State of Drosophila melanogaster Ovaries after a Full Cycle of Gametogenesis under Microgravity Modeling: Cellular Respiration and the Content of Cytoskeletal Proteins
Abstract
:1. Introduction
2. Results
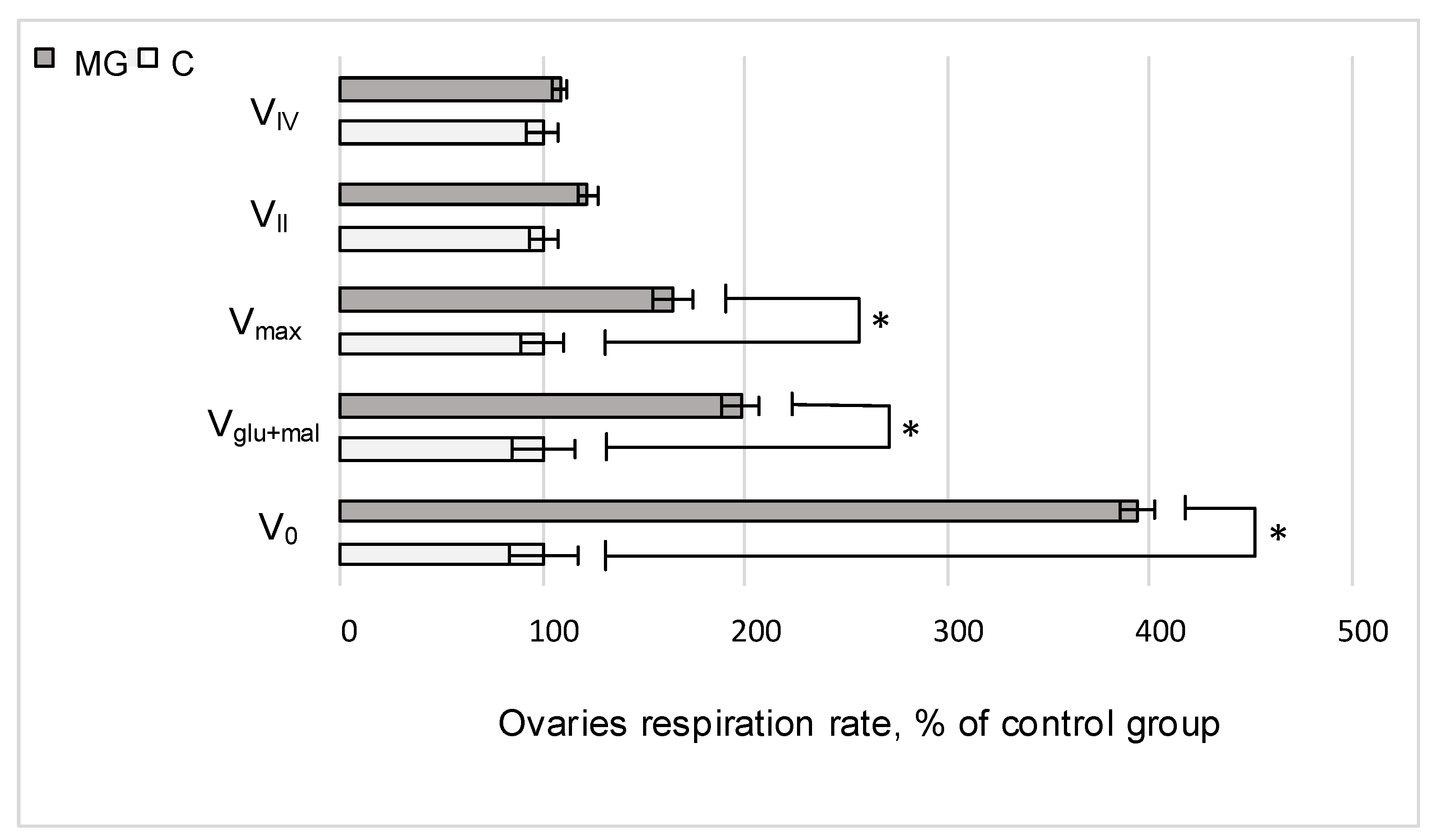
2.1. Cell Respiration
2.2. Protein Content
2.3. mRNA Relative Content
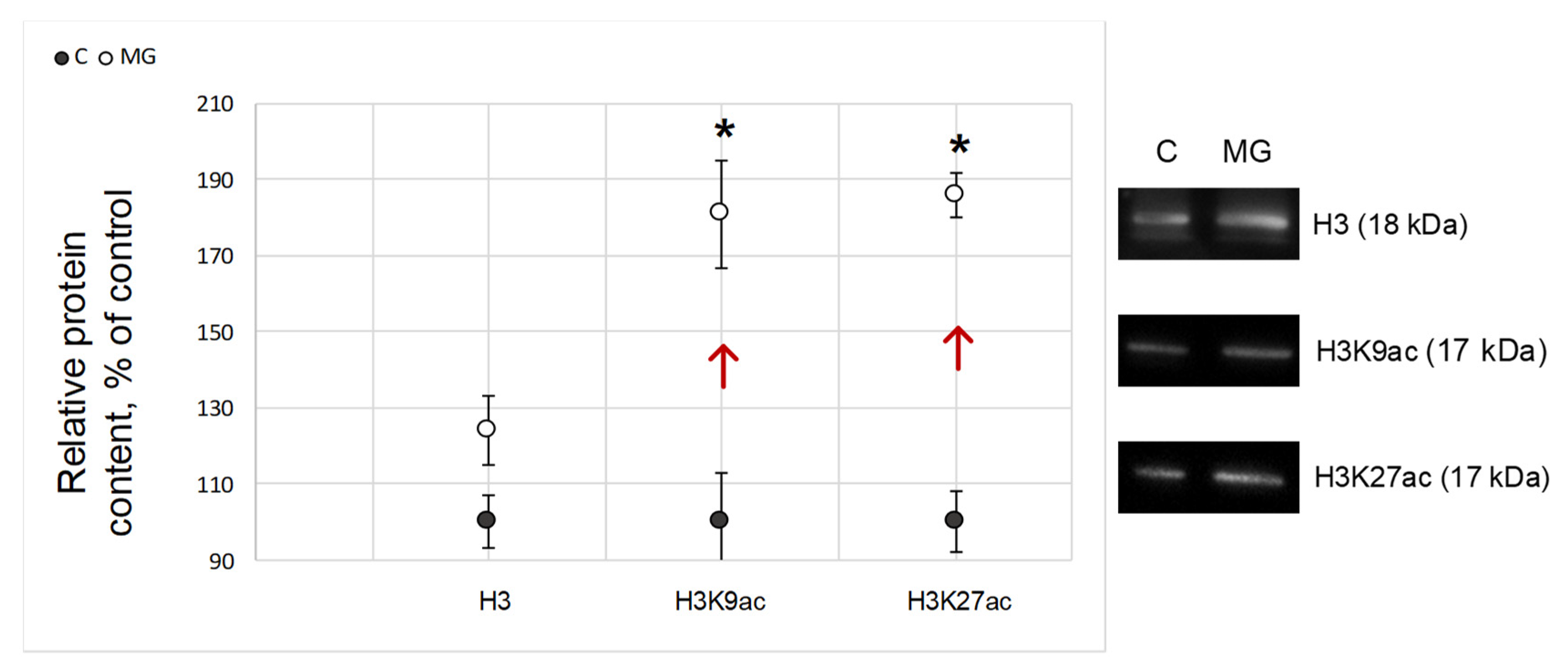
2.4. Histone Acetylation Relative Content
3. Discussion
4. Materials and Methods
4.1. Study Design
- −
- MG (simulated microgravity group), which was placed in conditions of simulated microgravity;
- −
- C (control group), which was placed next to the simulator platform to ensure identical containment conditions.
4.2. Measuring Cellular Respiration Using the Polarography Method
4.3. Estimation of the Relative Protein Content by Western Blotting
4.4. Estimation of the Relative Content of mRNA by Polymerase Chain Reaction (PCR)
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vernos, I.; Gonzalez-Jurado, J.; Calleja, M.; Marco, R. Microgravity effects on the oogenesis and development of embryos of Drosophila melanogaster laid in the Spaceshuttle during the Biorack experiment (ESA). Int. J. Dev. Biol. 1989, 33, 213–226. [Google Scholar]
- Ogneva, I.V.; Belyakin, S.N.; Sarantseva, S.V. The development of Drosophila melanogaster under different duration space flight and subsequent adaptation to Earth gravity. PLoS ONE 2016, 11, e0166885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijiri, K. Fish mating experiment in space-what it aimed at and how it was prepared. Biol. Sci. Space 1995, 9, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Aimar, C.; Bautz, A.; Durand, D.; Membre, H.; Chardard, D.; Gualandris-Parisot, L.; Husson, D.; Dournon, C. Microgravity and hypergravity effects on fertilization of the salamander Pleurodeles waltl (urodele amphibian). Biol. Reprod. 2000, 63, 551–558. [Google Scholar] [CrossRef]
- Schenker, E.; Forkheim, K. Mammalian mice embryo early development in weightlessness environment on STS 80 space flight. Isr. Aerosp. Med. Inst. Rep. 1998, 5. [Google Scholar]
- Kojima, Y.; Sasaki, S.; Kubota, Y.; Ikeuchi, T.; Hayashi, Y.; Kohri, K. Effects of simulated microgravity on mammalian fertilization and preimplantation embryonic development in vitro. Fertil. Steril. 2000, 74, 1142–1147. [Google Scholar] [CrossRef]
- Vaskivuo, T.E.; Tapanainen, J.S. Apoptosis in the human ovary. Reprod. Biomed. Online 2003, 6, 24–35. [Google Scholar] [CrossRef]
- Marcozzi, S.; Rossi, V.; Salustri, A.; De Felici, M.; Klinger, F.G. Programmed cell death in the human ovary. Minerva Ginecol. 2018, 70, 549–560. [Google Scholar] [CrossRef]
- Tanner, E.A.; Blute, T.A.; Brachmann, C.B.; McCall, K. Bcl-2 proteins and autophagy regulate mitochondrial dynamics during programmed cell death in the Drosophila ovary. Development 2011, 138, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Bolobolova, E.U.; Dorogova, N.V.; Fedorova, S.A. Major scenarios of genetically regulated cell death during oogenesis in Drosophila melanogaster. Russ. J. Genet. 2020, 56, 655–665. [Google Scholar] [CrossRef]
- Gourlay, C.W.; Ayscough, K.R. The actin cytoskeleton: A key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 2005, 6, 583–589. [Google Scholar] [CrossRef]
- Boldogh, I.R.; Pon, L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 2006, 1763, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Senning, E.N.; Marcus, A.H. Actin polymerization driven mitochondrial transport in mating S. cerevisiae. Proc. Natl. Acad. Sci. USA 2010, 107, 721–725. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Venit, T.; Drou, N.; Percipalle, P. In mitochondria beta-actin regulates mtDNA transcription and is required for mitochondrial quality control. iScience 2018, 3, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; He, J.; Mao, C.C.; Bailey, L.J.; Di Re, M.; Sembongi, H.; Kazak, L.; Dzionek, K.; Holmes, J.B.; Cluett, T.J.; et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011, 39, 5098–5108. [Google Scholar] [CrossRef]
- Schatten, H.; Lewis, M.L.; Chakrabarti, A. Spaceflight and clinorotation cause cytoskeleton and mitochondria changes and increases in apoptosis in cultured cells. Acta Astronaut. 2001, 49, 399–418. [Google Scholar] [CrossRef]
- Wilding, M.; De Placido, G.; De Matteo, L.; Marino, M.; Alviggi, C.; Dale, B. Chaotic mosaicism in human preimplantation embryos is correlated with a low mitochondrial membrane potential. Fertil. Steril. 2003, 79, 340–346. [Google Scholar] [CrossRef]
- Plett, P.A.; Abonour, R.; Frankovitz, S.M.; Orschell, C.M. Impact of modeled microgravity on migration, differentiation, and cell cycle control of primitive human hematopoietic progenitor cells. Exp. Hematol. 2004, 32, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Gershovich, P.M.; Gershovich, J.G.; Buravkova, L.B. Simulated microgravity alters actin cytoskeleton and integrin-mediated focal adhesions of cultured human mesenchymal stromal cells. J. Grav. Physiol. 2008, 15, 203–204. [Google Scholar]
- Ogneva, I.V.; Maximova, M.V.; Larina, I.M. Structure of cortical cytoskeleton in fibers of mouse muscle cells after being exposed to a 30-day space flight on board the BION-M1 biosatellite. J. Appl. Physiol. 2014, 116, 1315–1323. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009, 457, 405–412. [Google Scholar] [CrossRef]
- Miranda, T.B.; Jones, P.A. DNA methylation: The nuts and bolts of repression. J. Cell Physiol. 2007, 213, 384–390. [Google Scholar] [CrossRef]
- Rottach, A.; Leonhardt, H.; Spada, F. DNA methylation-mediated epigenetic control. J. Cell Biochem. 2009, 108, 43–51. [Google Scholar] [CrossRef]
- Grimaud, C.; Negre, N.; Cavalli, G. From genetics to epigenetics: The tale of Polycomb group and trithorax group genes. Chromosome Res. 2006, 14, 363–375. [Google Scholar] [CrossRef]
- Davie, J.R.; Spencer, V.A. Control of histone modifications. J. Cell Biochem. 1999, 75, 141–148. [Google Scholar] [CrossRef]
- Chen, T.; Dent, S.Y. Chromatin modifiers and remodellers: Regulators of cellular differentiation. Nat. Rev. Genet. 2014, 15, 93–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegrzyn, J.; Potla, R.; Chwae, Y.J.; Sepuri, N.B.; Zhang, Q.; Koeck, T.; Derecka, M.; Szczepanek, K.; Szelag, M.; Gornicka, A.; et al. Function of mitochondrial Stat3 in cellular respiration. Science 2009, 323, 793–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, O.; Henry, C.M.; Srinivasan, N.; Ahrens, S.; Franz, A.; Deddouche, S.; Chakravarty, P.; Phillips, D.; George, R.; Kjaer, S.; et al. α-actinin accounts for the bioactivity of actin preparations in inducing STAT target genes in Drosophila melanogaster. eLife 2018, 7, e38636. [Google Scholar] [CrossRef]
- Ogneva, I.V.; Usik, M.A. Mitochondrial respiration in Drosophila ovaries after a full cycle of oogenesis under simulated microgravity. Curr. Issues Mol. Biol. 2021, 43, 176–186. [Google Scholar] [CrossRef]
- Cox, R.T.; Spradling, A.C. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 2003, 130, 1579–1590. [Google Scholar] [CrossRef] [Green Version]
- Hurd, T.R.; Herrmann, B.; Sauerwald, J.; Sanny, J.; Grosch, M.; Lehmann, R. Long Oskar controls mitochondrial inheritance in Drosophila melanogaster. Dev. Cell. 2016, 39, 560–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandrioli, M.; Borsatti, F. DNA methylation of fly genes and transposons. Cell Mol. Life Sci. 2006, 63, 1933–1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paturi, S.; Deshmukh, M.V. A Glimpse of “Dicer Biology” through the Structural and Functional Perspective. Front. Mol. Biosci. 2021, 8, 643657. [Google Scholar] [CrossRef]
- Kavi, H.H.; Fernandez, H.R.; Xie, W.; Birchler, J.A. RNA silencing in Drosophila. FEBS Lett. 2005, 579, 5940–5949. [Google Scholar] [CrossRef] [Green Version]
- Simpson, R.T. Structure of chromatin containing extensively acetylated H3 and H4. Cell 1978, 13, 691–699. [Google Scholar] [CrossRef]
- Fenley, A.T.; Anandakrishnan, R.; Kidane, Y.H.; Onufriev, A.V. Modulation of nucleosomal DNA accessibility via charge-altering post-translational modifications in histone core. Epigenetics Chromatin 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Grotewold, E. Serial ChIP as a tool to investigate the co-localization or exclusion of proteins on plant genes. Plant Methods 2008, 4, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flouriot, G.; Huet, G.; Demay, F.; Pakdel, F.; Boujrad, N.; Michel, D. The actin/MKL1 signalling pathway influences cell growth and gene expression through large-scale chromatin reorganization and histone post-translational modifications. Biochem. J. 2014, 461, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yan, J.; Zhang, M.; Wang, Y.; Chen, Y.; Fu, X.; Wei, R.; Zheng, X.L.; Liu, Z.; Zhang, X.; et al. Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors. Cell 2018, 175, 186–199.e19. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, A.V.; Veksler, V.; Gellerich, F.N.; Saks, V.; Margreiter, R.; Kunz, W.S. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat. Protoc. 2008, 3, 965–976. [Google Scholar] [CrossRef]
- Ogneva, I.V.; Usik, M.A.; Biryukov, N.S.; Zhdankina, Y.S. Sperm motility of mice under simulated microgravity and hypergravity. Int. J. Mol. Sci. 2020, 21, 5054. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehlin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some application. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 2, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Primary Antibodies | Molecular Weight | Dilution | Producer | Catalog Number |
|---|---|---|---|---|
| Cytochrome c-1 | 13.5 kDa | 5 μg/mL | Abcam, UK, Cambridge | #ab13575 |
| Cytochrome c oxidase | 16 kDa | 1 μg/mL | Abcam, UK, Cambridge | #ab14744 |
| ATP synthase F1 (Blw) | 56 kDa | 1 μg/mL | Abcam, UK, Cambridge | #ab14748 |
| Alpha-tubulin | 50 kDa | 1:1000 | Abcam, UK, Cambridge | #ab52866 |
| Beta-tubulin | 50 kDa | 1:1000 | Abcam, UK, Cambridge | #ab179513 |
| Beta-actin | 42 kDa | 1:5000 | Abcam, UK, Cambridge | #ab227387 |
| Alpha-actinin | 102 kDa | 1 μg/mL | Abcam, UK, Cambridge | #ab50599 |
| Histone H3 | 11 kDa | 1:1000 | Abcam, UK, Cambridge | #ab10799 |
| Histone H3 Lys 9 acetylated | 17 kDa | 1:1000 | Abcam, UK, Cambridge | #ab4441 |
| Histone H3 Lys 27 acetylated | 17 kDa | 1 μg/mL | Abcam, UK, Cambridge | #ab4729 |
| Gene | Primer Sequence, Forward/Reverse (5′…3′) | Product Size, bp |
|---|---|---|
| CG4769 | GCAGCGACATTGCGAAGATT/ACTGCTCCAGGGCGTAGATA | 181 |
| CG10396 | ACTGCCGTCGAAATGAGCTT/TCACGTAGGGCACACAACTC | 227 |
| Blw | AATAGGAGTAGCGGTGCGTG/AACCACGGATTGAAGGCGAT | 201 |
| Act57B | GCCTAGCACCAACACTAGCA/CGCGAGCGATTAACAAGTGG | 288 |
| Act87E | CCGAATACCGAAAGCCCACT/CTGGGCCTCATCACCAACAT | 269 |
| Actn | ACAAGCCGAACATTGAGGAG/GCGTTTCCATCGTGTAGTTG | 96 |
| alphaTub84D | AAGGACTACGAGGAGGTCGG/ATGCGAGTGGGAGCGTATGA | 124 |
| alphaTub84B | CACTGGTACGTTGGTGAGGG/CCCATCGAGCGTTGAAGTGG | 166 |
| betaTub56D | AAGCGGACAGTTTGTGTTGTG/ACCAGCTTGGATGTGAACGA | 115 |
| Gapdh | ATACTCATCAACCCTCCCCC/GGCTGAGTTCCTGCTGTCTT | 142 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Usik, M.A.; Golubkova, M.A.; Ogneva, I.V. State of Drosophila melanogaster Ovaries after a Full Cycle of Gametogenesis under Microgravity Modeling: Cellular Respiration and the Content of Cytoskeletal Proteins. Int. J. Mol. Sci. 2021, 22, 9234. https://doi.org/10.3390/ijms22179234
Usik MA, Golubkova MA, Ogneva IV. State of Drosophila melanogaster Ovaries after a Full Cycle of Gametogenesis under Microgravity Modeling: Cellular Respiration and the Content of Cytoskeletal Proteins. International Journal of Molecular Sciences. 2021; 22(17):9234. https://doi.org/10.3390/ijms22179234
Chicago/Turabian StyleUsik, Maria A., Maria A. Golubkova, and Irina V. Ogneva. 2021. "State of Drosophila melanogaster Ovaries after a Full Cycle of Gametogenesis under Microgravity Modeling: Cellular Respiration and the Content of Cytoskeletal Proteins" International Journal of Molecular Sciences 22, no. 17: 9234. https://doi.org/10.3390/ijms22179234
APA StyleUsik, M. A., Golubkova, M. A., & Ogneva, I. V. (2021). State of Drosophila melanogaster Ovaries after a Full Cycle of Gametogenesis under Microgravity Modeling: Cellular Respiration and the Content of Cytoskeletal Proteins. International Journal of Molecular Sciences, 22(17), 9234. https://doi.org/10.3390/ijms22179234






