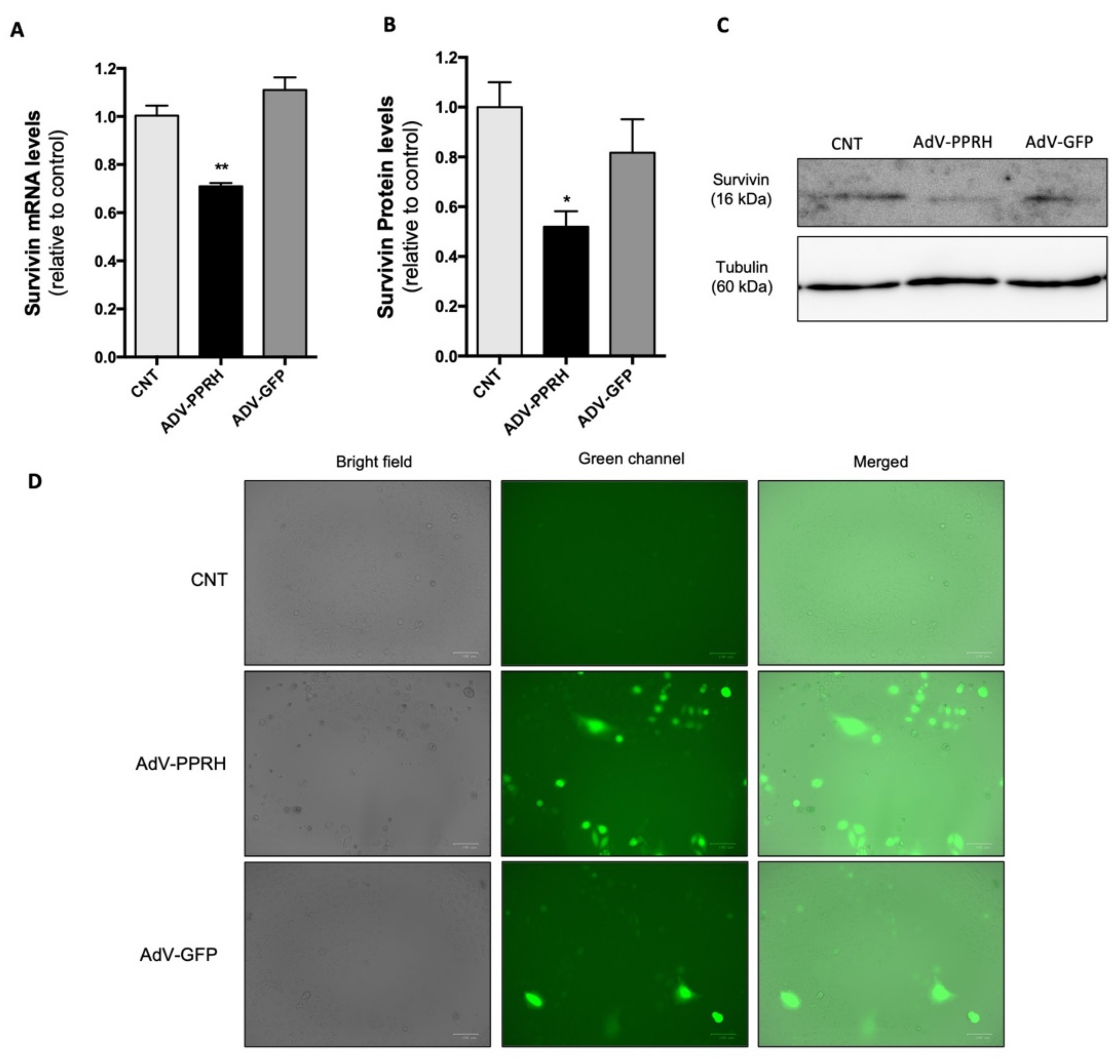
2.5. Effect of AdV-PPRH Infection on Survivin mRNA and Protein Levels
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. PPRHs Design
4.3. RNA-PPRH Binding Analyses
4.4. Plasmid Vector
4.5. Viral Vector Production
4.6. Transfection of PPRHs
4.7. Transfection of Vector
4.8. Transduction of Human Cells
4.9. Cell Viability Assays
4.10. Annexin V Apoptosis Detection Kit FITC
4.11. RNA Extraction
4.12. Reverse Transcription
4.13. Reverse Transcription
4.14. Western Blot Analyses for Survivin Detection
4.15. Statistical Analyses
Author Contributions
Funding
Conflicts of Interest
References
- Parsel, S.M.; Grandis, J.R.; Thomas, S.M. Nucleic acid targeting: Towards personalized therapy for head and neck cancer. Oncogene 2016, 35, 3217–3226. [Google Scholar] [CrossRef] [Green Version]
- Landmesser, U.; Poller, W.; Tsimikas, S.; Most, P.; Paneni, F.; Lüscher, T.F. From traditional pharmacological towards nucleic acid-based therapies for cardiovascular diseases. Eur. Heart J. 2020, 41, 3888–3899. [Google Scholar] [CrossRef]
- Nielsen, T.T.; Nielsen, J.E. Antisense Gene Silencing: Therapy for Neurodegenerative Disorders? Genes 2013, 4, 457–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazan-Halevy, I.; Landesman-Milo, D.; Rosenblum, D.; Mizrahy, S.; Ng, B.D.; Peer, D. Immunomodulation of hematological malignancies using oligonucleotides based-nanomedicines. J. Control. Release 2016, 244, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Ishiko, O.; Sumi, T.; Matsumoto, Y.; Ogita, S. Survivin, bcl-2 and matrix metalloproteinase-2 enhance progression of clear cell- and serous-type ovarian carcinomas. Int. J. Oncol. 2001, 19, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid (Reprinted from Nature, April 25, 1953). Nat. Cell Biol. 1969, 224, 470–471. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Iwamoto, S.; Gon, G.; Nohara, T.; Iwamoto, M.; Tanigawa, N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin. Cancer Res. 2000, 6, 127–134. [Google Scholar] [PubMed]
- Chou, T.C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Leventis, R.; Silvius, J.R. Interactions of mammalian cells with lipid dispersions containing novel metabolizable cationic amphiphiles. Biochim. Biophys. Acta (BBA)-Biomembr. 1990, 1023, 124–132. [Google Scholar] [CrossRef]
- Hakkak, A.M.; Keramatipour, M.; Talebi, S.; Brook, A.; Afshari, J.T.; Raazi, A.; Kianifar, H.R. Analysis of CFTR Gene Mutations in Children with Cystic Fibrosis, First Report from North-East of Iran. Iran J. Basic Med. Sci. 2013, 16, 917–921. [Google Scholar]
- Phear, G.; Armstrong, W.; Meuth, M. Molecular basis of spontaneous mutation at the aprt locus of hamster cells. J. Mol. Biol. 1989, 209, 577–582. [Google Scholar] [CrossRef]
- Scharner, J.; Qi, S.; Rigo, F.; Bennett, C.F.; Krainer, A.R. Delivery of GalNAc-Conjugated Splice-Switching ASOs to Non-hepatic Cells through Ectopic Expression of Asialoglycoprotein Receptor. Mol. Ther.-Nucleic Acids 2019, 16, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setoguchi, K.; Cui, L.; Hachisuka, N.; Obchoei, S.; Shinkai, K.; Hyodo, F.; Kato, K.; Wada, F.; Yamamoto, T.; Harada-Shiba, M.; et al. Antisense Oligonucleotides Targeting Y-Box Binding Protein-1 Inhibit Tumor Angiogenesis by Downregulating Bcl-xL-VEGFR2/-Tie Axes. Mol. Ther.-Nucleic Acids 2017, 9, 170–181. [Google Scholar] [CrossRef] [Green Version]
- Aguti, S.; Bolduc, V.; Ala, P.; Turmaine, M.; Bönnemann, C.G.; Muntoni, F.; Zhou, H. Exon-Skipping Oligonucleotides Restore Functional Collagen VI by Correcting a Common COL6A1 Mutation in Ullrich CMD. Mol. Ther.-Nucleic Acids 2020, 21, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Coma, S.; Noe, V.; Lavarino, C.; Adán, J.; Rivas, M.; López-Matas, M.; Pagan, R.; Mitjans, F.; Vilaró, S.; Piulats, J.; et al. Use of siRNAs and Antisense Oligonucleotides Against Survivin RNA to Inhibit Steps Leading to Tumor Angiogenesis. Oligonucleotides 2004, 14, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Kunze, R.; Kraemer, K.; Erdmann, K.; Froehner, M.; Wirth, M.P.; Fuessel, S. Simultaneous siRNA-mediated knockdown of antiapoptotic BCL2, Bcl-xL, XIAP and survivin in bladder cancer cells. Int. J. Oncol. 2012, 41, 1271–1277. [Google Scholar] [CrossRef]
- Shen, J.; Samul, R.; Silva, R.L.; Akiyama, H.; Liu, H.; Saishin, Y.; Hackett, S.F.; Zinnen, S.; Kossen, K.; Fosnaugh, K.; et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2005, 13, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Takei, Y.; Rofstad, E.K.; Mathiesen, B.; Galappathi, K. A Small Interfering RNA Targeting Vascular Endothelial Growth Factor as Cancer Therapeutics. Cancer Res. 2004, 64, 3365–3370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidel, J.D.; Liu, J.Y.-C.; Yen, Y.; Zhou, B.; Heale, B.S.; Rossi, J.J.; Bartlett, D.W.; Davis, M.E. Potent siRNA Inhibitors of Ribonucleotide Reductase Subunit RRM2 Reduce Cell ProliferationIn vitroandIn vivo. Clin. Cancer Res. 2007, 13, 2207–2215. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.P.; Cheok, C.F.; Lain, S. p53-based Cancer Therapy. Cold Spring Harb. Perspect. Biol. 2010, 2, a001222. [Google Scholar] [CrossRef] [Green Version]
- Stoleriu, M.G.; Steger, V.; Mustafi, M.; Michaelis, M.; Cinatl, J.; Schneider, W.; Nolte, A.; Kurz, J.; Wendel, H.P.; Schlensak, C.; et al. A new strategy in the treatment of chemoresistant lung adenocarcinoma via specific siRNA transfection of SRF, E2F1, Survivin, HIF and STAT3†. Eur. J. Cardio-Thoracic Surg. 2014, 46, 877–886. [Google Scholar] [CrossRef] [Green Version]
- Yanagi, T.; Tachikawa, K.; Wilkie-Grantham, R.; Hishiki, A.; Nagai, K.; Toyonaga, E.; Chivukula, P.; Matsuzawa, S.-I. Lipid Nanoparticle-mediated siRNA Transfer Against PCTAIRE1/PCTK1/Cdk16 Inhibits In Vivo Cancer Growth. Mol. Ther.-Nucleic Acids 2016, 5, e327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiroli, D.; Gómara, M.J.; Maurizi, E.; Atkinson, S.D.; Mairs, L.; Christie, K.A.; Cobice, D.F.; McCrudden, C.M.; Nesbit, M.A.; Haro, I.; et al. Effective In Vivo Topical Delivery of siRNA and Gene Silencing in Intact Corneal Epithelium Using a Modified Cell-Penetrating Peptide. Mol. Ther.-Nucleic Acids 2019, 17, 891–906. [Google Scholar] [CrossRef]
- Ng, B.; Cash-Mason, T.; Wang, Y.; Seitzer, J.; Burchard, J.; Brown, D.; Dudkin, V.; Davide, J.; Jadhav, V.; Sepp-Lorenzino, L.; et al. Intratracheal Administration of siRNA Triggers mRNA Silencing in the Lung to Modulate T Cell Immune Response and Lung Inflammation. Mol. Ther.-Nucleic Acids 2019, 16, 194–205. [Google Scholar] [CrossRef] [Green Version]
- Mencia, N.; Selga, E.; Noe, V.; Ciudad, C.J. Underexpression of miR-224 in methotrexate resistant human colon cancer cells. Biochem. Pharmacol. 2011, 82, 1572–1582. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.R.; Rebhan, M.A.E.; Crivelli, S.E.M.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014, 42, 609–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Bei, Y.; Huang, P.; Zhou, Q.; Shi, J.; Sun, Q.; Zhong, J.; Li, X.; Kong, X.; Xiao, J. Inhibition of miR-155 Protects Against LPS-induced Cardiac Dysfunction and Apoptosis in Mice. Mol. Ther.-Nucleic Acids 2016, 5, e374. [Google Scholar] [CrossRef] [Green Version]
- Reschke, C.R.; Silva, L.F.A.; Norwood, B.A.; Senthilkumar, K.; Morris, G.; Sanz-Rodriguez, A.; Conroy, R.M.; Costard, L.; Neubert, V.; Bauer, S.; et al. Potent Anti-seizure Effects of Locked Nucleic Acid Antagomirs Targeting miR-134 in Multiple Mouse and Rat Models of Epilepsy. Mol. Ther.-Nucleic Acids 2017, 6, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, Q.; Faleti, O.D.; Tsang, C.-M.; Zhao, M.; Wu, G.; Tsao, S.-W.; Fu, M.; Chen, Y.; Ding, T.; et al. Exosomal Delivery of AntagomiRs Targeting Viral and Cellular MicroRNAs Synergistically Inhibits Cancer Angiogenesis. Mol. Ther.-Nucleic Acids 2020, 22, 153–165. [Google Scholar] [CrossRef]
- Yan, H.; Wang, H.; Zhu, X.; Huang, J.; Li, Y.; Zhou, K.; Hua, Y.; Yan, F.; Wang, D.-Z.; Luo, Y. Adeno-associated virus-mediated delivery of anti-miR-199a tough decoys attenuates cardiac hypertrophy by targeting PGC-1alpha. Mol. Ther.-Nucleic Acids 2021, 23, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Sun, W.; Ling, S.; Wang, Y.; Wang, X.; Meng, H.; Li, Y.; Yuan, X.; Li, J.; Liu, R.; et al. AAV-Anti-miR-214 Prevents Collapse of the Femoral Head in Osteonecrosis by Regulating Osteoblast and Osteoclast Activities. Mol. Ther.-Nucleic Acids 2019, 18, 841–850. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Quijano, E.; Liu, Y.; Bahal, R.; Scanlon, S.E.; Song, E.; Hsieh, W.-C.; Braddock, D.E.; Ly, D.H.; Saltzman, W.M.; et al. Anti-tumor Activity of miniPEG-γ-Modified PNAs to Inhibit MicroRNA-210 for Cancer Therapy. Mol. Ther.-Nucleic Acids 2017, 9, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.N.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.; Engelman, D.M.; et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nat. Cell Biol. 2015, 518, 107–110. [Google Scholar] [CrossRef] [Green Version]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Gold, L. SELEX: How It Happened and Where It will Go. J. Mol. Evol. 2015, 81, 140–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Doggrell, S. Pegaptanib: The first antiangiogenic agent approved for neovascular macular degeneration. Expert Opin. Pharmacother. 2005, 6, 1421–1423. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Müller, S. Engineering of ribozymes with useful activities in the ancient RNA world. Ann. N. Y. Acad. Sci. 2015, 1341, 54–60. [Google Scholar] [CrossRef]
- Wang, T.; Tague, N.; Whelan, S.; Dunlop, M.J. Programmable gene regulation for metabolic engineering using decoy transcription factor binding sites. Nucleic Acids Res. 2021, 49, 1163–1172. [Google Scholar] [CrossRef]
- Cho-Chung, Y.S. CRE-Palindrome Oligonucleotide as a Transcription Factor Decoy and an Inhibitor of Tumor Growth. Antisense Nucleic Acid Drug Dev. 1998, 8, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.D.; Morishita, R.; Kaneda, Y.; Kim, H.S.; Chang, Y.-C.; Lee, K.-U.; Park, J.-Y.; Lee, H.W.; Kim, Y.-H.; Lee, I.-K. Novel E2F decoy oligodeoxynucleotides inhibit in vitro vascular smooth muscle cell proliferation and in vivo neointimal hyperplasia. Gene Ther. 2002, 9, 1682–1692. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef]
- Coma, S.; Noe, V.; Eritja, R.; Ciudad, C. Strand Displacement of Double-Stranded DNA by Triplex-Forming Antiparallel Purine-Hairpins. Oligonucleotides 2005, 15, 269–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoogsteen, K. The structure of crystals containing a hydrogen-bonded complex of 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1959, 12, 822–823. [Google Scholar] [CrossRef]
- Hoogsteen, K. The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 1963, 16, 907–916. [Google Scholar] [CrossRef]
- De Almagro, M.C.; Coma, S.; Noé, V.; Ciudad, C.J. Polypurine Hairpins Directed against the Template Strand of DNA Knock Down the Expression of Mammalian Genes. J. Biol. Chem. 2009, 284, 11579–11589. [Google Scholar] [CrossRef] [Green Version]
- De Almagro, M.C.; Mencia, N.; Noe, V.; Ciudad, C.J. Coding Polypurine Hairpins Cause Target-Induced Cell Death in Breast Cancer Cells. Hum. Gene Ther. 2011, 22, 451–463. [Google Scholar] [CrossRef]
- Rodríguez, L.; Villalobos, X.; Dakhel, S.; Padilla, L.; Hervas, R.; Hernández, J.L.; Ciudad, C.; Noé, V. Polypurine reverse Hoogsteen hairpins as a gene therapy tool against survivin in human prostate cancer PC3 cells in vitro and in vivo. Biochem. Pharmacol. 2013, 86, 1541–1554. [Google Scholar] [CrossRef]
- Villalobos, X.; Rodríguez, L.; Solé, A.; Lliberós, C.; Mencia, N.; Ciudad, C.; Noe, V. Effect of Polypurine Reverse Hoogsteen Hairpins on Relevant Cancer Target Genes in Different Human Cell Lines. Nucleic Acid Ther. 2015, 25, 198–208. [Google Scholar] [CrossRef]
- Rodríguez, L.; Villalobos, X.; Solé, A.; Lliberós, C.; Ciudad, C.J.; Noé, V. Improved Design of PPRHs for Gene Silencing. Mol. Pharm. 2015, 12, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Bener, G.; Félix, A.J.; De Diego, C.S.; Fabregat, I.P.; Ciudad, C.J.; Noé, V. Silencing of CD47 and SIRPα by Polypurine reverse Hoogsteen hairpins to promote MCF-7 breast cancer cells death by PMA-differentiated THP-1 cells. BMC Immunol. 2016, 17, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enríquez, M.M.M.; Félix, A.J.; Ciudad, C.J.; Noé, V. Cancer immunotherapy using PolyPurine Reverse Hoogsteen hairpins targeting the PD-1/PD-L1 pathway in human tumor cells. PLoS ONE 2018, 13, e0206818. [Google Scholar] [CrossRef]
- Ciudad, C.J.; Enriquez, M.M.M.; Félix, A.J.; Bener, G.; Noé, V. Silencing PD-1 and PD-L1: The potential of PolyPurine Reverse Hoogsteen hairpins for the elimination of tumor cells. Immunotherapy 2019, 11, 369–372. [Google Scholar] [CrossRef] [PubMed]
- Aubets, E.; Noé, V.; Ciudad, C.J. Targeting replication stress response using polypurine reverse hoogsteen hairpins directed against WEE1 and CHK1 genes in human cancer cells. Biochem. Pharmacol. 2020, 175, 113911. [Google Scholar] [CrossRef] [PubMed]
- Aubets, E.; Félix, A.J.; Garavís, M.; Reyes, L.; Aviñó, A.; Eritja, R.; Ciudad, C.J.; Noé, V. Detection of a G-Quadruplex as a Regulatory Element in Thymidylate Synthase for Gene Silencing Using Polypurine Reverse Hoogsteen Hairpins. Int. J. Mol. Sci. 2020, 21, 5028. [Google Scholar] [CrossRef]
- Félix, A.J.; Solé, A.; Noé, V.; Ciudad, C.J. Gene Correction of Point Mutations Using PolyPurine Reverse Hoogsteen Hairpins Technology. Front. Genome Ed. 2020, 2, 583577. [Google Scholar] [CrossRef]
- Félix, A.J.; Ciudad, C.J.; Noé, V. Correction of the aprt Gene Using Repair-Polypurine Reverse Hoogsteen Hairpins in Mammalian Cells. Mol. Ther.-Nucleic Acids 2020, 19, 683–695. [Google Scholar] [CrossRef]
- Solé, A.; Villalobos, X.; Ciudad, C.J.; Noe, V. Repair of Single-Point Mutations by Polypurine Reverse Hoogsteen Hairpins. Hum. Gene Ther. Methods 2014, 25, 288–302. [Google Scholar] [CrossRef] [Green Version]
- Noé, V.; Ciudad, C. Polypurine Reverse-Hoogsteen Hairpins as a Tool for Exon Skipping at the Genomic Level in Mammalian Cells. Int. J. Mol. Sci. 2021, 22, 3784. [Google Scholar] [CrossRef]
- Juliano, R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef]
- Nayerossadat, N.; Ali, P.A.; Maedeh, T. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1, 27. [Google Scholar] [CrossRef]
- Aubets, E.; Griera, R.; Felix, A.J.; Rigol, G.; Sikorski, C.; Limón, D.; Mastrorosa, C.; Busquets, M.A.; Pérez-García, L.; Noé, V.; et al. Synthesis and validation of DOPY: A new gemini dioleylbispyridinium based amphiphile for nucleic acid transfection. Eur. J. Pharm. Biopharm. 2021, 165, 279–292. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zeng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017, 4, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Ortiz, J.; Schaffer, D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Control. Release 2016, 240, 287–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, M.J.; Lee, K.H. Current Advances in Retroviral Gene Therapy. Curr. Gene Ther. 2011, 11, 218–228. [Google Scholar] [CrossRef]
- Milone, M.C.; O’Doherty, U. Clinical use of lentiviral vectors. Leukemia 2018, 32, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Manservigi, R. HSV Recombinant Vectors for Gene Therapy. Open Virol. J. 2010, 4, 123–156. [Google Scholar] [CrossRef] [Green Version]
- Ono, C.; Okamoto, T.; Abe, T.; Matsuura, Y. Baculovirus as a Tool for Gene Delivery and Gene Therapy. Viruses 2018, 10, 510. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, K.; Suda, T.; Zhang, G.; Liu, D. Advances in Gene Delivery Systems. Pharm. Med. 2011, 25, 293–306. [Google Scholar] [CrossRef]
- Lasaro, M.; Ertl, H.C. New Insights on Adenovirus as Vaccine Vectors. Mol. Ther. 2009, 17, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- De Santis, O.; Audran, R.; Pothin, E.; Warpelin-Decrausaz, L.; Vallotton, L.; Wuerzner, G.; Cochet, C.; Estoppey, D.; Steiner-Monard, V.; Lonchampt, S.; et al. Safety and immunogenicity of a chimpanzee adenovirus-vectored Ebola vaccine in healthy adults: A randomised, double-blind, placebo-controlled, dose-finding, phase 1/2a study. Lancet Infect. Dis. 2016, 16, 311–320. [Google Scholar] [CrossRef] [Green Version]
- Cerullo, V.; Pesonen, S.; Diaconu, I.; Escutenaire, S.; Arstila, P.T.; Ugolini, M.; Nokisalmi, P.; Raki, M.; Laasonen, L.; Särkioja, M.; et al. Oncolytic Adenovirus Coding for Granulocyte Macrophage Colony-Stimulating Factor Induces Antitumoral Immunity in Cancer Patients. Cancer Res. 2010, 70, 4297–4309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, I.-K.; Yun, C.-O. Recent developments in oncolytic adenovirus-based immunotherapeutic agents for use against metastatic cancers. Cancer Gene Ther. 2013, 20, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamm, I.; Wang, Y.; Sausville, E.; A Scudiero, D.; Vigna, N.; Oltersdorf, T.; Reed, J.C. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998, 58, 5315–5320. [Google Scholar] [PubMed]
- Falleni, M.; Pellegrini, C.; Marchetti, A.; Oprandi, B.; Buttitta, F.; Barassi, F.; Santambrogio, L.; Coggi, G.; Bosari, S. Survivin gene expression in early-stage non-small cell lung cancer. J. Pathol. 2003, 200, 620–626. [Google Scholar] [CrossRef]
- Ambrosini, G.; Adida, C.; Altieri, D.C. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 1997, 3, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.-H.; Joo, Y.-E.; Koh, Y.-S.; Chung, I.-J.; Park, Y.-K.; Lee, J.-H.; Kim, H.-S.; Choi, S.-K.; Rew, J.-S.; Park, C.-S.; et al. Expression of survivin in gastric cancer and its relationship with tumor angiogenesis. Eur. J. Gastroenterol. Hepatol. 2006, 18, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Farma, J.M.; Coppola, D.; Hakam, A.; Fulp, W.J.; Chen, D.-T.; Siegel, E.M.; Yeatman, T.J.; Shibata, D. Expression of the Antiapoptotic Protein Survivin in Colon Cancer. Clin. Color. Cancer 2011, 10, 188–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiu, K.-T.; Hwang, T.I.; Hsieh, H.-Y.; Shen, C.-H.; Wang, Y.-H.; Juang, G.-D. Expression of survivin in bladder cancer cell lines using quantitative real-time polymerase chain reaction. Urol. Sci. 2014, 25, 19–21. [Google Scholar] [CrossRef] [Green Version]
- Nasu, S.; Yagihashi, A.; Izawa, A.; Saito, K.; Asanuma, K.; Nakamura, M.; Kobayashi, D.; Okazaki, M.; Watanabe, N. Survivin mRNA expression in patients with breast cancer. Anticancer. Res. 2002, 22, 1839–1843. [Google Scholar] [PubMed]
- Gu, X.; Lin, H.; Shao, J.; Zhang, M.; Liang, H. Analysis of Survivin Expression in the Subtypes of Lymphoma. Chin.-Ger. J. Clin. Oncol. 2005, 4, 238–243. [Google Scholar] [CrossRef]
- Azuhata, T.; Scott, D.; Takamizawa, S.; Wen, J.; Davidoff, A.; Fukuzawa, M.; Sandler, A. The inhibitor of apoptosis protein survivin is associated with high-risk behavior of neuroblastoma. J. Pediatr. Surg. 2001, 36, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, P.K.; Goel, A.; Mittal, R.D. Survivin: A molecular biomarker in cancer. Indian J. Med. Res. 2015, 141, 389–397. [Google Scholar] [CrossRef]
- Ito, R.; Asami, S.; Motohashi, S.; Ootsuka, S.; Yamaguchi, Y.; Chin, M.; Shichino, H.; Yoshida, Y.; Nemoto, N.; Mugishima, H.; et al. Significance of Survivin mRNA Expression in Prognosis of Neuroblastoma. Biol. Pharm. Bull. 2005, 28, 565–568. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Coen, J.J.; Suzuki, Y.; Siedow, M.R.; Niemierko, A.; Khor, L.-Y.; Pollack, A.; Zhang, Y.; Zietman, A.L.; Shipley, W.U.; et al. Survivin Is a Potential Mediator of Prostate Cancer Metastasis. Int. J. Radiat. Oncol. 2010, 78, 1095–1103. [Google Scholar] [CrossRef] [Green Version]
- Gianani, R.; Jarboe, E.; Orlicky, D.; Frost, M.; Bobak, J.; Lehner, R.; Shroyer, K.R. Expression of survivin in normal, hyperplastic, and neoplastic colonic mucosa. Hum. Pathol. 2001, 32, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Carter, B.Z.; Milella, M.; Altieri, D.C.; Andreeff, M. Cytokine-regulated expression of survivinin myeloid leukemia. Blood 2001, 97, 2784–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciudad, C.J.; Rodríguez, L.; Villalobos, X.; Félix, A.J.; Noé, V.; Ciudad, L.R.C.J. Polypurine Reverse Hoogsteen Hairpins as a Gene Silencing Tool for Cancer. Curr. Med. Chem. 2017, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Noé, V.; Aubets, E.; Félix, A.J.; Ciudad, C.J. Nucleic acids therapeutics using PolyPurine Reverse Hoogsteen hairpins. Biochem. Pharmacol. 2020, 189, 114371. [Google Scholar] [CrossRef]
- Villalobos, X.; Rodríguez, L.; Prévot, J.; Oleaga, C.; Ciudad, C.; Noé, V. Stability and Immunogenicity Properties of the Gene-Silencing Polypurine Reverse Hoogsteen Hairpins. Mol. Pharm. 2014, 11, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Félix, A.J.; Ciudad, C.J.; Noé, V. Functional pharmacogenomics and toxicity of PolyPurine Reverse Hoogsteen hairpins directed against survivin in human cells. Biochem. Pharmacol. 2018, 155, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Pearson, S.; Jia, H.; Kandachi, K. China approves first gene therapy. Nat. Biotechnol. 2004, 22, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z. Current Status of Gendicine in China: Recombinant Human Ad-p53 Agent for Treatment of Cancers. Hum. Gene Ther. 2005, 16, 1016–1027. [Google Scholar] [CrossRef] [PubMed]
- Johnson & Johnson. Available online: https://www.jnj.com/johnson-johnson-announces-european-commission-approval-for-janssens-preventive-ebola-vaccine (accessed on 1 April 2021).
- Nakagami, H. Development of COVID-19 vaccines utilizing gene therapy technology. Int. Immunol. 2021. [Google Scholar] [CrossRef]
- Sharma, P.; Chawla, M.; Sharma, S.; Mitra, A. On the role of Hoogsteen:Hoogsteen interactions in RNA: Ab initio investigations of structures and energies. RNA 2010, 16, 942–957. [Google Scholar] [CrossRef] [Green Version]
- McDonald, C.D.; Maher, L.J. Recognition of duplex DNA by RNA polynucleotides. Nucleic Acids Res. 1995, 23, 500–506. [Google Scholar] [CrossRef] [Green Version]
- Sui, G.; Soohoo, C.; Affar, E.B.; Gay, F.; Shi, Y.; Forrester, W.C. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 5515–5520. [Google Scholar] [CrossRef] [Green Version]
- Paddison, P.J.; Caudy, A.; Bernstein, E.; Hannon, G.J.; Conklin, D. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef] [Green Version]
- Devroe, E.; A Silver, P. Retrovirus-delivered siRNA. BMC Biotechnol. 2002, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Tomar, R.S.; Matta, H.; Chaudhary, P.M. Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene 2003, 22, 5712–5715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, G.; Medzhitov, R. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14943–14945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, M.I.; Mohuczy-Dominiak, D.; Coffey, M.; Galli, S.M.; Kimura, B.; Wu, P.; Zelles, T. Prolonged Reduction of High Blood Pressure With an In Vivo, Nonpathogenic, Adeno-Associated Viral Vector Delivery of AT 1 -R mRNA Antisense. Hypertension 1997, 29, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raper, S.E.; Chirmule, N.; Lee, F.; Wivel, N.A.; Bagg, A.; Gao, G.-P.; Wilson, J.; Batshaw, M.L. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003, 80, 148–158. [Google Scholar] [CrossRef]
- Kremer, E.J.; Boutin, S.; Chillon, M.; Danos, O. Canine Adenovirus Vectors: An Alternative for Adenovirus-Mediated Gene Transfer. J. Virol. 2000, 74, 505–512. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.Z.; Hyattb, A.; Boyle, D.B.; Both, G.W. Construction of Ovine Adenovirus Recombinants by Gene Insertion or Deletion of Related Terminal Region Sequences. Virology 1997, 230, 62–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, K.; Da Costa, A.; Yamamoto, A.; Berry, D.; Lindsay, R.W.B.; Darrah, P.A.; Wang, L.; Cheng, C.; Kong, W.-P.; Gall, J.G.D.; et al. Comparative Analysis of the Magnitude, Quality, Phenotype, and Protective Capacity of Simian Immunodeficiency Virus Gag-Specific CD8+T Cells following Human-, Simian-, and Chimpanzee-Derived Recombinant Adenoviral Vector Immunization. J. Immunol. 2013, 190, 2720–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ersching, J.; Hernandez, M.; Cezarotto, F.S.; Ferreira, J.D.; Martins, A.B.; Switzer, W.M.; Xiang, Z.; Ertl, H.C.; Zanetti, C.R.; Pinto, A.R. Neutralizing antibodies to human and simian adenoviruses in humans and New-World monkeys. Virology 2010, 407, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, P.N.; Zinn, K.; Gavrilyuk, V.D.; Balyasnikova, I.V.; Rogers, B.E.; Buchsbaum, D.J.; Wang, M.H.; Miletich, D.J.; Grizzle, W.E.; Douglas, J.T.; et al. A Targetable, Injectable Adenoviral Vector for Selective Gene Delivery to Pulmonary Endothelium in Vivo. Mol. Ther. 2000, 2, 562–578. [Google Scholar] [CrossRef]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Douglas, J.T.; Rogers, B.E.; Rosenfeld, M.E.; Michael, S.I.; Feng, M.; Curiel, D.T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat. Biotechnol. 1996, 14, 1574–1578. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.N.; Nicklin, S.; Kaliberova, L.; Boatman, B.G.; Grizzle, W.E.; Balyasnikova, I.V.; Baker, A.H.; Danilov, S.M.; Curiel, D.T. Combined transductional and transcriptional targeting improves the specificity of transgene expression in vivo. Nat. Biotechnol. 2001, 19, 838–842. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Samulski, R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020, 21, 255–272. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aubets, E.; Chillon, M.; Ciudad, C.J.; Noé, V. PolyPurine Reverse Hoogsteen Hairpins Work as RNA Species for Gene Silencing. Int. J. Mol. Sci. 2021, 22, 10025. https://doi.org/10.3390/ijms221810025
Aubets E, Chillon M, Ciudad CJ, Noé V. PolyPurine Reverse Hoogsteen Hairpins Work as RNA Species for Gene Silencing. International Journal of Molecular Sciences. 2021; 22(18):10025. https://doi.org/10.3390/ijms221810025
Chicago/Turabian StyleAubets, Eva, Miguel Chillon, Carlos J. Ciudad, and Véronique Noé. 2021. "PolyPurine Reverse Hoogsteen Hairpins Work as RNA Species for Gene Silencing" International Journal of Molecular Sciences 22, no. 18: 10025. https://doi.org/10.3390/ijms221810025
APA StyleAubets, E., Chillon, M., Ciudad, C. J., & Noé, V. (2021). PolyPurine Reverse Hoogsteen Hairpins Work as RNA Species for Gene Silencing. International Journal of Molecular Sciences, 22(18), 10025. https://doi.org/10.3390/ijms221810025