Proteomic and Biochemical Analyses of the Mechanism of Tolerance in Mutant Soybean Responding to Flooding Stress
Abstract
:1. Introduction
2. Results
2.1. Morphological Analysis of Mutant Soybean under Flooding Stress
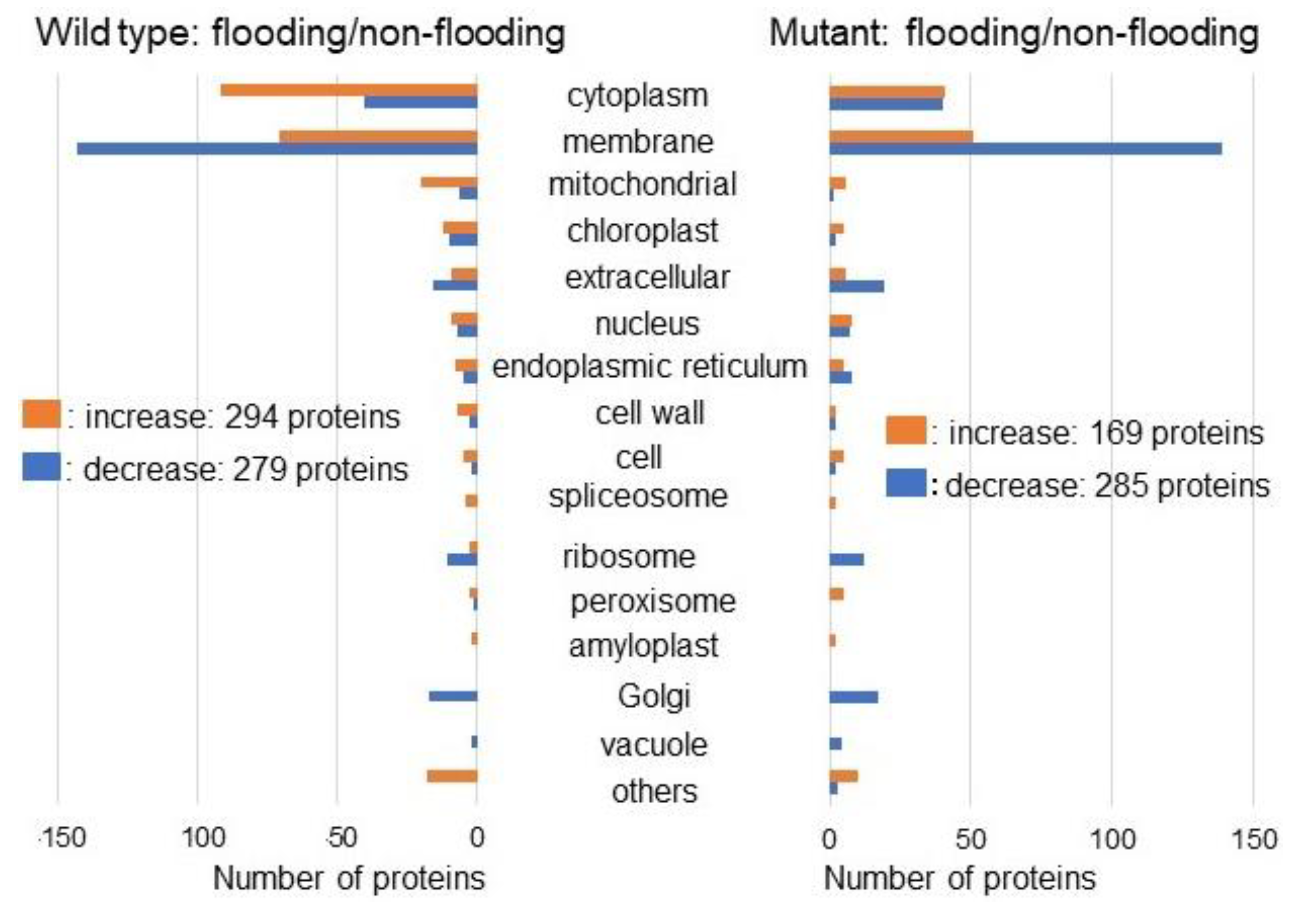
2.2. Identification and Functional Investigation of Proteins in Mutant Soybean under Flooding Stress
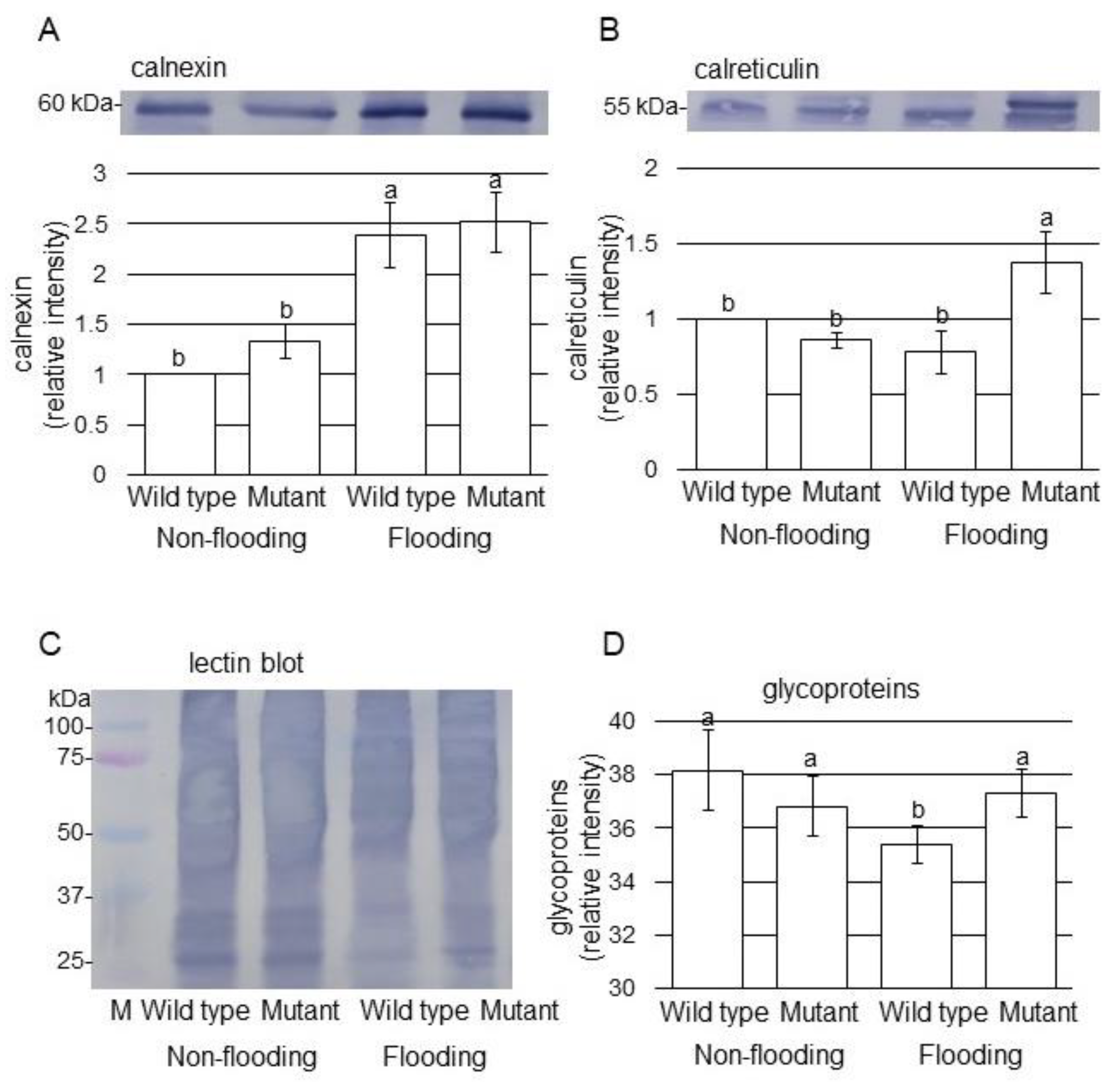
2.3. Immunoblot Analysis of Proteins Involved in Endoplasmic Reticulum in Mutant Soybean under Flooding Stress
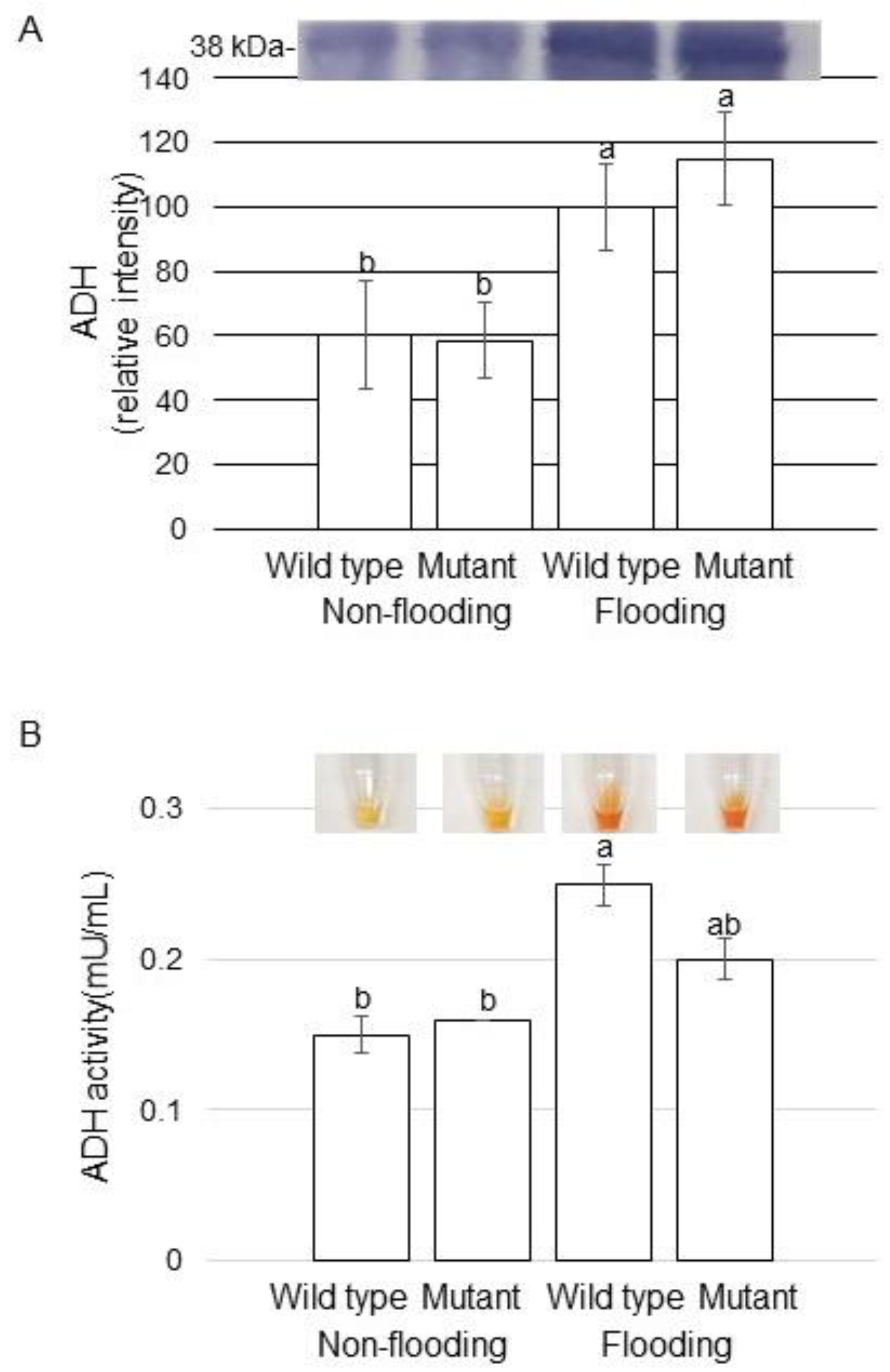
2.4. Alcohol Dehydrogenase (ADH) Accumulation and Activity in Mutant Soybean under Flooding Stress
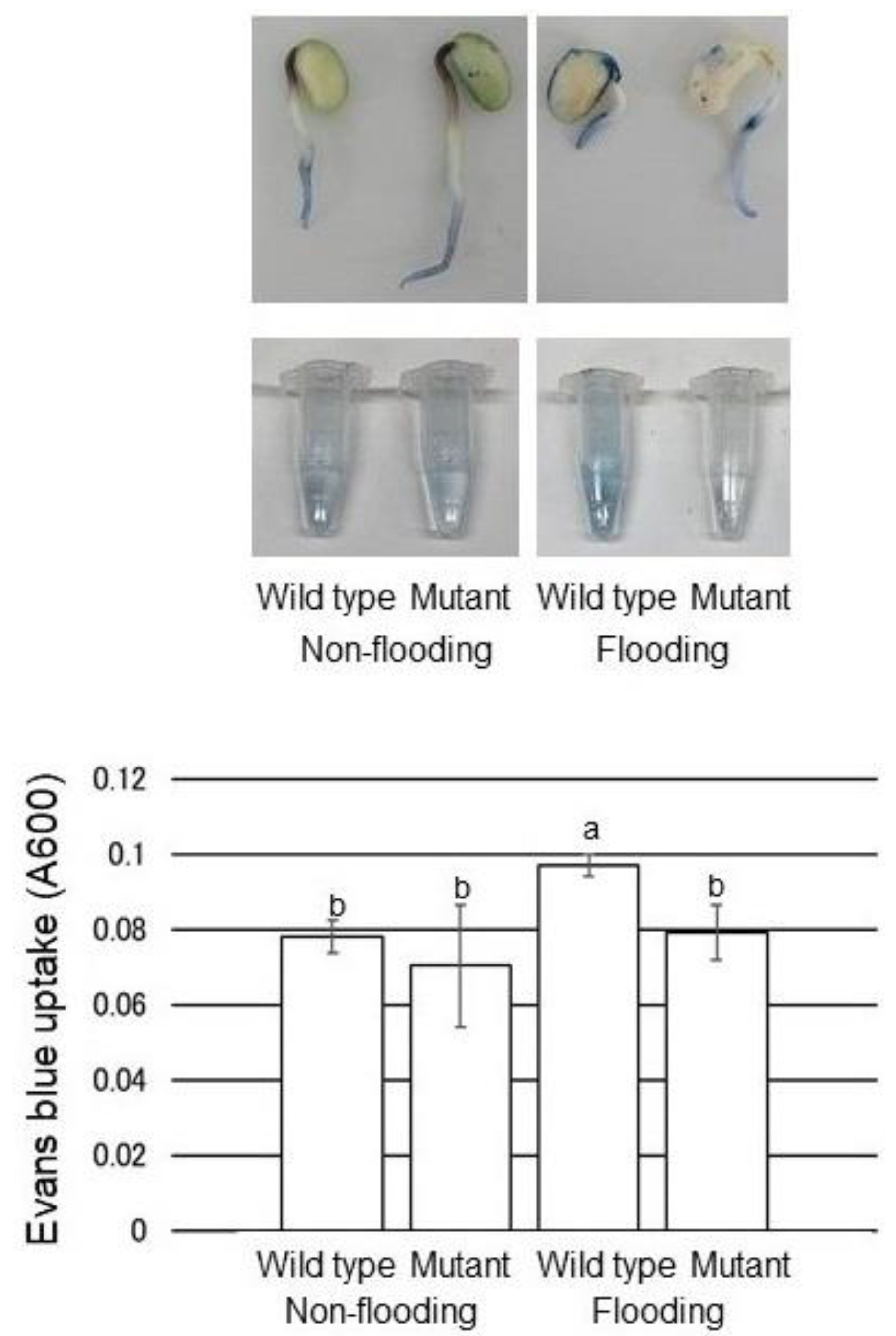
2.5. Evans-Blue Staining with Mutant Soybean under Flooding Stress
3. Discussion
3.1. Mutation in Flooding-Tolerant Soybean Has Positive Effects on the Growth of Plant under Flooding Stress
3.2. Regulation of Alcohol-Formation Pathway Is Related to the Mechanism of Flooding Tolerance
3.3. Glycoprotein Folding Is Related to the Mechanism of Flooding Tolerance
3.4. Cell Death in Soybean Root Is Related to Flooding Tolerance
4. Materials and Methods
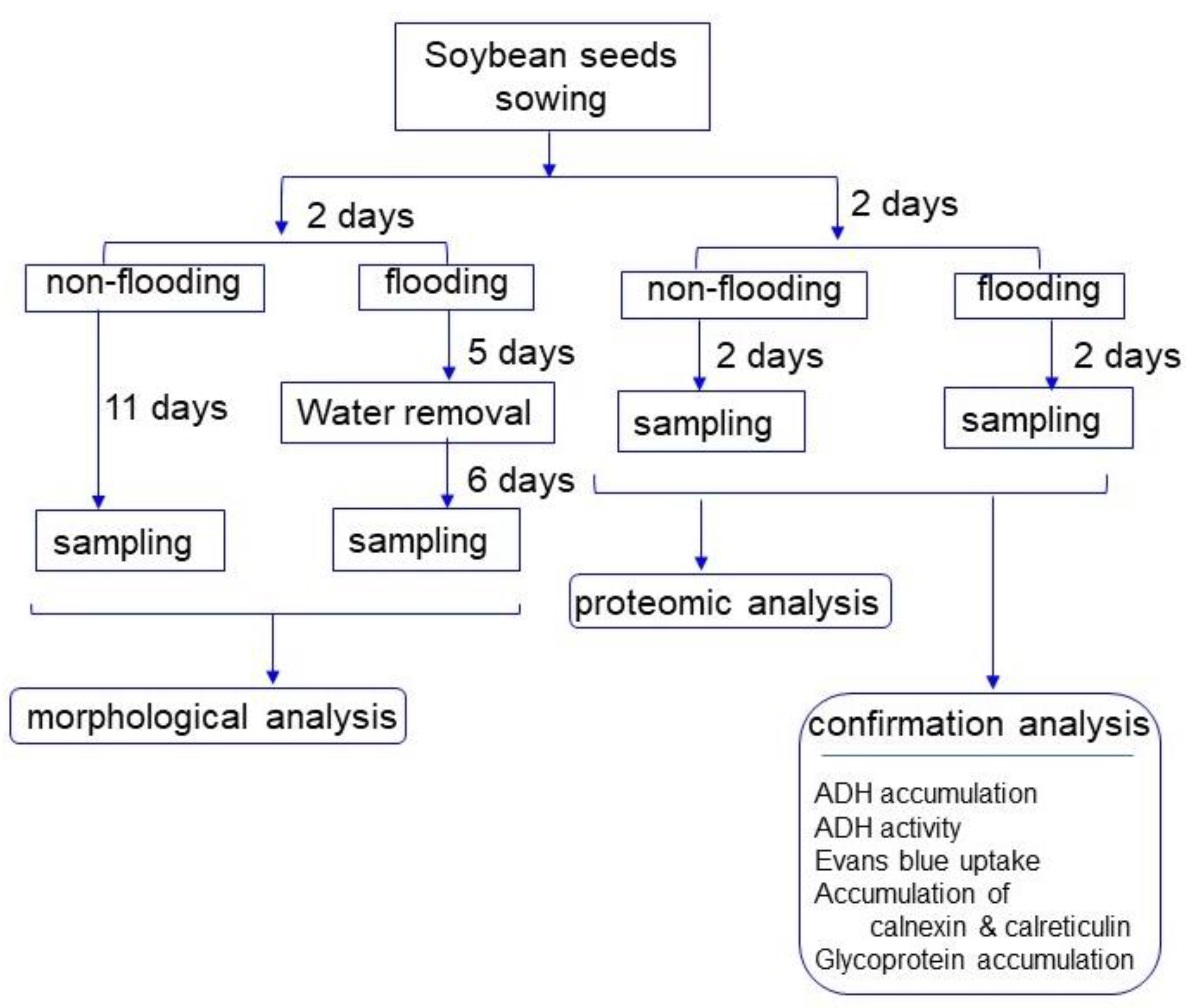
4.1. Plant Material and Experimental Design
4.2. Protein Extraction, Protein Enrichment, Reduction, Alkylation, and Digestion
4.3. Protein Identification Using Nano-Liquid Chromatography Mass Spectrometry
4.4. Mass Spectrometry Data Analysis
4.5. Differential Analysis of Proteins Using Mass Spectrometry Data
4.6. Bioinformatic Analyses of Protein Functional Categorization
4.7. Immunoblot Analysis
4.8. ADH Activity Assay
4.9. Evaluation of Cell Death Using Evans-Blue Dye
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tanoue, M.; Hirabayashi, Y.; Ikeuchi, H. Global-scale river flood vulnerability in the last 50 years. Sci. Rep. 2016, 6, 36021. [Google Scholar] [CrossRef]
- Fukao, T.; Barrera-Figueroa, B.E.; Juntawong, P.; Pena-Castro, J.M. Submergence and waterlogging stress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 2019, 10, 340. [Google Scholar] [CrossRef]
- Setter, T.L.; Waters, I. Review of prospects for germplasm improvement for waterlogging tolerance in wheat, barley and oats. Plant Soil 2013, 253, 1–34. [Google Scholar] [CrossRef]
- Jia, W.; Ma, M.; Chen, J.; Wu, S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int. J. Mol. Sci. 2021, 22, 1088. [Google Scholar] [CrossRef]
- Ma, M.; Cen, W.; Li, R.; Wang, S.; Luo, J. The molecular regulatory pathways and metabolic adaptation in the seed germination and early seedling growth of rice in response to low O2 stress. Plants 2020, 9, 1363. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Fukao, T.; Xu, K.; Ronald, P.C.; Bailey-Serres, J. A variable cluster of ethylene response factor-like genes regulates metabolic and developmental acclimation responses to submergence in rice. Plant Cell 2006, 18, 2021–2034. [Google Scholar] [CrossRef] [Green Version]
- Githiri, S.M.; Watanabe, S.; Harada, K.; Takahashi, R. QTL analysis of flooding tolerance in soybean at an early vegetative growth stage. Plant Breed. 2006, 125, 613–618. [Google Scholar] [CrossRef]
- Oosterhuis, D.M.; Scott, H.D.; Hampton, R.E.; Wullschleter, S.D. Physiological response of two soybean (Glycine max L. Merr) cultivars to short-term flooding. Environ. Exp. Bot. 1990, 30, 85–92. [Google Scholar] [CrossRef]
- Shimamura, S.; Mochizuki, T.; Nada, Y.; Fukuyama, M. Formation and function of secondary aerenchyma in hypocotyl, roots and nodules of soybean (Glycine max) under flooded conditions. Plant Soil 2003, 251, 351–359. [Google Scholar] [CrossRef]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteome Res. 2012, 11, 68–78. [Google Scholar] [CrossRef]
- Komatsu, S.; Shirasaka, N.; Sakata, K. ‘Omics’ techniques for identifying flooding-response mechanisms in soybean. J. Proteom. 2013, 93, 169–178. [Google Scholar] [CrossRef]
- Komatsu, S.; Sakata, K.; Nanjo, Y. ‘Omics’ techniques and their use to identify how soybean responds to flooding. J. Anal. Sci. Technol. 2015, 6, 9. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Komatsu, S. Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteom. 2018, 172, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Review: Proteomic techniques for the development of flood-tolerant soybean. Int. J. Mol. Sci. 2020, 21, 7497. [Google Scholar] [CrossRef]
- Komatsu, S.; Nanjo, Y.; Nishimura, M. Proteomic analysis of the flooding tolerance mechanism in mutant soybean. J. Proteom. 2013, 79, 231–250. [Google Scholar] [CrossRef]
- Yin, X.; Nishimura, M.; Hajika, M.; Komatsu, S. Quantitative proteomics reveals the flooding-tolerance mechanism in mutant and abscisic acid-treated soybean. J. Proteome Res. 2016, 15, 2008–2025. [Google Scholar] [CrossRef]
- Yin, X.; Hiraga, S.; Hajika, M.; Nishimura, M.; Komatsu, S. Transcriptomic analysis reveals the flooding progeny in flooding tolerant line and abscisic acid treated soybean. Plant Mol. Biol. 2017, 93, 479–496. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, W.; Hashiguchi, A.; Nishimura, M.; Tian, J.; Komatsu, S. Metabolic profiles of flooding-tolerant mechanism in early-stage soybean responding to initial stress. Plant Mol. Biol. 2017, 94, 669–685. [Google Scholar] [CrossRef]
- Nanjo, Y.; Jang, H.Y.; Kim, H.S.; Hiraga, S.; Woo, S.H.; Komatsu, S. Analyses of flooding tolerance of soybean varieties at emergence and varietal di_erences in their proteomes. Phytochemistry 2014, 106, 25–36. [Google Scholar] [CrossRef]
- Lin, Y.; Li, W.; Zhang, Y.; Xia, C.; Liu, Y.; Wang, C.; Xu, R.; Zhang, L. Identification of Genes/Proteins Related to Submergence Tolerance by Transcriptome and Proteome Analyses in Soybean. Sci. Rep. 2019, 9, 14688. [Google Scholar] [CrossRef] [Green Version]
- Nagai, K.; Hattori, Y.; Ashikari, M. Stunt or elongate? Two opposite strategies by which rice adapts to floods. J. Plant Res. 2010, 123, 303–309. [Google Scholar] [CrossRef]
- Xu, K.; Xu, X.; Fukao, T.; Canlas, P.; Maghirang-Rodriguez, R.; Heuer, S.; Ismail, A.M.; Bailey-Serres, J.; Ronald, P.C.; Mackill, D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 2006, 442, 705–708. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, D. Genetic control of alcohol dehydrogenase—A competition model for regulation of gene action. Genetics 1971, 67, 411–425. [Google Scholar] [CrossRef]
- Ventura, I.; Brunello, L.; Iacopino, S.; Valeri, M.C.; Novi, G.; Dornbusch, T.; Perata, P.; Loreti, E. Arabidopsis phenotyping reveals the importance of alcohol dehydrogenase and pyruvate decarboxylase for aerobic plant growth. Sci. Rep. 2020, 10, 16669. [Google Scholar] [CrossRef]
- Xuan, L.; Hua, J.; Zhang, F.; Wang, Z.; Pei, X.; Yang, Y.; Yin, Y.; Creech, D.L. Identification and functional analysis of ThADH1 and ThADH4 genes involved in tolerance to waterlogging stress in taxodium hybrid ‘Zhongshanshan 406′. Genes 2021, 12, 225. [Google Scholar] [CrossRef]
- Tougou, M.; Hashiguchi, A.; Yukawa, K.; Nanjo, Y.; Hiraga, S.; Nakamura, T.; Nishizawa, K.; Komatsu, S. Responses to flooding stress in soybean seedling with the alcohol dehydrogenase transgene. Plant Biotech. 2012, 29, 301–306. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Greenway, H.; Matsumura, H.; Tsutsumi, N.; Nakazono, M. Rice alcohol dehydrogenase 1 promotes survival and has a major impact on carbohydrate metabolism in the embryo and endosperm when seeds are germinated in partially oxygenated water. Ann. Bot. 2014, 113, 851–859. [Google Scholar] [CrossRef] [Green Version]
- Healy, S.J.; Verfaillie, T.; Jäger, R.; Agostinis, P.; Samali, A. Biology of the endoplasmic reticulum. In Endoplasmic Reticulum Stress in Health and Disease; Agostinis, P., Samali, A., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 3–22. [Google Scholar]
- Bergeron, J.J.; Brenner, M.B.; Thomas, D.Y.; Williams, D.B. Calnexin: A membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem. Sci. 1994, 19, 124–128. [Google Scholar] [CrossRef]
- Michalak, M.; Robert Parker, J.M.; Opas, M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium 2002, 32, 269–278. [Google Scholar] [CrossRef]
- Tanaka, N.; Mitsui, S.; Nobori, H.; Yanagi, K.; Komatsu, S. Expression and Function of Proteins during Development of the Basal Region in Rice Seedlings. Mol. Cell. Proteom. 2005, 4, 796–808. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Komatsu, S. Gel-free/label-free proteomic analysis of endoplasmic reticulum proteins in soybean root tips under flooding and drought stresses. J. Proteome Res. 2016, 15, 2211–2227. [Google Scholar] [CrossRef]
- Hashimoto, T.; Mustafa, G.; Nishiuchi, T.; Komatsu, S. Comparative analysis of the effect of inorganic and organic chemicals with silver nanoparticles on soybean under flooding stress. Int. J. Mol. Sci. 2020, 21, 1300. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Kobayashi, Y.; Nishizawa, K.; Nanjo, Y.; Furukawa, K. Comparative proteomics analysis of differentially expressed proteins in soybean cell wall during flooding stress. Amino Acids 2010, 39, 1435–1449. [Google Scholar] [CrossRef]
- Wang, X.; Sakata, K.; Komatsu, S. An integrated approach of proteomics and computational genetic modification effectiveness analysis to uncover the mechanisms of flood tolerance in soybeans. Int. J. Mol Sci. 2018, 19, 1301. [Google Scholar] [CrossRef] [Green Version]
- Mitch, W.E.; Goldberg, A.L. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N. Engl. J. Med. 1996, 335, 1897–1905. [Google Scholar] [CrossRef]
- Dielen, A.S.; Badaoui, S.; Candresse, T.; German-Retana, S. The ubiquitin/26S proteasome system in plant-pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef]
- Subbaiah, C.C.; Sachs, M.M. Molecular and cellular adaptations of maize to flooding stress. Ann. Bot. 2003, 91, 119–127. [Google Scholar] [CrossRef]
- Williams, B.; Verchot, J.; Dickman, M.B. When supply does not meet demand-ER stress and plant programmed cell death. Front. Plant Sci. 2014, 5, 211. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Han, C.; Nanjo, Y.; Altaf-Un-Nahar, M.; Wang, K.; He, D.; Yang, P. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013, 12, 4769–4784. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Li, X.; Rehman, S.U.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Yamaguchi, T.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Proteomic analysis of the effect of plant-derived smoke on soybean during recovery from flooding stress. J. Proteom. 2018, 181, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Komatsu, S.; Deschamps, T.; Hiraga, S.; Kato, M.; Chiba, M.; Hashiguchi, A.; Tougou, M.; Shimamura, S.; Yasue, H. Characterization of a novel flooding stress-responsive alcohol dehydrogenase expressed in soybean roots. Plant Mol. Biol. 2011, 77, 309–322. [Google Scholar] [CrossRef]
- Nouri, M.Z.; Hiraga, S.; Yanagawa, Y.; Sunohara, Y.; Matsumoto, H.; Komatsu, S. Characterization of calnexin in soybean roots and hypocotyls under osmotic stress. Phytochemistry 2012, 74, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Masuda, T.; Abe, K. Phosphorylation of a protein (pp56) is related to the regeneration of rice cultured suspension cells. Plant Cell Physiol. 1996, 37, 748–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacyn Baker, C.; Mock, N.M. An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 1994, 39, 7–12. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Côté, R.G.; Csordas, A.; Dianes, J.A.; Fabregat, A.; Foster, J.M.; Griss, J.; Alpi, E.; Birim, M.; Contell, J.; et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013, 41, D1063–D1069. [Google Scholar] [CrossRef]
- Okuda, S.; Watanabe, Y.; Moriya, Y.; Kawano, S.; Yamamoto, T.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Araki, N.; Yoshizawa, A.C.; et al. jPOSTTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017, 45, D1107–D1111. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komatsu, S.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Kono, Y.; Nishimura, M. Proteomic and Biochemical Analyses of the Mechanism of Tolerance in Mutant Soybean Responding to Flooding Stress. Int. J. Mol. Sci. 2021, 22, 9046. https://doi.org/10.3390/ijms22169046
Komatsu S, Yamaguchi H, Hitachi K, Tsuchida K, Kono Y, Nishimura M. Proteomic and Biochemical Analyses of the Mechanism of Tolerance in Mutant Soybean Responding to Flooding Stress. International Journal of Molecular Sciences. 2021; 22(16):9046. https://doi.org/10.3390/ijms22169046
Chicago/Turabian StyleKomatsu, Setsuko, Hisateru Yamaguchi, Keisuke Hitachi, Kunihiro Tsuchida, Yuhi Kono, and Minoru Nishimura. 2021. "Proteomic and Biochemical Analyses of the Mechanism of Tolerance in Mutant Soybean Responding to Flooding Stress" International Journal of Molecular Sciences 22, no. 16: 9046. https://doi.org/10.3390/ijms22169046
APA StyleKomatsu, S., Yamaguchi, H., Hitachi, K., Tsuchida, K., Kono, Y., & Nishimura, M. (2021). Proteomic and Biochemical Analyses of the Mechanism of Tolerance in Mutant Soybean Responding to Flooding Stress. International Journal of Molecular Sciences, 22(16), 9046. https://doi.org/10.3390/ijms22169046







