Multistep Signaling in Nature: A Close-Up of Geobacter Chemotaxis Sensing
Abstract
1. Introduction
2. The Topology of the Two-Component Systems
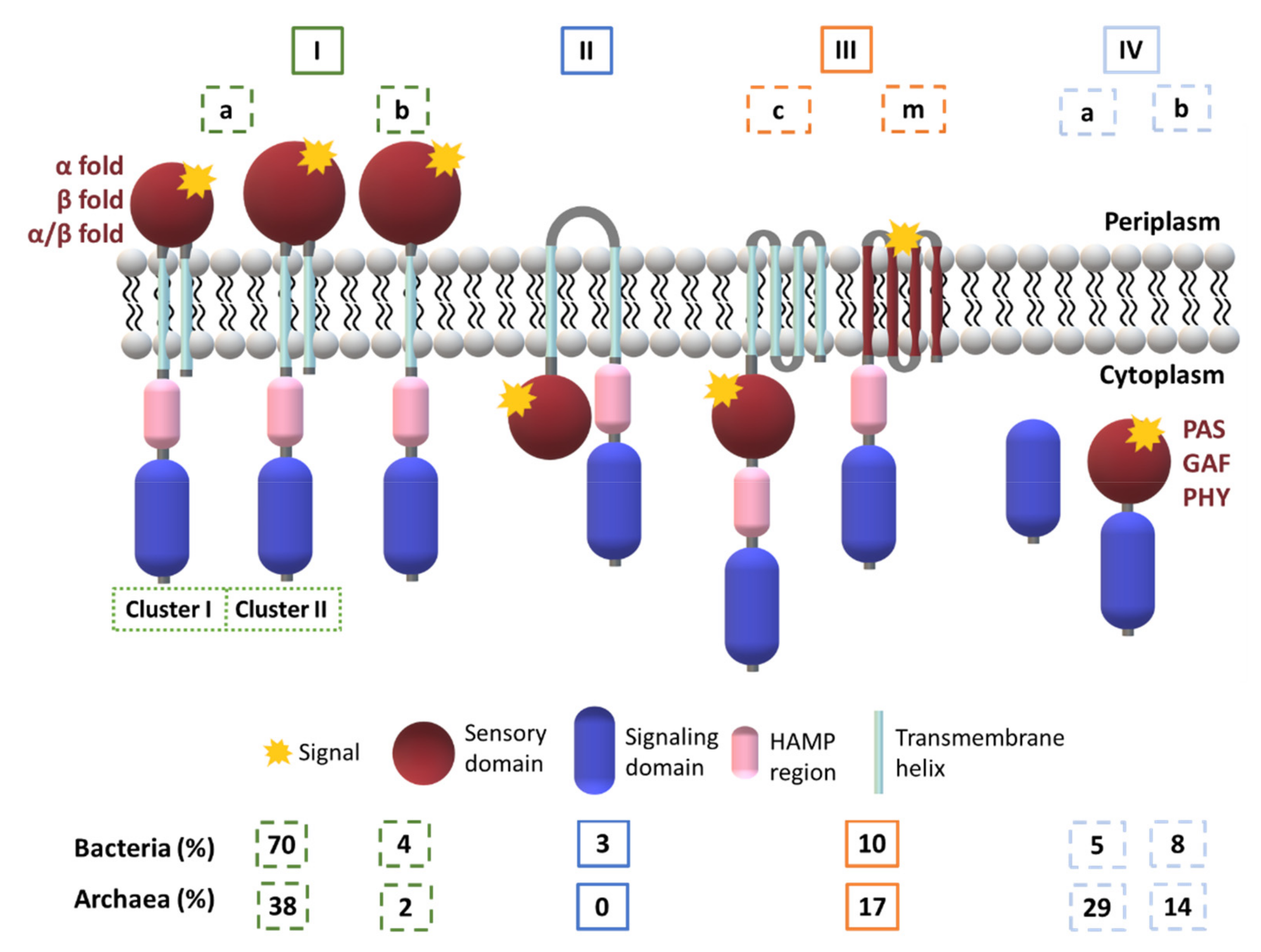
2.1. Diversity in the Sensor Domains
2.1.1. Cytoplasmic Sensor Domains
2.1.2. Membrane-Embedded Sensor Domains
2.1.3. Extracytoplasmic Sensor Domains
2.1.4. Heme-Based Sensors
b-Type Heme Sensor Domains
c-Type Heme Sensor Domains
2.2. Diversity in the Response Regulator Domains
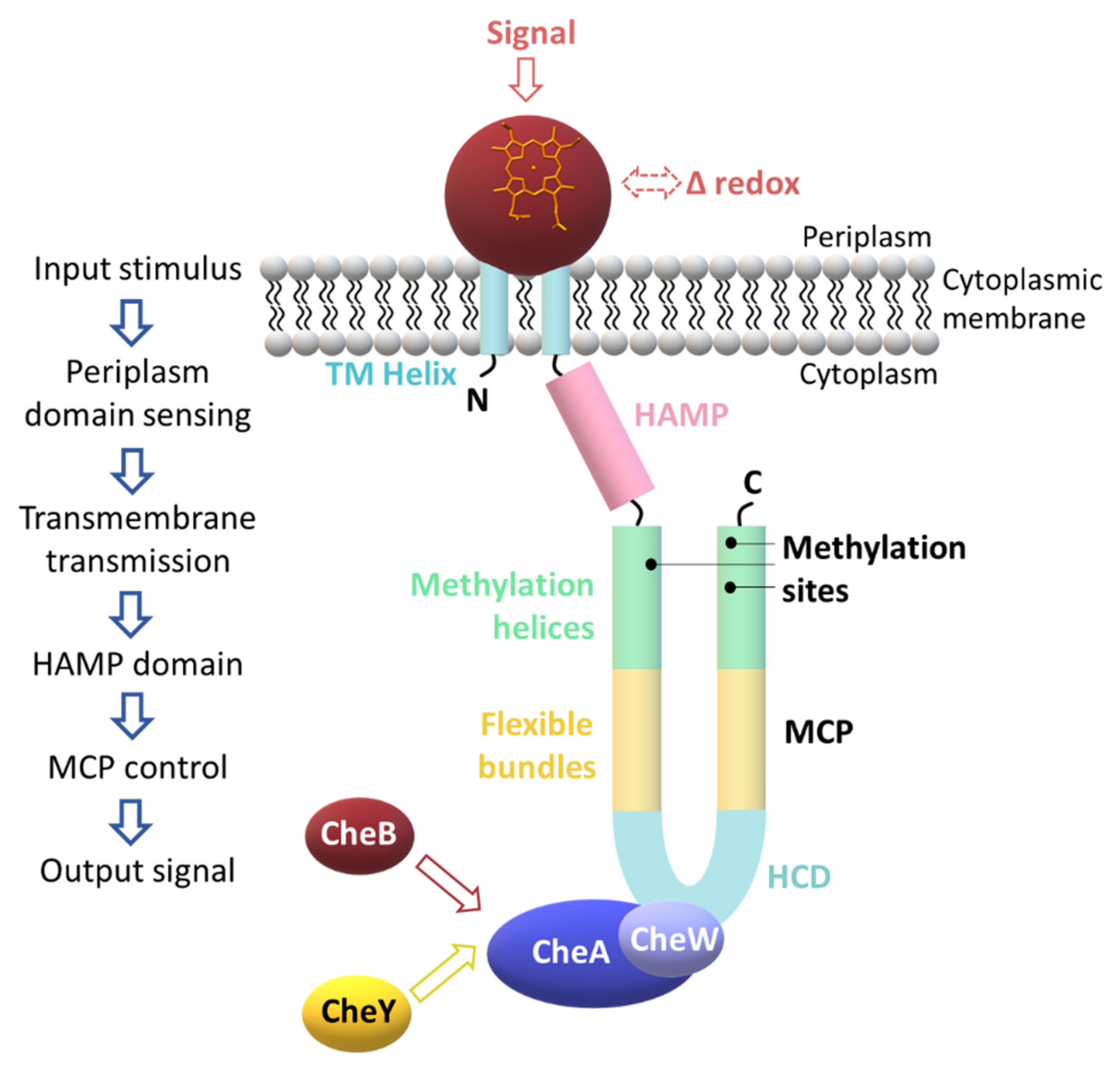
2.3. Signaling Mechanisms
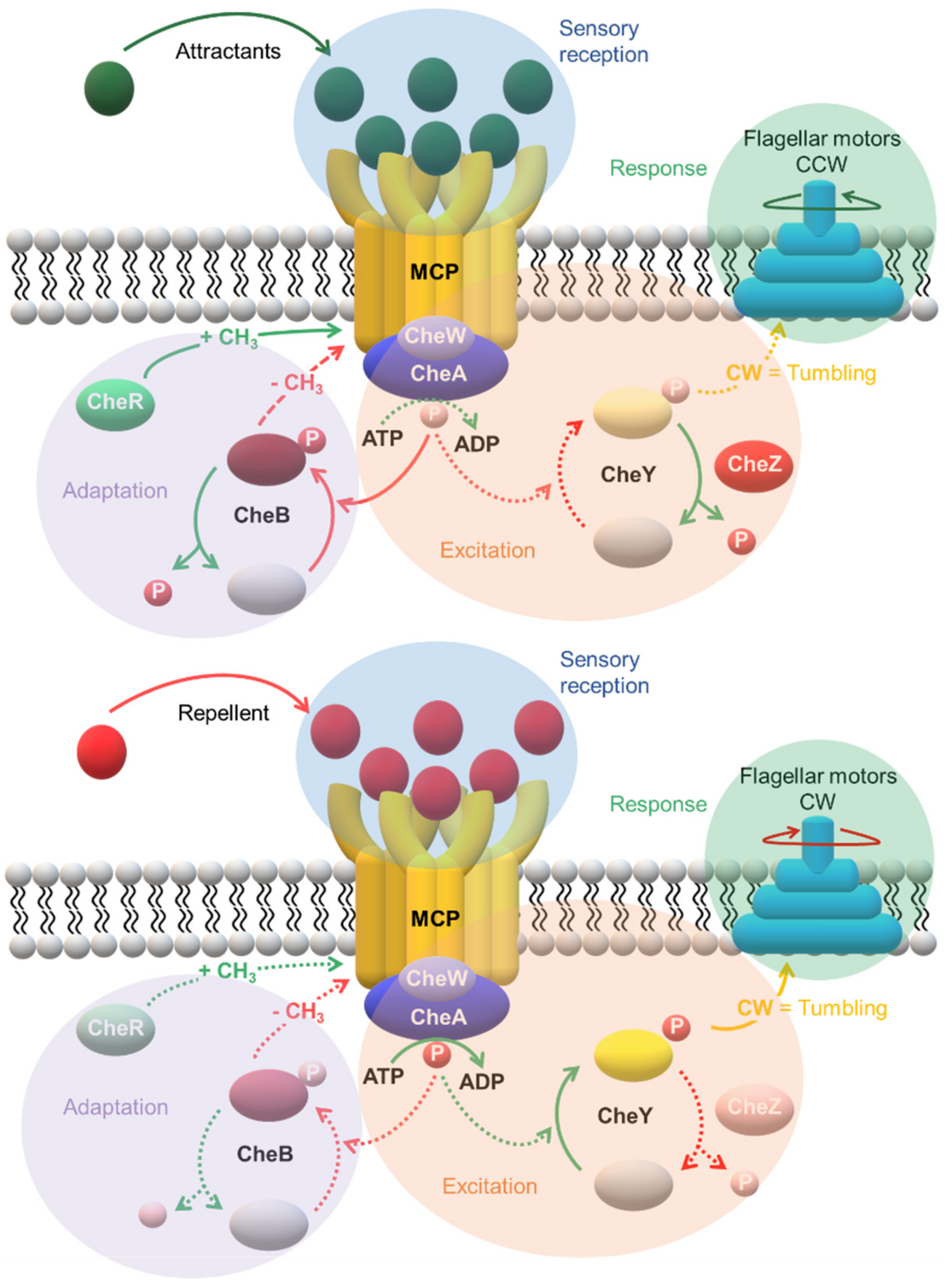
3. A Specialized Two-Component System for Chemotaxis
4. The Multiple Chemosensory Systems in Geobacter Bacteria
4.1. Topology of Heme MCP in G. sulfurreducens
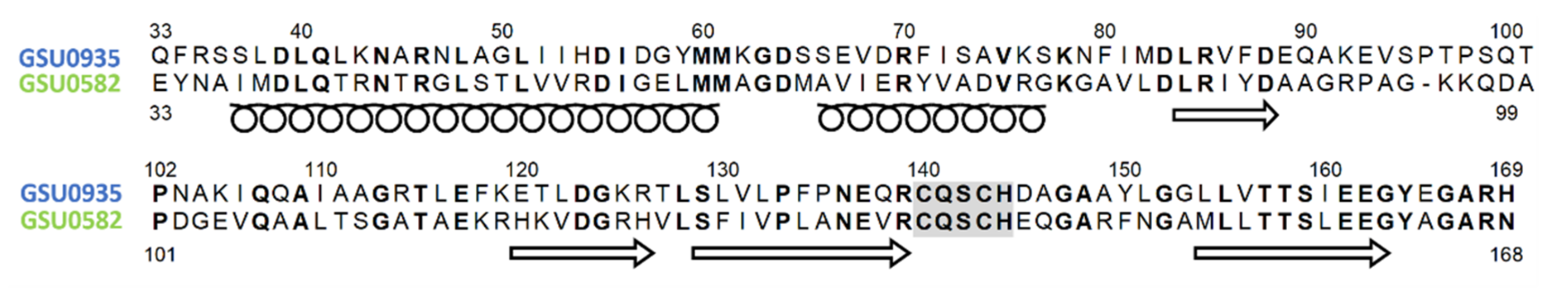
4.2. Structural Features of MCP Containing Heme Groups in G. sulfurreducens
4.3. Spectroscopic Features of Heme MCP in G. sulfurreducens
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Mizuno, T. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 1997, 4, 161–168. [Google Scholar] [CrossRef]
- Parkinson, J.S.; Kofoid, E.C. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 1992, 26, 71–112. [Google Scholar] [CrossRef]
- Hoch, J.A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000, 3, 165–170. [Google Scholar] [CrossRef]
- Wurgler-Murphy, S.M.; Saito, H. Two-component signal transducers and MAPK cascades. Trends Biochem. Sci. 1997, 22, 172–176. [Google Scholar] [CrossRef]
- Li, S.; Ault, A.; Malone, C.L.; Raitt, D.; Dean, S.; Johnston, L.H.; Deschenes, R.J.; Fassler, J.S. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998, 17, 6952–6962. [Google Scholar] [CrossRef] [PubMed]
- Srikantha, T.; Tsai, L.; Daniels, K.; Enger, L.; Highley, K.; Soll, D.R. The two-component hybrid kinase regulator CaNIK1 of Candida albicans. Microbiology 1998, 144, 2715–2729. [Google Scholar] [CrossRef]
- Calera, J.A.; Zhao, X.J.; Calderone, R. Defective hyphal development and avirulence caused by a deletion of the SSK1 response regulator gene in Candida albicans. Infect. Immun. 2000, 68, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Thomason, P.; Traynor, D.; Kay, R. Taking the plunge. Terminal differentiation in Dictyostelium. Trends Genet. 1999, 15, 15–19. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two-component systems in plant signal transduction. Trends Plant. Sci. 2000, 5, 67–74. [Google Scholar] [CrossRef]
- Capra, E.J.; Laub, M.T. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012, 66, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, P.M.; Thomason, P.A.; Stock, J.B. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002, 3, REVIEWS3013. [Google Scholar] [CrossRef]
- Gislason, A.S.; Choy, M.; Bloodworth, R.A.; Qu, W.; Stietz, M.S.; Li, X.; Zhang, C.; Cardona, S.T. Competitive Growth Enhances Conditional Growth Mutant Sensitivity to Antibiotics and Exposes a Two-Component System as an Emerging Antibacterial Target in Burkholderia cenocepacia. Antimicrob. Agents Chemother. 2017, 61, e00790-16. [Google Scholar] [CrossRef] [PubMed]
- West, A.H.; Stock, A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001, 26, 369–376. [Google Scholar] [CrossRef]
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef]
- Bijlsma, J.J.; Groisman, E.A. Making informed decisions: Regulatory interactions between two-component systems. Trends Microbiol. 2003, 11, 359–366. [Google Scholar] [CrossRef]
- Cheung, J.; Hendrickson, W.A. Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 2010, 13, 116–123. [Google Scholar] [CrossRef]
- Miller, M.B.; Skorupski, K.; Lenz, D.H.; Taylor, R.K.; Bassler, B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 2002, 110, 303–314. [Google Scholar] [CrossRef]
- Jiang, M.; Shao, W.; Perego, M.; Hoch, J.A. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000, 38, 535–542. [Google Scholar] [CrossRef]
- Paul, R.; Weiser, S.; Amiot, N.C.; Chan, C.; Schirmer, T.; Giese, B.; Jenal, U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004, 18, 715–727. [Google Scholar] [CrossRef]
- Falke, J.J.; Bass, R.B.; Butler, S.L.; Chervitz, S.A.; Danielson, M.A. The two-component signaling pathway of bacterial chemotaxis: A molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997, 13, 457–512. [Google Scholar] [CrossRef]
- Iuchi, S.; Weiner, L. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J. Biochem. 1996, 120, 1055–1063. [Google Scholar] [CrossRef]
- Unden, G.; Bongaerts, J. Alternative respiratory pathways of Escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1997, 1320, 217–234. [Google Scholar] [CrossRef]
- Hoch, J.A. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 1993, 47, 441–465. [Google Scholar] [CrossRef] [PubMed]
- Perego, M. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 1998, 6, 366–370. [Google Scholar] [CrossRef]
- Domian, I.J.; Quon, K.C.; Shapiro, L. The control of temporal and spatial organization during the Caulobacter cell cycle. Curr. Opin. Genet. Dev. 1996, 6, 538–544. [Google Scholar] [CrossRef]
- Wu, J.; Newton, A. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 1997, 24, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, H.B.; Plamann, L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol. Lett. 1996, 139, 89–95. [Google Scholar] [CrossRef]
- Ward, M.J.; Zusman, D.R. Regulation of directed motility in Myxococcus xanthus. Mol. Microbiol. 1997, 24, 885–893. [Google Scholar] [CrossRef]
- Krell, T.; Lacal, J.; Busch, A.; Silva-Jimenez, H.; Guazzaroni, M.E.; Ramos, J.L. Bacterial sensor kinases: Diversity in the recognition of environmental signals. Annu. Rev. Microbiol. 2010, 64, 539–559. [Google Scholar] [CrossRef]
- Taylor, B.L.; Zhulin, I.B. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Hefti, M.H.; Francoijs, K.J.; de Vries, S.C.; Dixon, R.; Vervoort, J. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 2004, 271, 1198–1208. [Google Scholar] [CrossRef]
- Scheu, P.D.; Kim, O.B.; Griesinger, C.; Unden, G. Sensing by the membrane-bound sensor kinase DcuS: Exogenous versus endogenous sensing of C(4)-dicarboxylates in bacteria. Future Microbiol. 2010, 5, 1383–1402. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016, 12, e1004862. [Google Scholar] [CrossRef]
- Chang, C.; Tesar, C.; Gu, M.; Babnigg, G.; Joachimiak, A.; Pokkuluri, P.R.; Szurmant, H.; Schiffer, M. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J. Bacteriol. 2010, 192, 1156–1159. [Google Scholar] [CrossRef] [PubMed]
- Moglich, A.; Ayers, R.A.; Moffat, K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 2009, 17, 1282–1294. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Ferrin, T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007, 157, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Burden, L.M.; Hurley, J.H. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000, 19, 5288–5299. [Google Scholar] [CrossRef]
- Yang, X.; Kuk, J.; Moffat, K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. USA 2008, 105, 14715–14720. [Google Scholar] [CrossRef]
- Yang, X.; Kuk, J.; Moffat, K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. USA 2009, 106, 15639–15644. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Zhang, J.; Brunzelle, J.S.; Vierstra, R.D.; Forest, K.T. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 2007, 282, 12298–12309. [Google Scholar] [CrossRef]
- Yang, X.; Stojkovic, E.A.; Kuk, J.; Moffat, K. Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc. Natl. Acad. Sci. USA 2007, 104, 12571–12576. [Google Scholar] [CrossRef] [PubMed]
- Essen, L.-O.; Mailliet, J.; Hughes, J. The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. USA 2008, 105, 14709–14714. [Google Scholar] [CrossRef]
- Albanesi, D.; Martin, M.; Trajtenberg, F.; Mansilla, M.C.; Haouz, A.; Alzari, P.M.; de Mendoza, D.; Buschiazzo, A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. USA 2009, 106, 16185–16190. [Google Scholar] [CrossRef]
- Martin, M.; Albanesi, D.; Alzari, P.M.; de Mendoza, D. Functional in vitro assembly of the integral membrane bacterial thermosensor DesK. Protein Expr. Purif. 2009, 66, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Bogel, G.; Schrempf, H.; de Orue Lucana, D.O. The heme-binding protein HbpS regulates the activity of the Streptomyces reticuli iron-sensing histidine kinase SenS in a redox-dependent manner. Amino Acids 2009, 37, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Voet-van-Vormizeele, J.; Groth, G. Ethylene controls autophosphorylation of the histidine kinase domain in ethylene receptor ETR1. Mol. Plant 2008, 1, 380–387. [Google Scholar] [CrossRef][Green Version]
- Geisinger, E.; George, E.A.; Chen, J.; Muir, T.W.; Novick, R.P. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J. Biol. Chem. 2008, 283, 8930–8938. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.O.; Winzer, K.; Clarke, S.R.; Chan, W.C.; Williams, P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 2008, 381, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Gordeliy, V.I.; Labahn, J.; Moukhametzianov, R.; Efremov, R.; Granzin, J.; Schlesinger, R.; Buldt, G.; Savopol, T.; Scheidig, A.J.; Klare, J.P.; et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 2002, 419, 484–487. [Google Scholar] [CrossRef]
- Lacal, J.; Garcia-Fontana, C.; Munoz-Martinez, F.; Ramos, J.L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Hendrickson, W.A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265. [Google Scholar] [CrossRef]
- Gao, R.; Stock, A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009, 63, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.J.; Ulrich, L.E.; Zhulin, I.B. The NIT domain: A predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 2003, 28, 121–124. [Google Scholar] [CrossRef]
- Emami, K.; Topakas, E.; Nagy, T.; Henshaw, J.; Jackson, K.A.; Nelson, K.E.; Mongodin, E.F.; Murray, J.W.; Lewis, R.J.; Gilbert, H.J. Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J. Biol. Chem. 2009, 284, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Nan, B.; Nan, J.; Ma, Q.; Panjikar, S.; Liang, Y.H.; Wang, Y.; Su, X.D. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008, 383, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Yeh, J.I.; Biemann, H.P.; Prive, G.G.; Pandit, J.; Koshland, D.E., Jr.; Kim, S.H. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J. Mol. Biol. 1996, 262, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Aravind, L. Cache—A signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 2000, 25, 535–537. [Google Scholar] [CrossRef]
- Girvan, H.M.; Munro, A.W. Heme sensor proteins. J. Biol. Chem. 2013, 288, 13194–13203. [Google Scholar] [CrossRef]
- Farhana, A.; Saini, V.; Kumar, A.; Lancaster, J.R., Jr.; Steyn, A.J. Environmental heme-based sensor proteins: Implications for understanding bacterial pathogenesis. Antioxid. Redox. Signal. 2012, 17, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Gonzalez, M.A.; Gonzalez, G. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 2004, 96, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Gonzalez, M.A.; Gonzalez, G.; Perutz, M.F.; Kiger, L.; Marden, M.C.; Poyart, C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 1994, 33, 8067–8073. [Google Scholar] [CrossRef]
- Tomita, T.; Gonzalez, G.; Chang, A.L.; Ikeda-Saito, M.; Gilles-Gonzalez, M.A. A comparative resonance Raman analysis of heme-binding PAS domains: Heme iron coordination structures of the BjFixL, AxPDEA1, EcDos, and MtDos proteins. Biochemistry 2002, 41, 4819–4826. [Google Scholar] [CrossRef]
- Gilles-Gonzalez, M.A.; Gonzalez, G. Heme-based sensors: Defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 2005, 99, 1–22. [Google Scholar] [CrossRef]
- Crosson, S.; McGrath, P.T.; Stephens, C.; McAdams, H.H.; Shapiro, L. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl. Acad. Sci. USA 2005, 102, 8018–8023. [Google Scholar] [CrossRef]
- David, M.; Daveran, M.L.; Batut, J.; Dedieu, A.; Domergue, O.; Ghai, J.; Hertig, C.; Boistard, P.; Kahn, D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 1988, 54, 671–683. [Google Scholar] [CrossRef]
- Rey, F.E.; Harwood, C.S. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol. Microbiol. 2010, 75, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Gilles-Gonzalez, M.A.; Ditta, G.S.; Helinski, D.R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 1991, 350, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Nixon, V.M.; Gonzalez, G.; Gilles-Gonzalez, M.A. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 2000, 39, 2685–2691. [Google Scholar] [CrossRef]
- Tanaka, A.; Takahashi, H.; Shimizu, T. Critical role of the heme axial ligand, Met95, in locking catalysis of the phosphodiesterase from Escherichia coli (Ec DOS) toward Cyclic diGMP. J. Biol. Chem. 2007, 282, 21301–21307. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Sousa, E.H.; Wan, X.; Saito, J.A.; Alam, M.; Gilles-Gonzalez, M.A. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 2009, 48, 9764–9774. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Cho, H.J.; Kim, Y.M.; Oh, J.I.; Kang, B.S. Structural insight into the heme-based redox sensing by DosS from Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 13057–13067. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Ioanoviciu, A.; Ortiz de Montellano, P.R. 2.3 A X-ray structure of the heme-bound GAF domain of sensory histidine kinase DosT of Mycobacterium tuberculosis. Biochemistry 2008, 47, 12523–12531. [Google Scholar] [CrossRef] [PubMed]
- Molitor, B.; Stassen, M.; Modi, A.; El-Mashtoly, S.F.; Laurich, C.; Lubitz, W.; Dawson, J.H.; Rother, M.; Frankenberg-Dinkel, N. A heme-based redox sensor in the methanogenic archaeon Methanosarcina acetivorans. J. Biol. Chem. 2013, 288, 18458–18472. [Google Scholar] [CrossRef]
- Motomura, T.; Suga, M.; Hienerwadel, R.; Nakagawa, A.; Lai, T.L.; Nitschke, W.; Kuma, T.; Sugiura, M.; Boussac, A.; Shen, J.R. Crystal structure and redox properties of a novel cyanobacterial heme protein with a His/Cys heme axial ligation and a Per-Arnt-Sim (PAS)-like domain. J. Biol. Chem. 2017, 292, 9599–9612. [Google Scholar] [CrossRef]
- Yoshioka, S.; Kobayashi, K.; Yoshimura, H.; Uchida, T.; Kitagawa, T.; Aono, S. Biophysical properties of a c-type heme in chemotaxis signal transducer protein DcrA. Biochemistry 2005, 44, 15406–15413. [Google Scholar] [CrossRef]
- Pokkuluri, P.R.; Pessanha, M.; Londer, Y.Y.; Wood, S.J.; Duke, N.E.; Wilton, R.; Catarino, T.; Salgueiro, C.A.; Schiffer, M. Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: Implications for signal transduction. J. Mol. Biol. 2008, 377, 1498–1517. [Google Scholar] [CrossRef]
- Londer, Y.Y.; Dementieva, I.S.; D’Ausilio, C.A.; Pokkuluri, P.R.; Schiffer, M. Characterization of a c-type heme-containing PAS sensor domain from Geobacter sulfurreducens representing a novel family of periplasmic sensors in Geobacteraceae and other bacteria. FEMS Microbiol. Lett. 2006, 258, 173–181. [Google Scholar] [CrossRef][Green Version]
- Saito, H. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem. Rev. 2001, 101, 2497–2509. [Google Scholar] [CrossRef]
- Martinez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Milani, M.; Leoni, L.; Rampioni, G.; Zennaro, E.; Ascenzi, P.; Bolognesi, M. An active-like structure in the unphosphorylated StyR response regulator suggests a phosphorylation- dependent allosteric activation mechanism. Structure 2005, 13, 1289–1297. [Google Scholar] [CrossRef]
- Batchelor, J.D.; Doucleff, M.; Lee, C.J.; Matsubara, K.; De Carlo, S.; Heideker, J.; Lamers, M.H.; Pelton, J.G.; Wemmer, D.E. Structure and regulatory mechanism of Aquifex aeolicus NtrC4: Variability and evolution in bacterial transcriptional regulation. J. Mol. Biol. 2008, 384, 1058–1075. [Google Scholar] [CrossRef]
- Sidote, D.J.; Barbieri, C.M.; Wu, T.; Stock, A.M. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure 2008, 16, 727–735. [Google Scholar] [CrossRef]
- O’Hara, B.P.; Norman, R.A.; Wan, P.T.; Roe, S.M.; Barrett, T.E.; Drew, R.E.; Pearl, L.H. Crystal structure and induction mechanism of AmiC-AmiR: A ligand-regulated transcription antitermination complex. EMBO J. 1999, 18, 5175–5186. [Google Scholar] [CrossRef]
- Guhaniyogi, J.; Wu, T.; Patel, S.S.; Stock, A.M. Interaction of CheY with the C-terminal peptide of CheZ. J. Bacteriol. 2008, 190, 1419–1428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okamura, H.; Hanaoka, S.; Nagadoi, A.; Makino, K.; Nishimura, Y. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J. Mol. Biol. 2000, 295, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.S.A.; Saeki, A.; Hikima, T.; Nishizono, Y.; Hisano, T.; Kamaya, M.; Nukina, K.; Nishitani, H.; Nakamura, H.; Yamamoto, M.; et al. Architecture of the complete oxygen-sensing FixL-FixJ two-component signal transduction system. Sci. Signal. 2018, 11, eaaq0825. [Google Scholar] [CrossRef]
- Boudes, M.; Lazar, N.; Graille, M.; Durand, D.; Gaidenko, T.A.; Stewart, V.; van Tilbeurgh, H. The structure of the NasR transcription antiterminator reveals a one-component system with a NIT nitrate receptor coupled to an ANTAR RNA-binding effector. Mol. Microbiol. 2012, 85, 431–444. [Google Scholar] [CrossRef] [PubMed]
- West, A.H.; Martinez-Hackert, E.; Stock, A.M. Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB. J. Mol. Biol. 1995, 250, 276–290. [Google Scholar] [CrossRef] [PubMed]
- Tabib-Salazar, A.; Liu, B.; Barker, D.; Burchell, L.; Qimron, U.; Matthews, S.J.; Wigneshweraraj, S. T7 phage factor required for managing RpoS in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, E5353–E5362. [Google Scholar] [CrossRef]
- Sevvana, M.; Vijayan, V.; Zweckstetter, M.; Reinelt, S.; Madden, D.R.; Herbst-Irmer, R.; Sheldrick, G.M.; Bott, M.; Griesinger, C.; Becker, S. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J. Mol. Biol. 2008, 377, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Hendrickson, W.A. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure 2009, 17, 190–201. [Google Scholar] [CrossRef]
- Cheung, J.; Le-Khac, M.; Hendrickson, W.A. Crystal structure of a histidine kinase sensor domain with similarity to periplasmic binding proteins. Proteins 2009, 77, 235–241. [Google Scholar] [CrossRef]
- Etzkorn, M.; Kneuper, H.; Dunnwald, P.; Vijayan, V.; Kramer, J.; Griesinger, C.; Becker, S.; Unden, G.; Baldus, M. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat. Struct. Mol. Biol. 2008, 15, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.O.; Hendrickson, W.A. Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure 2009, 17, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Neiditch, M.B.; Federle, M.J.; Pompeani, A.J.; Kelly, R.C.; Swem, D.L.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 2006, 126, 1095–1108. [Google Scholar] [CrossRef]
- Ward, S.M.; Delgado, A.; Gunsalus, R.P.; Manson, M.D. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 2002, 44, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Phadtare, S.; Inouye, M. The design and development of Tar-EnvZ chimeric receptors. Methods Enzymol. 2007, 423, 166–183. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Bingman, C.A.; Reyngold, M.; Hendrickson, W.A.; Waldburger, C.D. Crystal structure of a functional dimer of the PhoQ sensor domain. J. Biol. Chem. 2008, 283, 13762–13770. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.D.; Soto, C.S.; Waldburger, C.D.; Degrado, W.F. Determination of the physiological dimer interface of the PhoQ sensor domain. J. Mol. Biol. 2008, 379, 656–665. [Google Scholar] [CrossRef]
- Hulko, M.; Berndt, F.; Gruber, M.; Linder, J.U.; Truffault, V.; Schultz, A.; Martin, J.; Schultz, J.E.; Lupas, A.N.; Coles, M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 2006, 126, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Tomchick, D.R.; Brautigam, C.A.; Machius, M.; Kort, R.; Hellingwerf, K.J.; Gardner, K.H. Changes at the KinA PAS-A dimerization interface influence histidine kinase function. Biochemistry 2008, 47, 4051–4064. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Hao, B.; Mansy, S.S.; Gonzalez, G.; Gilles-Gonzalez, M.A.; Chan, M.K. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. USA 1998, 95, 15177–15182. [Google Scholar] [CrossRef] [PubMed]
- Hao, B.; Isaza, C.; Arndt, J.; Soltis, M.; Chan, M.K. Structure-based mechanism of O2 sensing and ligand discrimination by the FixL heme domain of Bradyrhizobium japonicum. Biochemistry 2002, 41, 12952–12958. [Google Scholar] [CrossRef]
- Lee, J.M.; Cho, H.Y.; Cho, H.J.; Ko, I.J.; Park, S.W.; Baik, H.S.; Oh, J.H.; Eom, C.Y.; Kim, Y.M.; Kang, B.S.; et al. O2- and NO-sensing mechanism through the DevSR two-component system in Mycobacterium smegmatis. J. Bacteriol. 2008, 190, 6795–6804. [Google Scholar] [CrossRef] [PubMed]
- Anantharaman, V.; Balaji, S.; Aravind, L. The signaling helix: A common functional theme in diverse signaling proteins. Biol. Direct. 2006, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Silverman, M.; Simon, M. Chemotaxis in Escherichia coli: Methylation of che gene products. Proc. Natl. Acad. Sci. USA 1977, 74, 3317–3321. [Google Scholar] [CrossRef] [PubMed]
- Macnab, R.M.; Koshland, D.E., Jr. The gradient-sensing mechanism in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 1972, 69, 2509–2512. [Google Scholar] [CrossRef]
- Adler, J.; Dahl, M.M. A method for measuring the motility of bacteria and for comparing random and non-random motility. J. Gen. Microbiol. 1967, 46, 161–173. [Google Scholar] [CrossRef]
- Faguy, D.M.; Jarrell, K.F. A twisted tale: The origin and evolution of motility and chemotaxis in prokaryotes. Microbiology 1999, 145, 279–281. [Google Scholar] [CrossRef][Green Version]
- Kort, E.N.; Goy, M.F.; Larsen, S.H.; Adler, J. Methylation of a membrane protein involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 1975, 72, 3939–3943. [Google Scholar] [CrossRef]
- Bibikov, S.I.; Biran, R.; Rudd, K.E.; Parkinson, J.S. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 1997, 179, 4075–4079. [Google Scholar] [CrossRef] [PubMed]
- Rebbapragada, A.; Johnson, M.S.; Harding, G.P.; Zuccarelli, A.J.; Fletcher, H.M.; Zhulin, I.B.; Taylor, B.L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. USA 1997, 94, 10541–10546. [Google Scholar] [CrossRef] [PubMed]
- Kleene, S.J.; Toews, M.L.; Adler, J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J. Biol. Chem. 1977, 252, 3214–3218. [Google Scholar] [CrossRef]
- Kleene, S.J.; Hobson, A.C.; Adler, J. Attractants and repellents influence methylation and demethylation of methyl-accepting chemotaxis proteins in an extract of Escherichia coli. Proc. Natl. Acad. Sci. USA 1979, 76, 6309–6313. [Google Scholar] [CrossRef]
- Alexander, R.P.; Zhulin, I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 2007, 104, 2885–2890. [Google Scholar] [CrossRef]
- Methe, B.A.; Nelson, K.E.; Eisen, J.A.; Paulsen, I.T.; Nelson, W.; Heidelberg, J.F.; Wu, D.; Wu, M.; Ward, N.; Beanan, M.J.; et al. Genome of Geobacter sulfurreducens: Metal reduction in subsurface environments. Science 2003, 302, 1967–1969. [Google Scholar] [CrossRef]
- Milburn, M.V.; Prive, G.G.; Milligan, D.L.; Scott, W.G.; Yeh, J.; Jancarik, J.; Koshland, D.E., Jr.; Kim, S.H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 1991, 254, 1342–1347. [Google Scholar] [CrossRef]
- Tran, H.T.; Krushkal, J.; Antommattei, F.M.; Lovley, D.R.; Weis, R.M. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genom. 2008, 9, 471. [Google Scholar] [CrossRef]
- Zhulin, I.B. The superfamily of chemotaxis transducers: From physiology to genomics and back. Adv. Microb. Physiol. 2001, 45, 157–198. [Google Scholar] [CrossRef]
- Wuichet, K.; Alexander, R.P.; Zhulin, I.B. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007, 422, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Hazelbauer, G.L.; Falke, J.J.; Parkinson, J.S. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 2008, 33, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Khursigara, C.M.; Wu, X.; Zhang, P.; Lefman, J.; Subramaniam, S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc. Natl. Acad. Sci. USA 2008, 105, 16555–16560. [Google Scholar] [CrossRef] [PubMed]
- Zhulin, I.B. A novel phototaxis receptor hidden in the cyanobacterial genome. J. Mol. Microbiol. Biotechnol. 2000, 2, 491–493. [Google Scholar] [PubMed]
- Wuichet, K.; Zhulin, I.B. Molecular evolution of sensory domains in cyanobacterial chemoreceptors. Trends Microbiol. 2003, 11, 200–203. [Google Scholar] [CrossRef]
- Anand, G.S.; Goudreau, P.N.; Stock, A.M. Activation of methylesterase CheB: Evidence of a dual role for the regulatory domain. Biochemistry 1998, 37, 14038–14047. [Google Scholar] [CrossRef]
- Hess, J.F.; Oosawa, K.; Kaplan, N.; Simon, M.I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 1988, 53, 79–87. [Google Scholar] [CrossRef]
- Kato, J.; Nakamura, T.; Kuroda, A.; Ohtake, H. Cloning and characterization of chemotaxis genes in Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 1999, 63, 155–161. [Google Scholar] [CrossRef]
- Manson, M.D. The tie that binds the dynamic duo: The connector between AS1 and AS2 in the HAMP domain of the Escherichia coli Tsr chemoreceptor. J. Bacteriol. 2008, 190, 6544–6547. [Google Scholar] [CrossRef]
- Welch, M.; Oosawa, K.; Aizawa, S.; Eisenbach, M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 1993, 90, 8787–8791. [Google Scholar] [CrossRef]
- Toker, A.S.; Macnab, R.M. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 1997, 273, 623–634. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, M.M.; Bren, A.; Eisenbach, M.; Dahlquist, F.W. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein fliM. J. Mol. Biol. 1999, 289, 1423–1433. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R.; Holmes, D.E.; Nevin, K.P. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 2004, 49, 219–286. [Google Scholar] [CrossRef] [PubMed]
- Nevin, K.P.; Kim, B.C.; Glaven, R.H.; Johnson, J.P.; Woodard, T.L.; Methe, B.A.; Didonato, R.J.; Covalla, S.F.; Franks, A.E.; Liu, A.; et al. Anode biofilm transcriptomics reveals outer surface components essential for high density current production in Geobacter sulfurreducens fuel cells. PLoS ONE 2009, 4, e5628. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Bug juice: Harvesting electricity with microorganisms. Nat. Rev. Microbiol. 2006, 4, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Lovley, D.R. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat. Rev. Microbiol. 2003, 1, 35–44. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Zhou, S.; Liu, Y. Genome sequence of a dissimilatory Fe(III)-reducing bacterium Geobacter soli type strain GSS01(T). Stand. Genom. Sci. 2015, 10, 118. [Google Scholar] [CrossRef]
- Coppi, M.V.; Leang, C.; Sandler, S.J.; Lovley, D.R. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 2001, 67, 3180–3187. [Google Scholar] [CrossRef]
- Butler, J.E.; Young, N.D.; Aklujkar, M.; Lovley, D.R. Comparative genomic analysis of Geobacter sulfurreducens KN400, a strain with enhanced capacity for extracellular electron transfer and electricity production. BMC Genom. 2012, 13, 471. [Google Scholar] [CrossRef]
- Aklujkar, M.; Krushkal, J.; DiBartolo, G.; Lapidus, A.; Land, M.L.; Lovley, D.R. The genome sequence of Geobacter metallireducens: Features of metabolism, physiology and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 2009, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc 2009, 4, 363–371. [Google Scholar] [CrossRef]
- Catarino, T.; Pessanha, M.; De Candia, A.G.; Gouveia, Z.; Fernandes, A.P.; Pokkuluri, P.R.; Murgida, D.; Marti, M.A.; Todorovic, S.; Salgueiro, C.A. Probing the chemotaxis periplasmic sensor domains from Geobacter sulfurreducens by combined resonance Raman and molecular dynamic approaches: NO and CO sensing. J. Phys. Chem. B 2010, 114, 11251–11260. [Google Scholar] [CrossRef]
- Mao, J.; Hauser, K.; Gunner, M.R. How cytochromes with different folds control heme redox potentials. Biochemistry 2003, 42, 9829–9840. [Google Scholar] [CrossRef] [PubMed]
- Sourjik, V.; Berg, H.C. Functional interactions between receptors in bacterial chemotaxis. Nature 2004, 428, 437–441. [Google Scholar] [CrossRef]
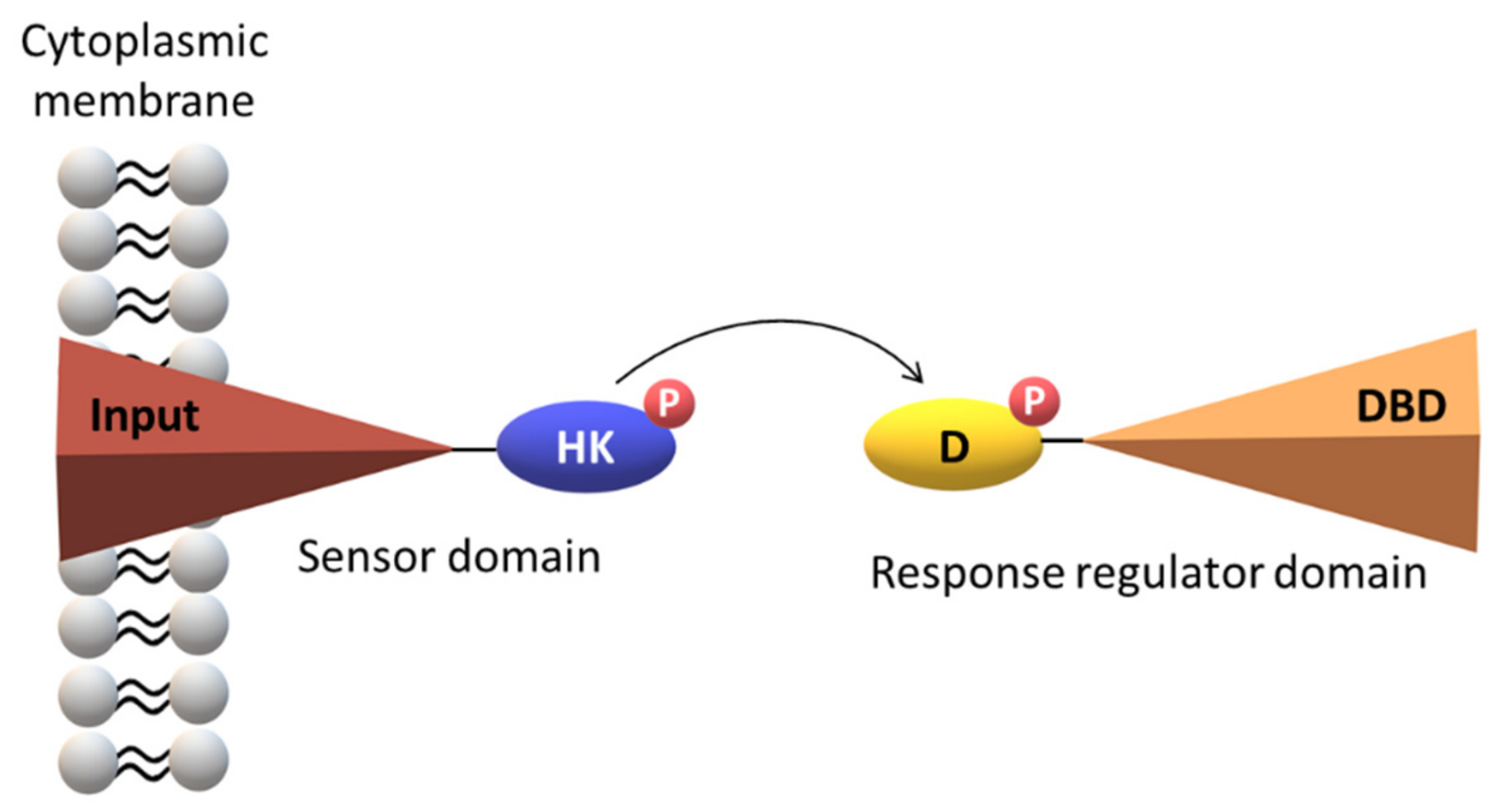
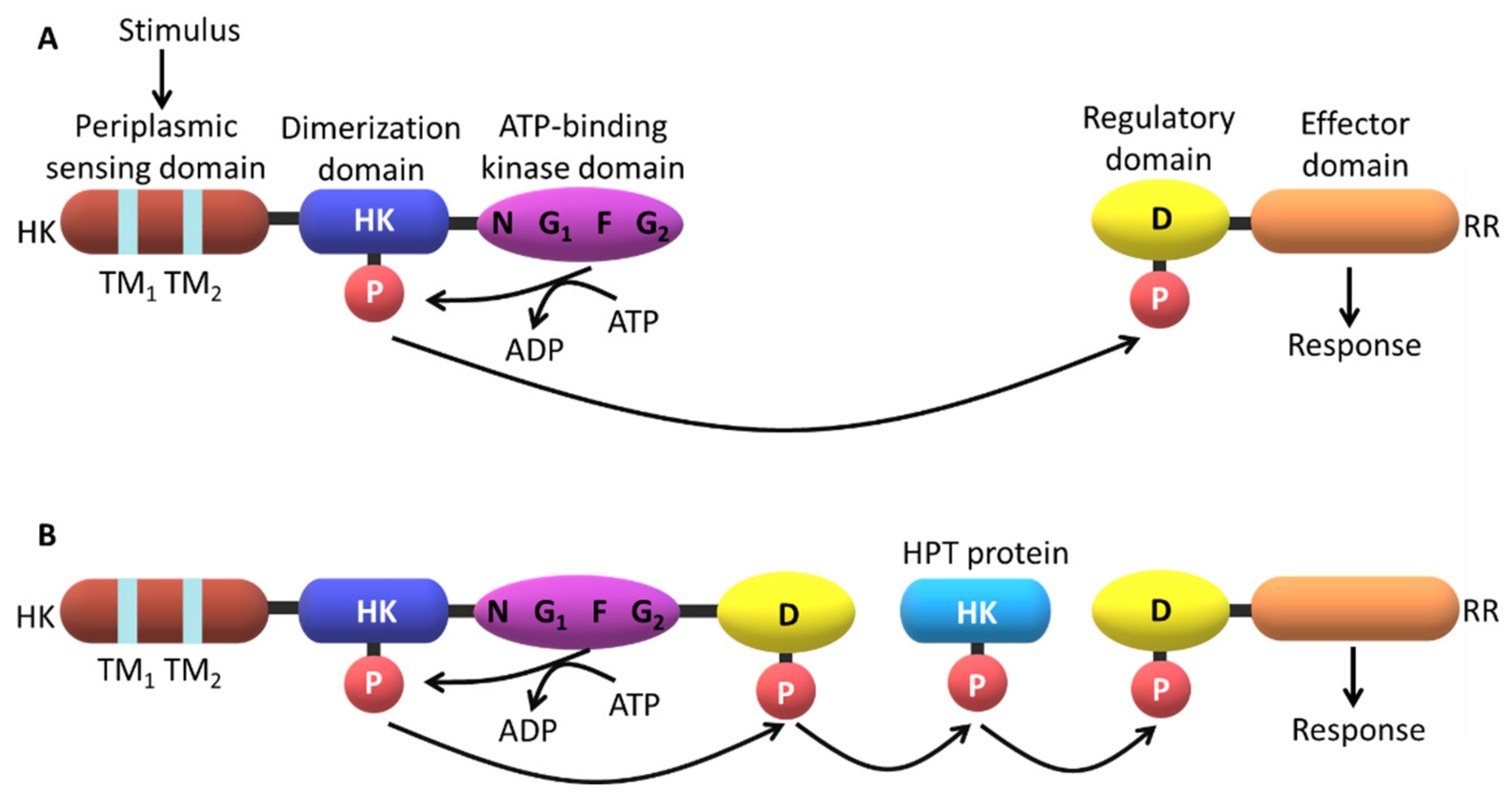
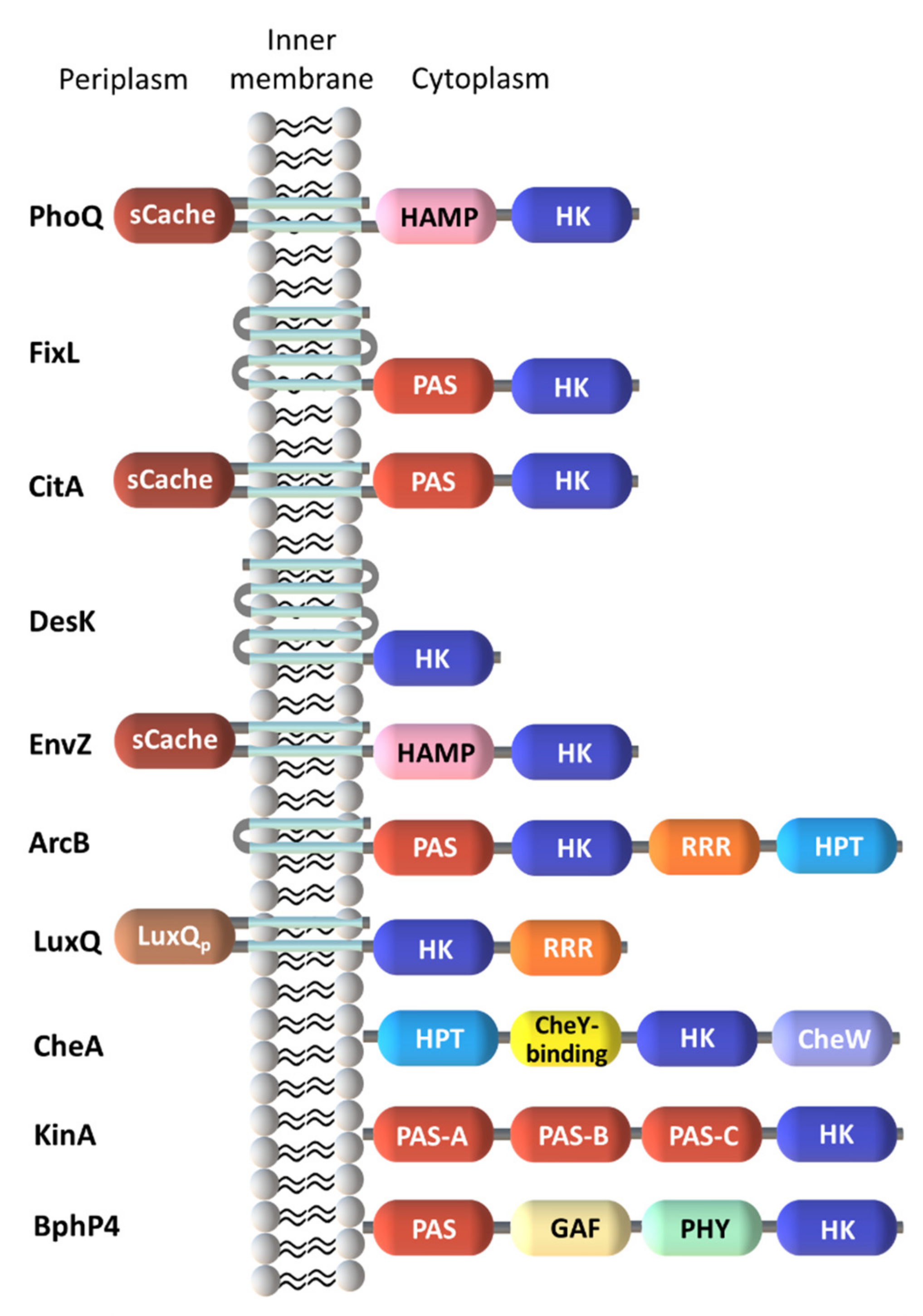
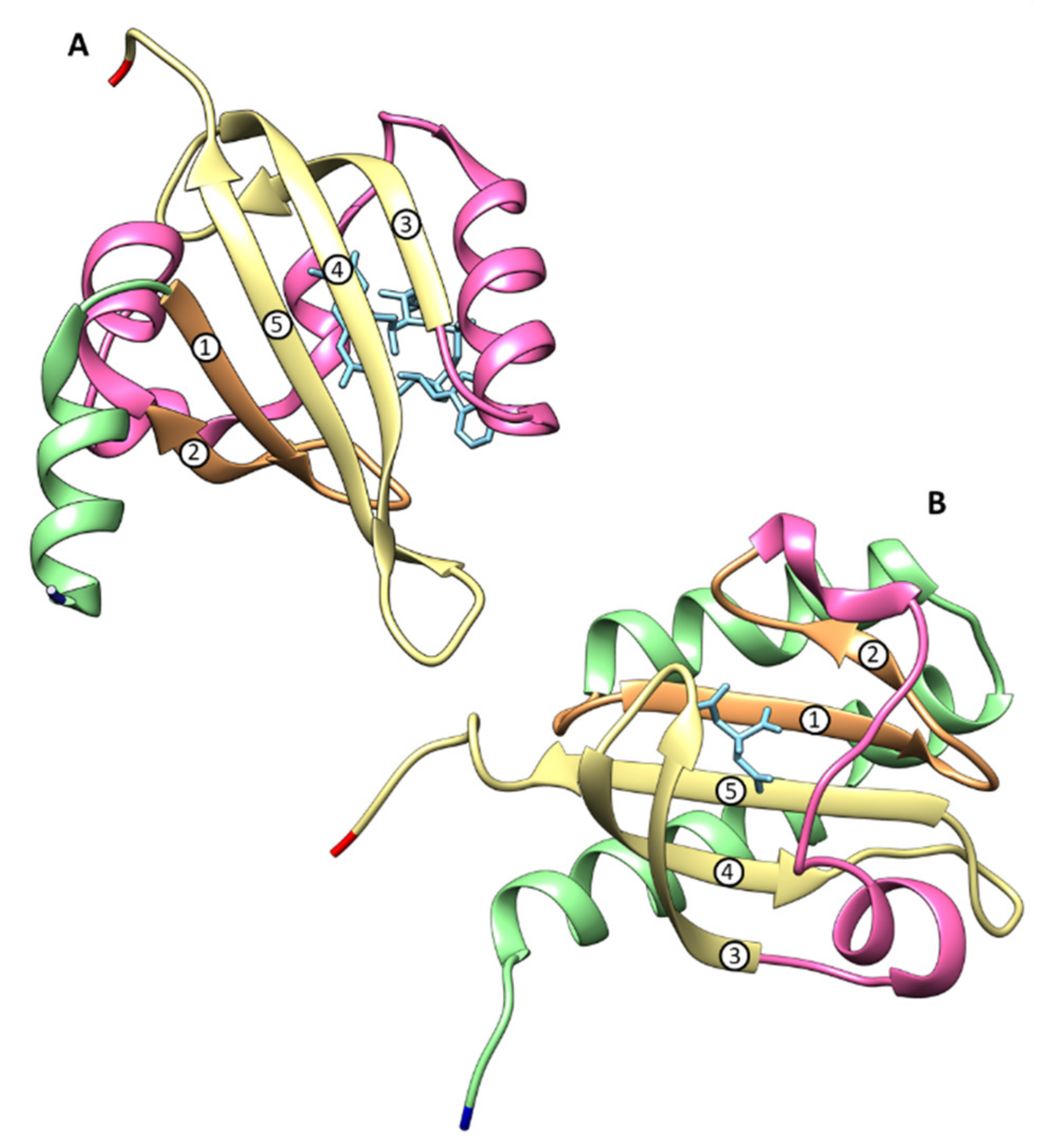
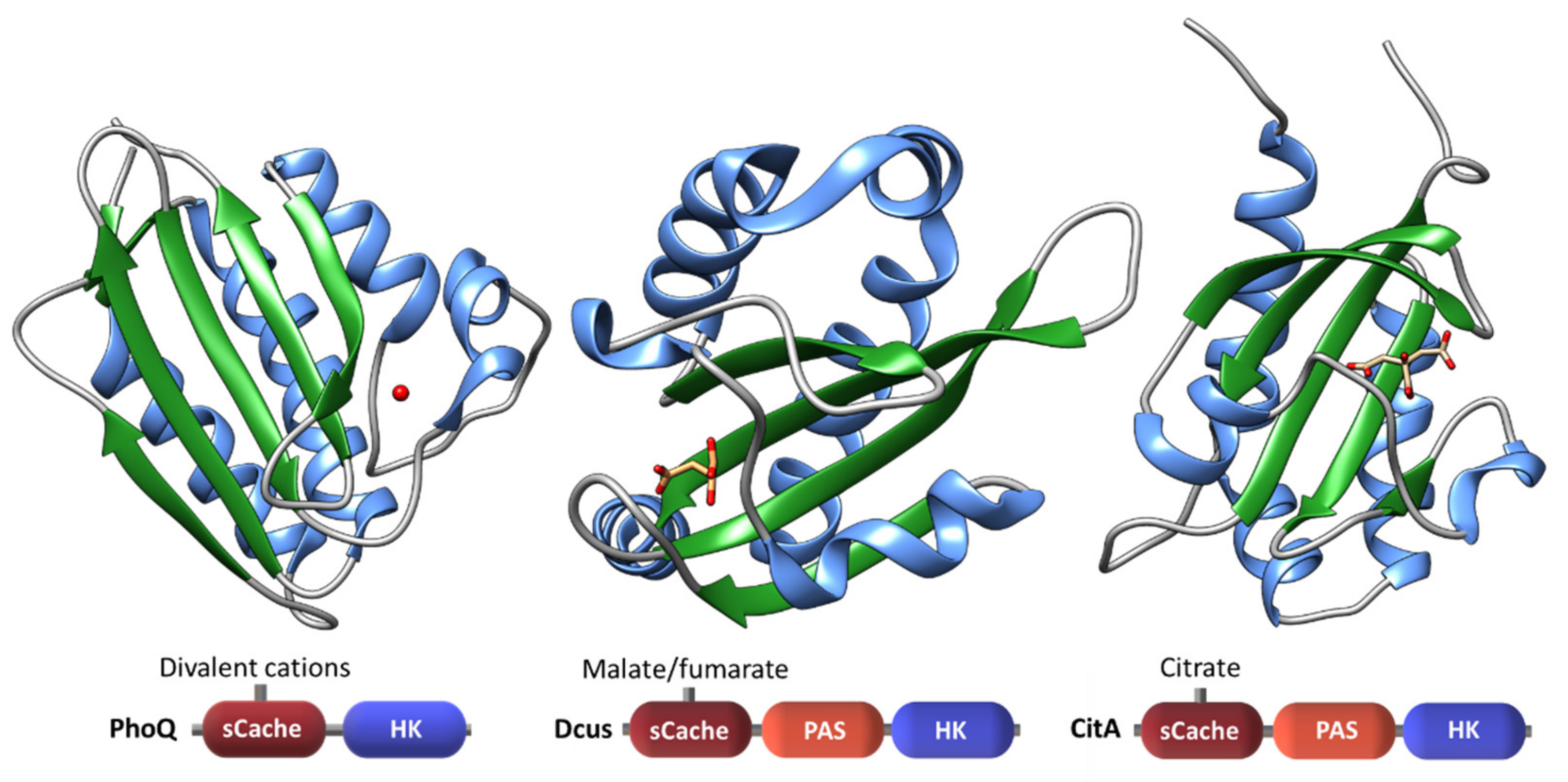
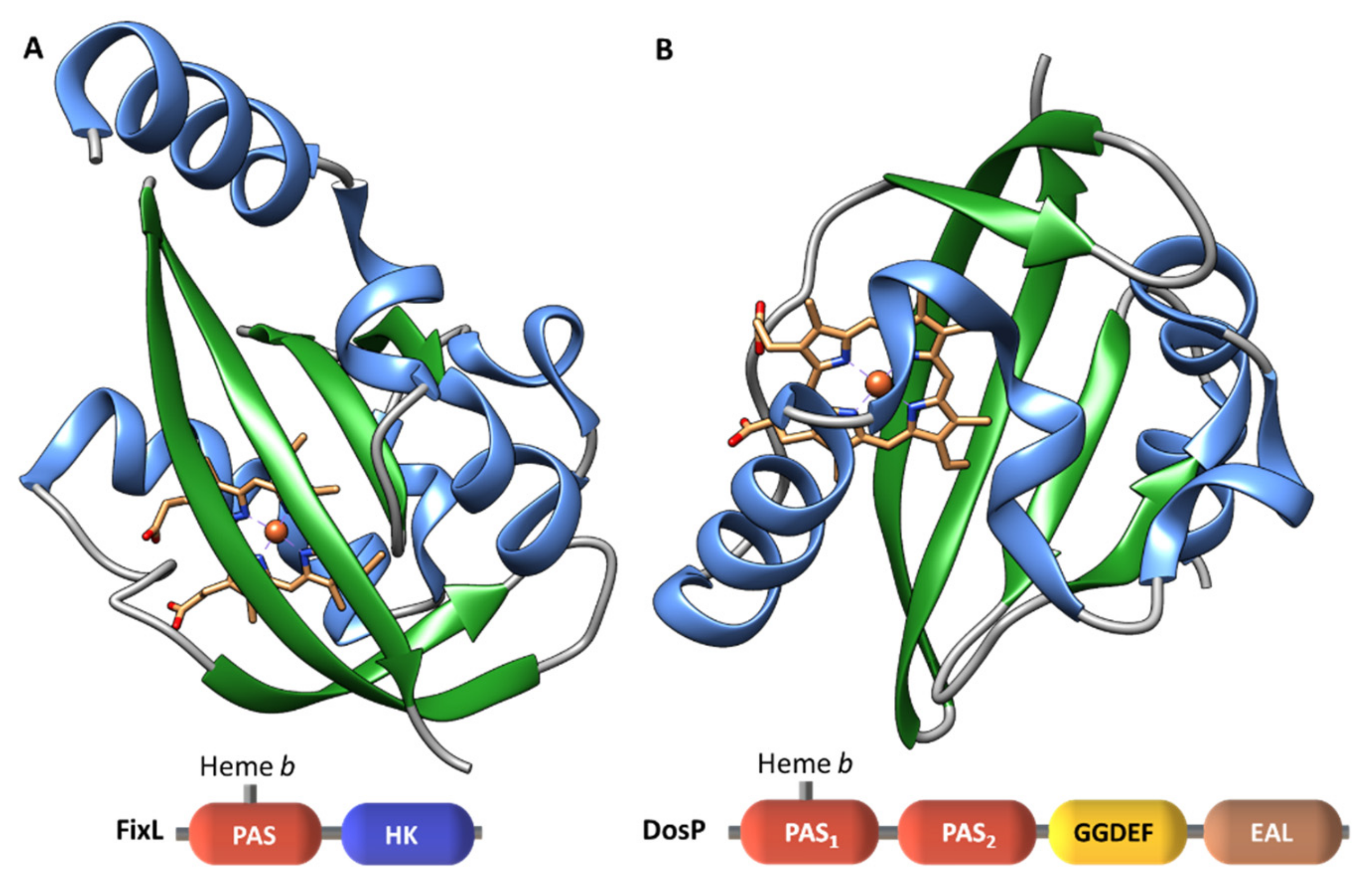
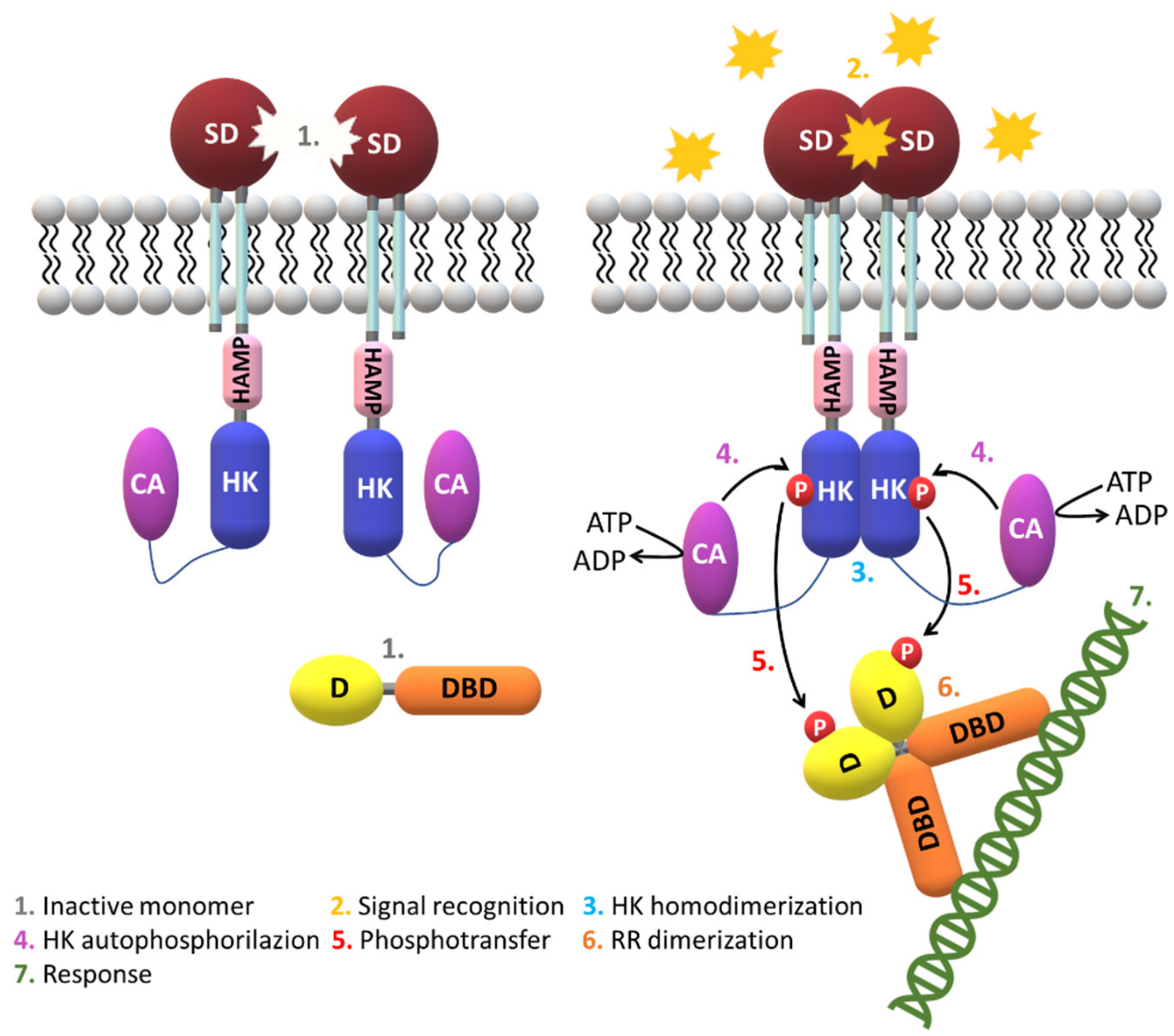
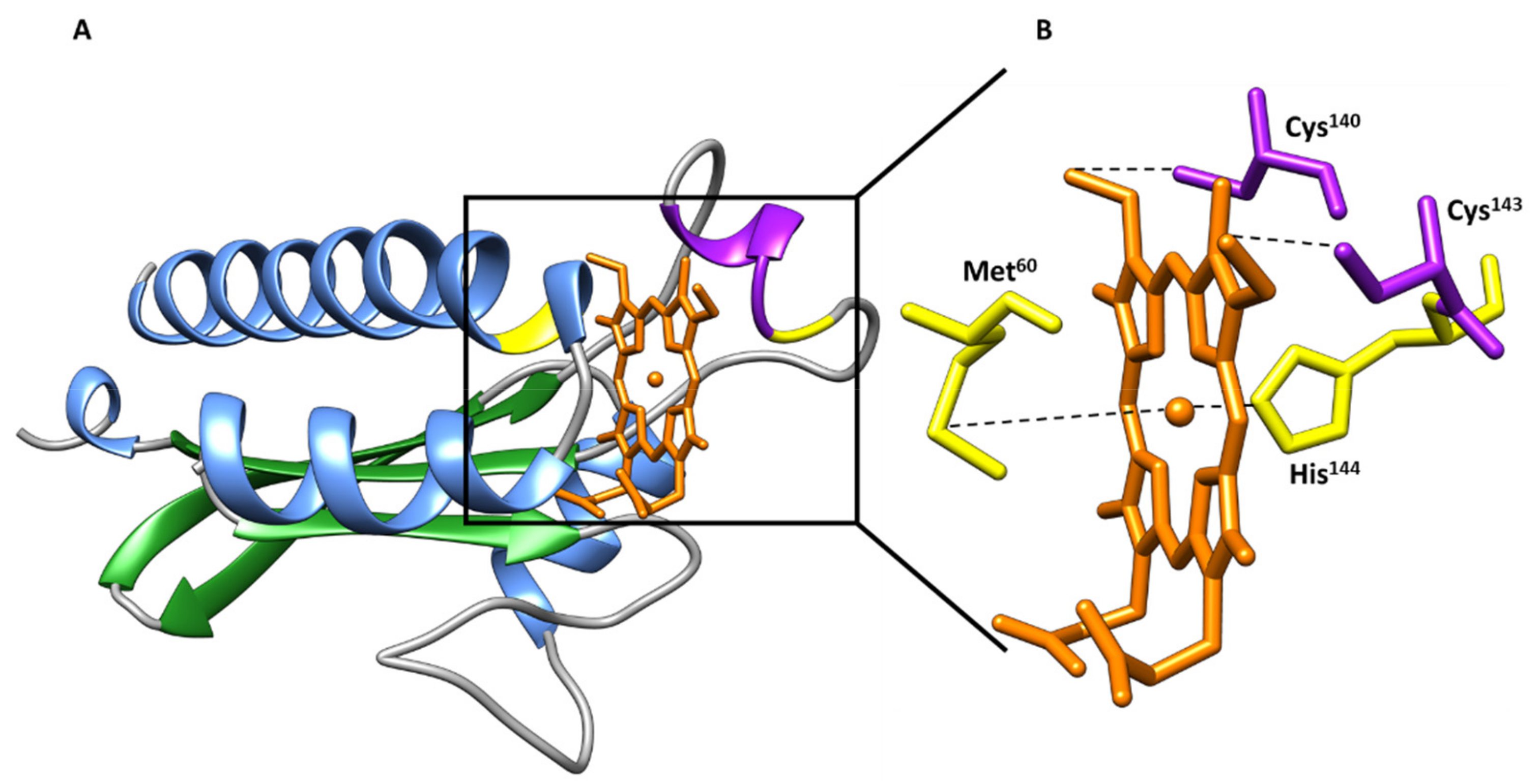
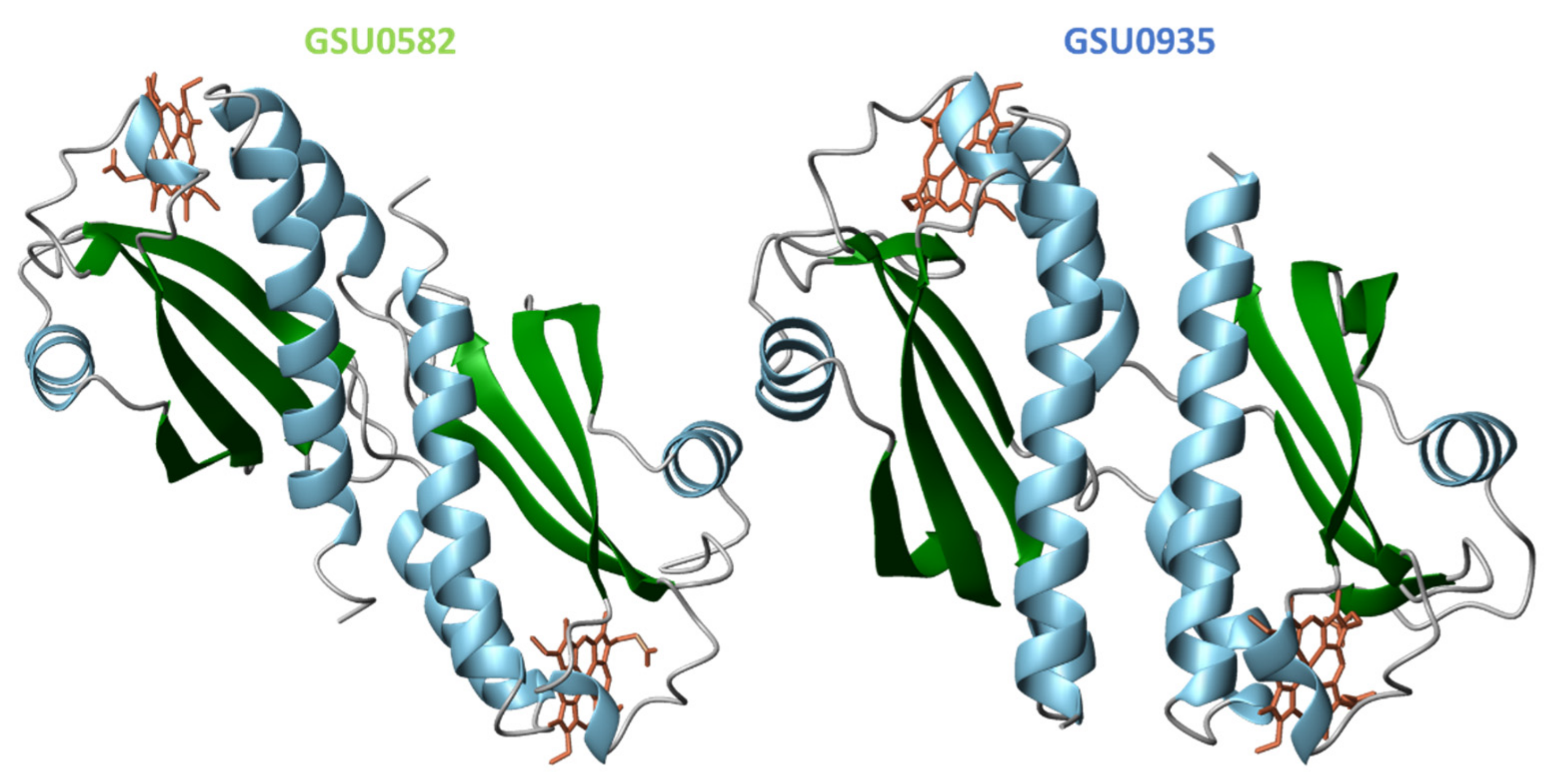
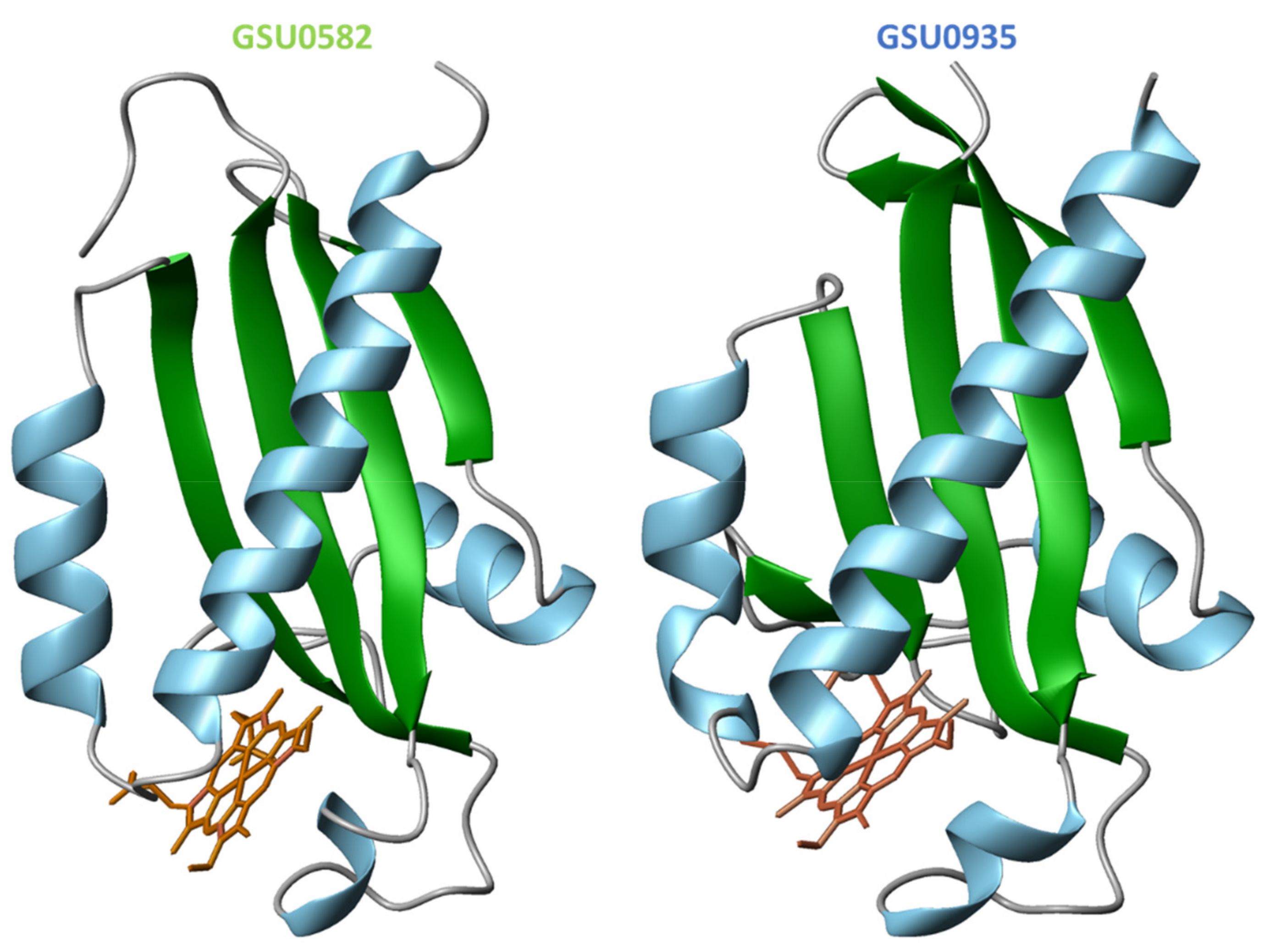
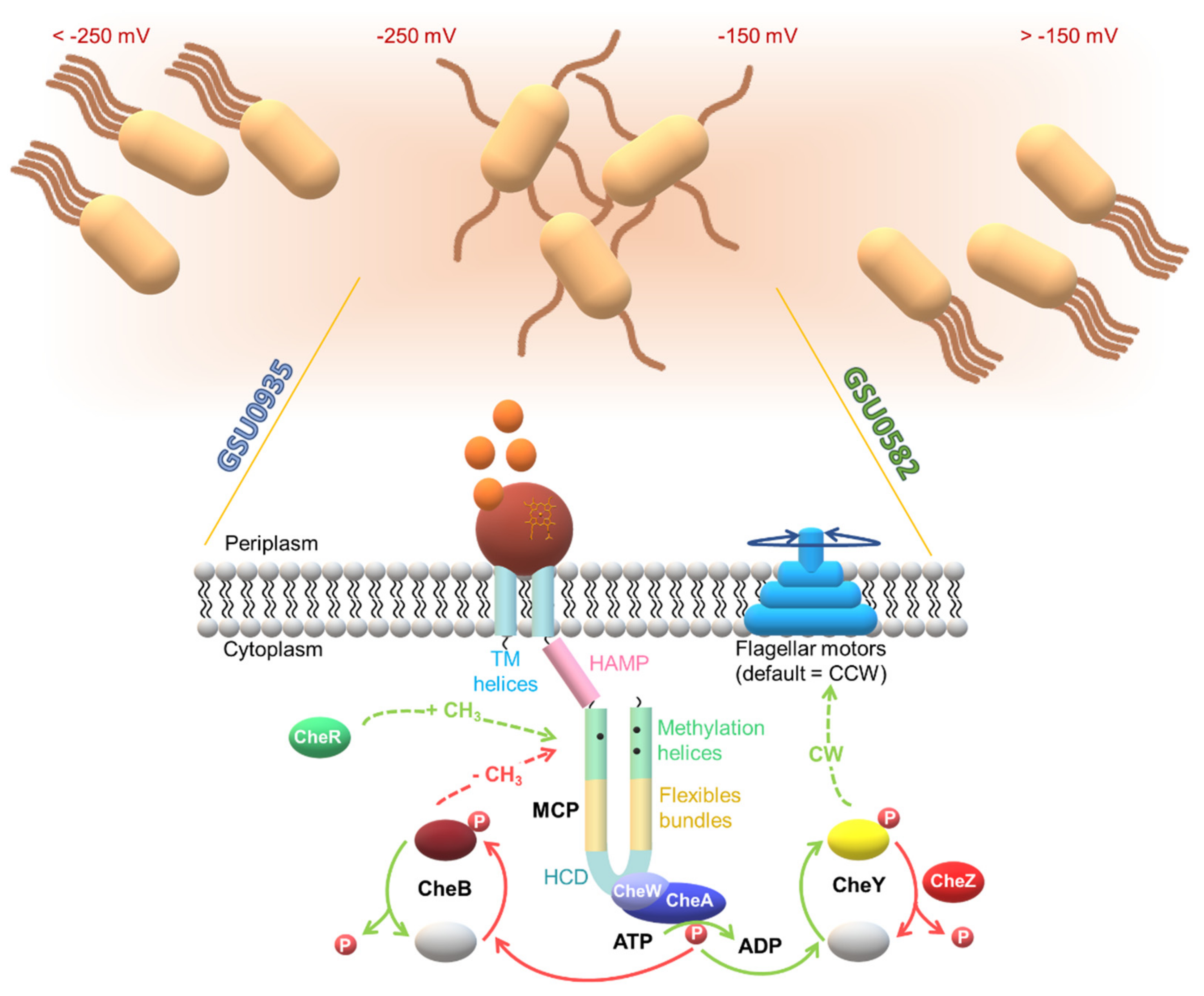
| Protein | Class | Role in Chemotaxis |
|---|---|---|
| MCP | Chemoreceptor | Receptor for chemotaxis stimulus. |
| CheA | Histidine protein kinase | Autophosphorylates in response to MCP. Phosphorylates CheY and CheB. |
| CheW | Adaptor protein | Involved in the formation of the MCP-CheA-CheW complex. |
| CheY | Response regulator | Binds FliM when phosphorylated to alter flagellar rotation. Multiple CheY may act as phosphate sinks. |
| CheB | Response regulator, methylesterase | Removes methyl groups from MCP. Activated upon phosphorylation. |
| CheR | Methyltransferase | Constitutively methylates MCP. |
| CheZ | Phosphatase | Stimulates dephosphorylation of CheY and signal termination. |
| CheV | CheW-CheY fusion | May function as a phosphate sink and signal termination. Alternatively, may function in adaptation. |
| FliM | Motor switch protein | Alters flagellar rotation in response to CheY~P binding. |
| Strain | Chemotaxis Genes | MCP | cheA | cheB | cheR | cheW | cheY | cheC | cheD | cheX | cheV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| G. sulfurreducens PCA | 79 | 35 | 4 | 4 | 4 | 10 | 10 | 1 | 3 | 4 | 1 |
| G. sulfurreducens KN400 | 77 | 34 | 4 | 4 | 4 | 8 | 11 | 1 | 3 | 4 | 1 |
| G. metallireducens | 68 | 19 | 5 1 | 8 2 | 8 2 | 8 | 11 1 | 1 | 3 | 3 | 1 |
| G. uraniireducens | 73 | 27 | 7 | 6 | 9 | 10 | 6 | 1 | 2 | 1 | 1 |
| G. lovleyi | 68 | 29 | 6 | 5 2 | 7 2 | 7 | 7 | 2 | 1 | 1 | 2 |
| G. bemidjiensis | 90 | 35 | 8 | 7 | 8 | 11 | 11 | 1 | 3 | 3 | 1 |
| G. daltonii FRC-32 | 75 | 27 | 7 | 8 2 | 11 2 | 10 | 6 | - | 1 | 1 | 1 |
| M21 | 94 | 37 | 9 | 7 | 10 | 12 | 7 | 2 | 3 | 3 | 1 |
| M18 | 97 | 32 | 10 | 10 | 11 | 14 | 9 | 2 | 2 | 3 | 1 |
| G. pickeringii | 70 | 18 | 5 | 2 | 6 | 7 | 25 | - | 3 | 3 | 1 |
| G. anodireducens | 57 | 28 | 4 | 1 | 5 | 10 | - | 1 | 3 | 4 | 1 |
| G. bremensis | 109 | 38 | 19 | 9 2 | 12 2 | 1 | 8 | 1 | 3 | 3 | 1 |
| G. soli | 85 3 | - | - | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, M.A.; Salgueiro, C.A. Multistep Signaling in Nature: A Close-Up of Geobacter Chemotaxis Sensing. Int. J. Mol. Sci. 2021, 22, 9034. https://doi.org/10.3390/ijms22169034
Silva MA, Salgueiro CA. Multistep Signaling in Nature: A Close-Up of Geobacter Chemotaxis Sensing. International Journal of Molecular Sciences. 2021; 22(16):9034. https://doi.org/10.3390/ijms22169034
Chicago/Turabian StyleSilva, Marta A., and Carlos A. Salgueiro. 2021. "Multistep Signaling in Nature: A Close-Up of Geobacter Chemotaxis Sensing" International Journal of Molecular Sciences 22, no. 16: 9034. https://doi.org/10.3390/ijms22169034
APA StyleSilva, M. A., & Salgueiro, C. A. (2021). Multistep Signaling in Nature: A Close-Up of Geobacter Chemotaxis Sensing. International Journal of Molecular Sciences, 22(16), 9034. https://doi.org/10.3390/ijms22169034







