Modulation of PTPN2/22 Function by Spermidine in CRISPR-Cas9-Edited T-Cells Associated with Crohn’s Disease and Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. CRISPR-Cas9 Transfection, Stimulation, and Treatment of Jurkat T-Cells
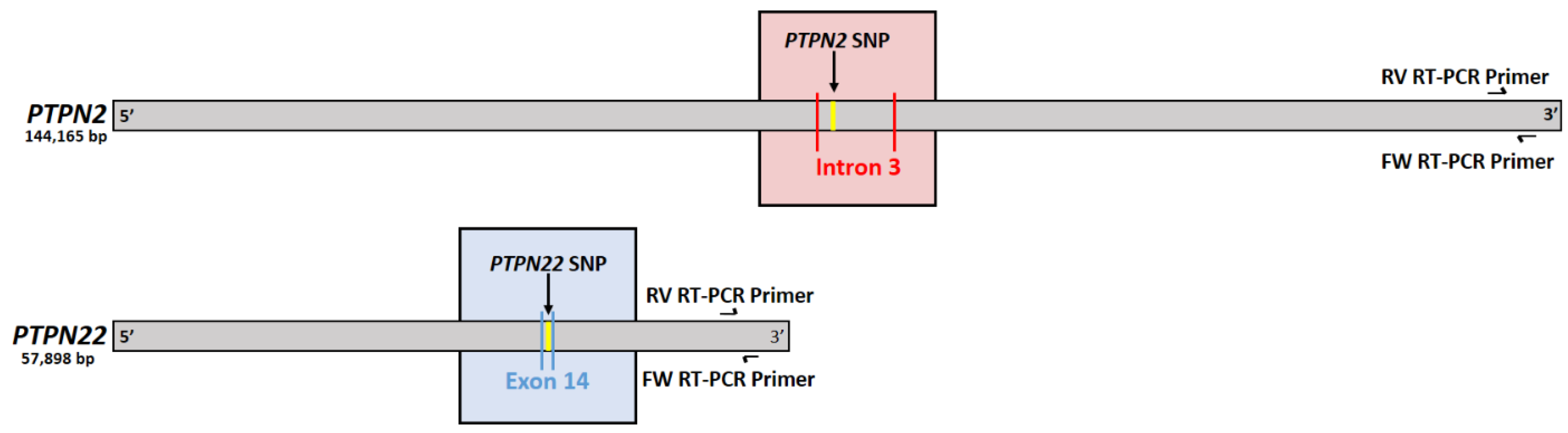
2.2. Confirming Induction of PTPN2/22 SNPs
2.3. Measurement of PTPN2/22 Expression in Treated Jurkat T-Cells
2.4. T-Cell Proliferation Assay of Treated Jurkat T-Cells
2.5. Measurement of IFN-γ and TNF-α Secretion in Treated Jurkat T-Cells
2.6. CD25 (IL-2RA) Fluorescent Immunostaining Assay
2.7. Statistical Analysis
3. Results
3.1. PTPN2/22 SNP Sequencing
3.2. PTPN2/22 SNPs Decrease Gene Expression upon Induction in Jurkat T-Cells Using CRISPR-Cas9
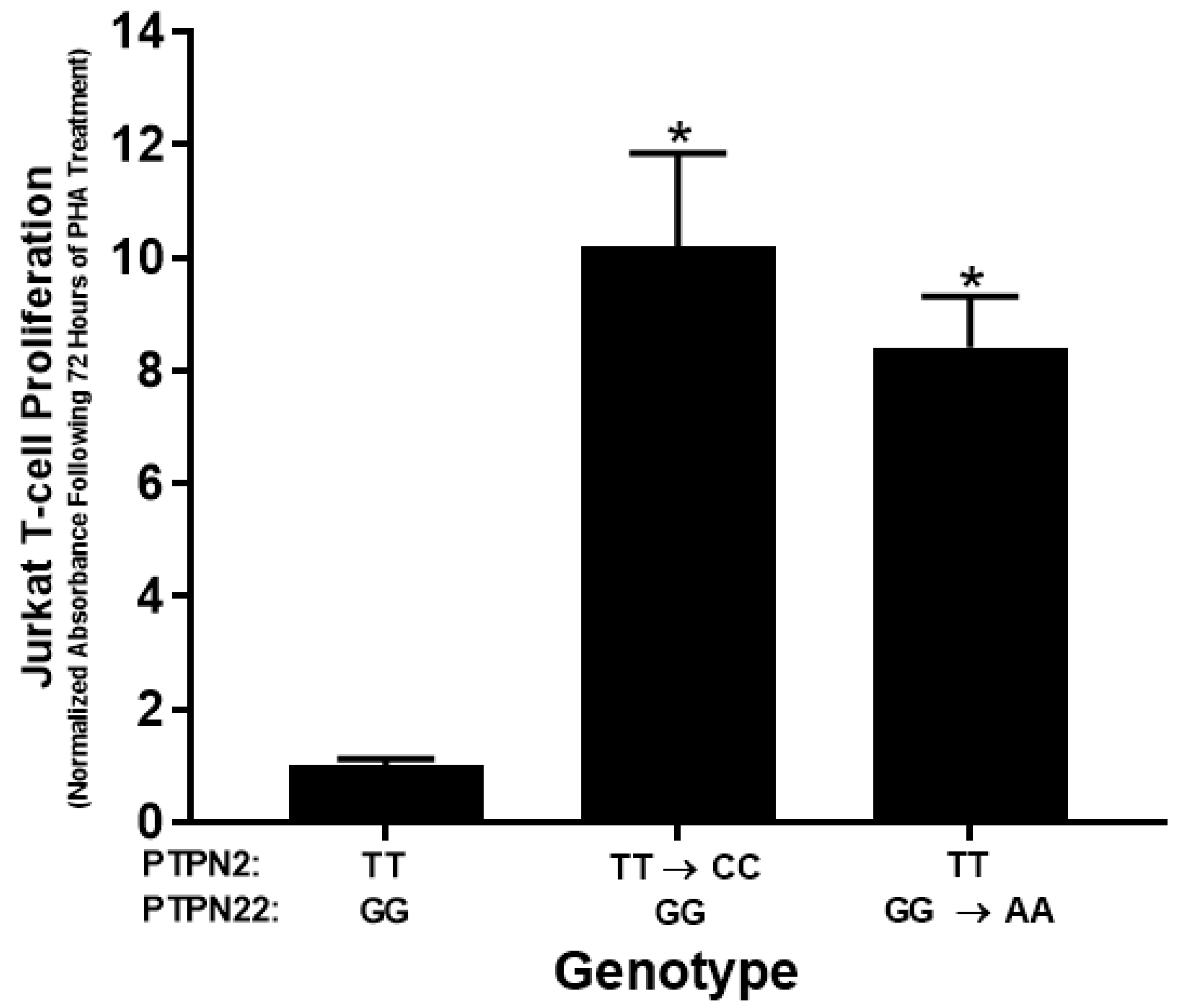
3.3. PTPN2/22 SNPs Increase T-Cell Proliferation upon PHA Induction in Jurkat T-Cells Using CRISPR-Cas9
3.4. PTPN2/22 SNPs Increase Pro-Inflammatory Cytokine Secretion upon Induction in Jurkat T-Cells Using CRISPR-Cas9
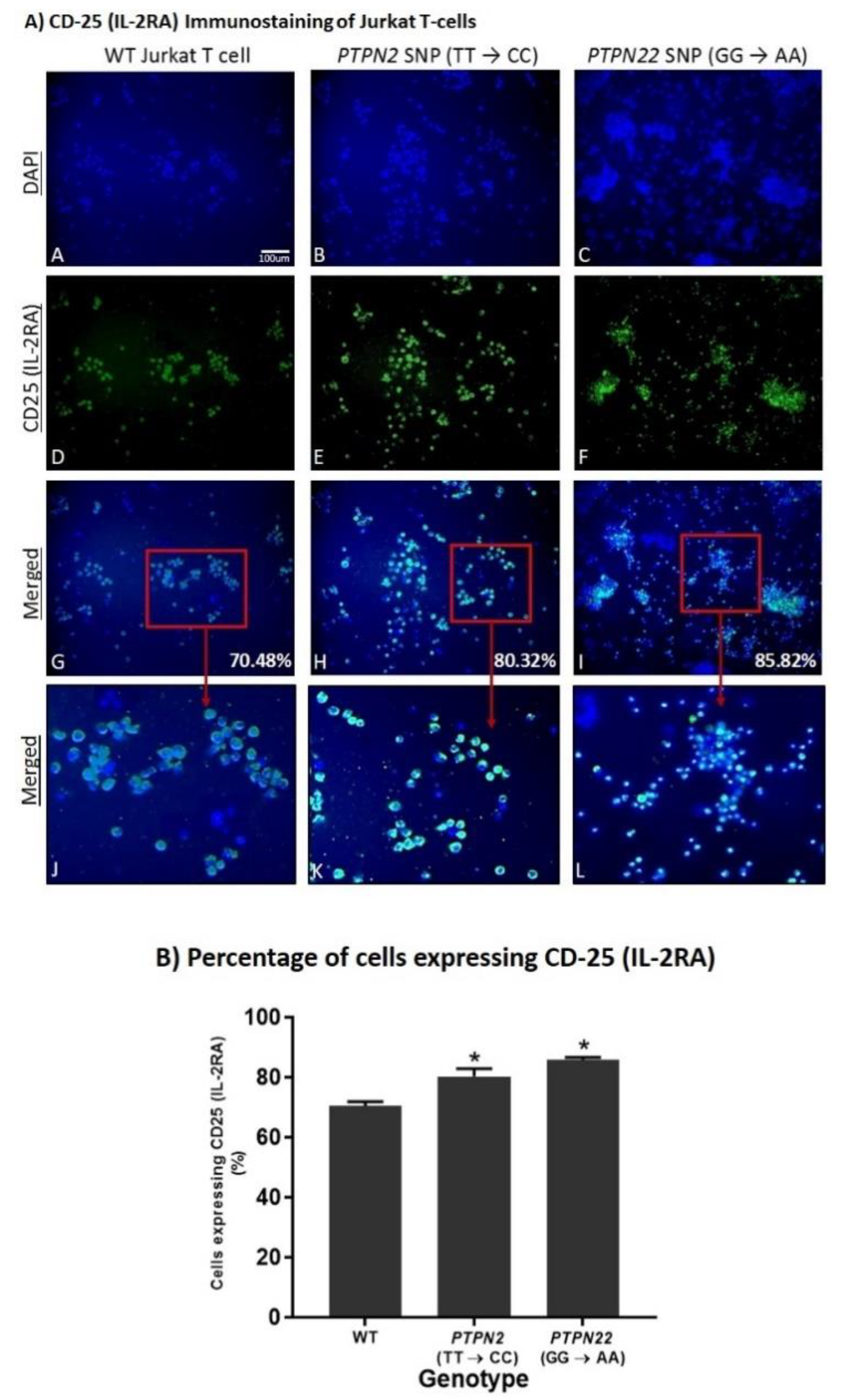
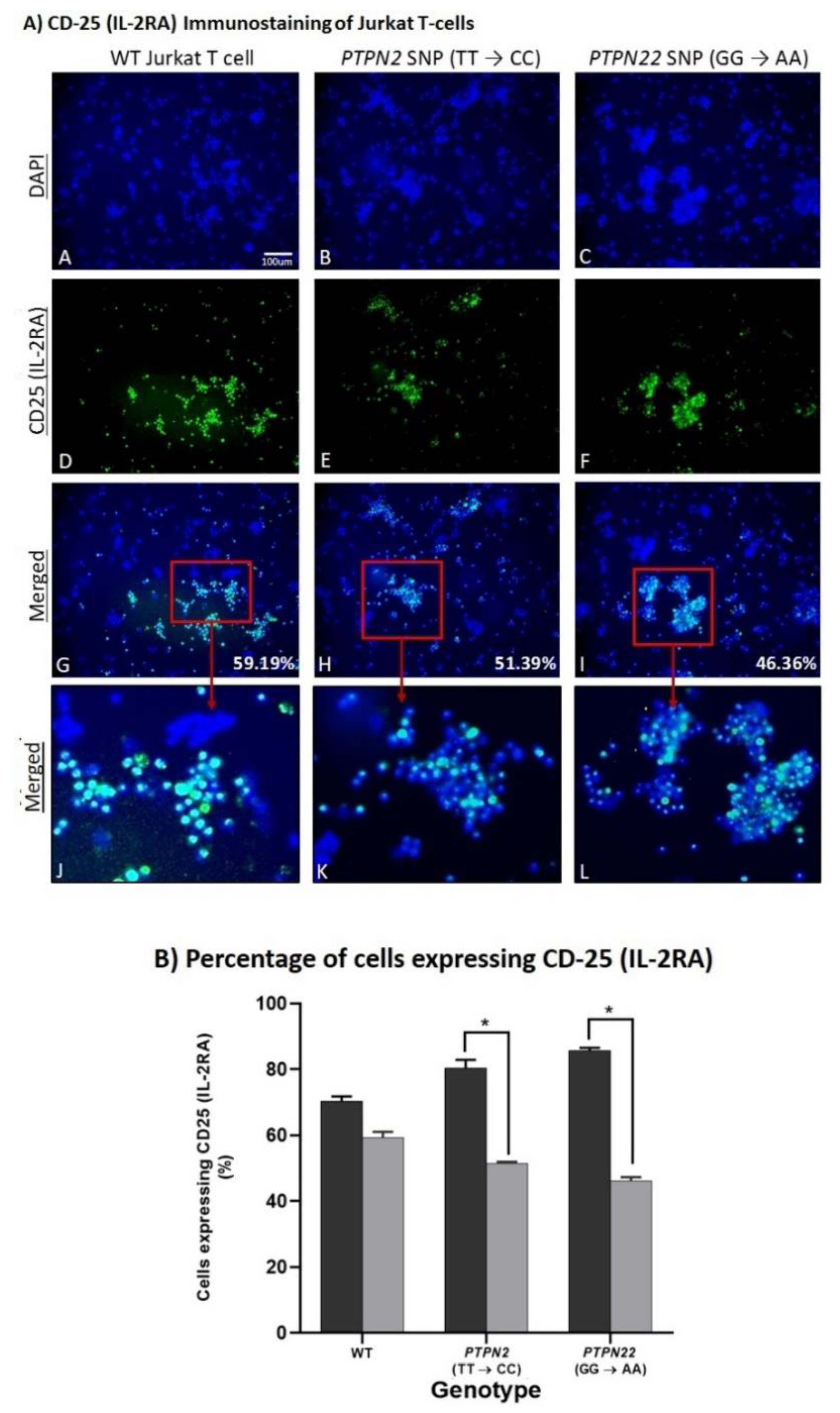
3.5. PTPN2/22 SNPs Increase T-Cell Activation upon Induction in Jurkat T-Cells Using CRISPR-Cas9
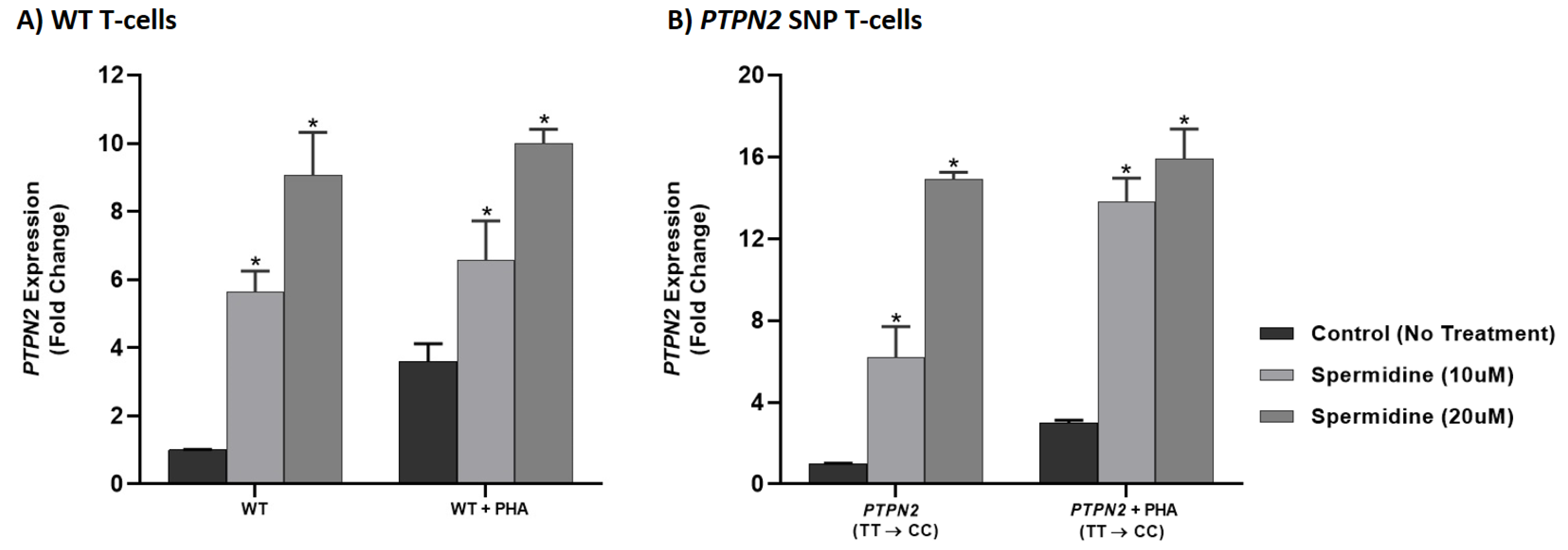
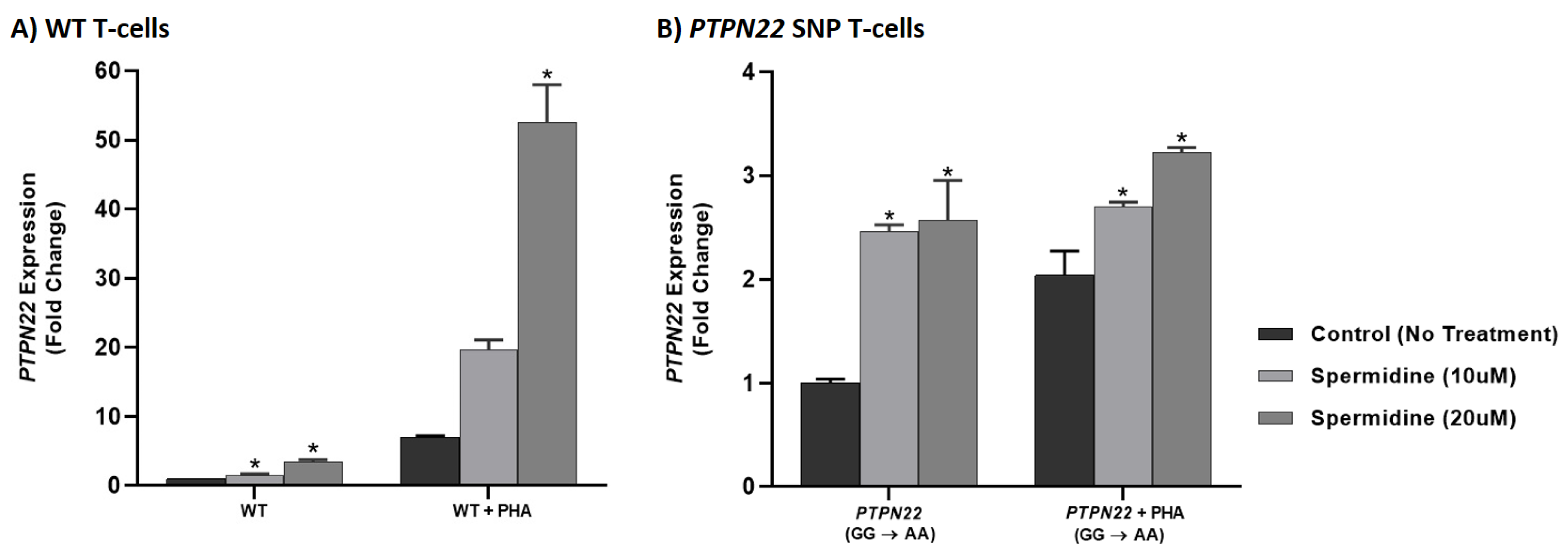
3.6. Spermidine Increased PTPN2/22 Expression in CRISPR-Cas9-Edited Jurkat T-Cells with PTPN2/22 SNPs
3.7. Spermidine Decreases T-Cell Proliferation in CRISPR-Cas9-Edited Jurkat T-Cells with PTPN2/22 SNPs
3.8. Spermidine Decreases Pro-Inflammatory Cytokine Secretion in CRISPR-Cas9-Edited Jurkat T-Cells with PTPN2/22 SNPs
4. Spermidine Decreases T-Cell Activation in CRISPR-Cas9-Edited Jurkat T-Cells with PTPN2/22 SNPs
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qasem, A.; Elkamel, E.; Naser, S.A. Anti-MAP Triple Therapy Supports Immunomodulatory Therapeutic Response in Crohn’s Disease through Downregulation of NF-κB Activation in the Absence of MAP Detection. Biomedicines 2020, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Naser, S.A.; Sagramsingh, S.R.; Naser, A.S.; Thanigachalam, S. Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J. Gastroenterol. 2014, 20, 7403–7415. [Google Scholar] [CrossRef] [PubMed]
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence review of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.; Mehandru, S.; Colombel, J.-F.; Peyrin-Biroulet, L. Crohn’s Disease. Lancet 2017, 389, 1741–1755. [Google Scholar] [CrossRef]
- Sharp, R.C.; Beg, S.A.; Naser, S.A. Polymorphisms in protein tyrosine phosphatase non-receptor type 2 and 22 (PTPN2/22) are linked to hyper-proliferative T-cells and susceptibility to Mycobacteria and Rheumatoid Arthritis. Front. Cell. Infect. Microbiol. 2018, 8, 11. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef]
- van der Woude, D.; van der Helm-van Mil, A.H.M. Update on the epidemiology, risk factors, and disease outcomes of Rheumatoid Arthritis. Best Pract. Res. Clin. Rheumatol. 2018, 32, 174–187. [Google Scholar] [CrossRef]
- Sharp, R.C.; Abdulrahim, M.; Naser, E.S.; Naser, S.A. Genetic variations of PTPN2 and PTPN22: Role in the pathogenesis of type 1 diabetes and Crohn’s disease. Front. Cell. Infect. Microbiol. 2015, 5, 95. [Google Scholar] [CrossRef]
- Qasem, A.; Naser, A.E.; Naser, S.A. The alternate effects of anti-TNFα therapeutics and their role in mycobacterial granulomatous infection in Crohn’s disease. Expert Rev. Anti Infect. Ther. 2017, 15, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Sharp, R.C.; Beg, S.A.; Naser, S.A. Role of PTPN2/22 polymorphisms in pathophysiology of Crohn’s disease. World J. Gastroenterol. 2018, 24, 657–670. [Google Scholar] [CrossRef]
- Qasem, A.; Ramesh, S.; Naser, S.A. Genetic polymorphisms in tumour necrosis factor receptors (TNFRSF1A/1B) illustrate differential treatment response to TNFα inhibitors in patients with Crohn’s disease. BMJ Open Gastroenterol. 2019, 6, e000246. [Google Scholar] [CrossRef]
- Naser, A.; Odeh, A.K.; Sharp, R.C.; Qasem, A.; Beg, S.; Naser, S.A. Polymorphisms in TNF receptor superfamily 1B (TNFRSF1B: rs3397) are linked to Mycobacterium avium paratuberculosis infection and osteoporosis in Rheumatoid Arthritis. Microorganisms 2019, 7, 646. [Google Scholar] [CrossRef] [PubMed]
- Bottini, N.; Musumeci, L.; Alonso, A.; Rahmouni, S.; Nika, K.; Rostamkhani, M.; MacMurray, J.; Meloni, G.F.; Lucarelli, P.; Pellecchia, M.; et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 2004, 36, 337–338. [Google Scholar] [CrossRef] [PubMed]
- Gjorloff-Wingren, A.; Saxena, M.; Williams, S.; Hammi, D.; Mustelin, T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur. J. Immunol. 1999, 29, 3845–3854. [Google Scholar] [CrossRef]
- Mustelin, T.; Abraham, R.T.; Rudd, C.E.; Alonso, A.; Merlo, J.J. Protein tyrosine phosphorylation in T cell signaling. Front. Biosci 2002, 7, d918–d969. [Google Scholar] [CrossRef]
- Espino-Paisan, L.; de la Calle, H.; Fernandez-Arquero, M.; Figuredo, M.A.; de la Concha, E.G.; Urcelay, E.; Santiago, J.L. A polymorphism in PTPN2 gene is associated with an earlier onset of type 1 diabetes. Immunogenetics 2011, 63, 255–258. [Google Scholar] [CrossRef]
- Hendriks, W.J.; Pulido, R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys Acta 2013, 1832, 1673–1696. [Google Scholar] [CrossRef]
- Vang, T.; Miletic, A.V.; Bottini, N.; Mustelin, T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity 2007, 40, 453–461. [Google Scholar] [CrossRef]
- Zikherman, J.; Weiss, A. Unraveling the functional implications of GWAS: How T cell protein tyrosine phosphatase drives autoimmune disease. J. Clin. Investig. 2011, 121, 4618–4621. [Google Scholar] [CrossRef]
- Qu, H.; Tessier, M.C.; Hudson, T.J.; Polychronakos, C.; Qu, H. Confirmation of the association of the R620W polymorphism in the protein tyrosine phosphatase PTPN22 with type 1 diabetes in a family based study. J. Med. Genet. 2005, 42, 266–270. [Google Scholar] [CrossRef][Green Version]
- NCBI. National Center for Biotechnology Information. 2020. Available online: https://www.ncbi.nlm.nih.gov (accessed on 5 October 2019).
- Smolen, J.S.; Aletaha, D.; Koeller, M.; Weisman, M.H.; Emery, P. New therapies for treatment of Rheumatoid Arthritis. Lancet 2007, 3701, 861–1874. [Google Scholar] [CrossRef]
- Smolen, J.S.; Aletha, D.; McInnes, I.B. Rheumatoid Arthritis. Lancet 2016, 388, 22–28. [Google Scholar] [CrossRef]
- Fakhrudin, N.; Hastuti, S.; Andriani, A.; Widyarini, S.; Nurrochmad, A. Study on the antiinflammatory activity of Artocarpus altilis leaves extract in mice. Int. J. Pharmacogn. Phytochem. Res. 2015, 7, 1080–1085. [Google Scholar]
- Bartelds, G.M.; Wijbrandts, C.A.; Nurmohamed, M.T.; Stapel, S.; Lems, W.F.; Aarden, L.; Dijkmans, B.A.C.; Tak, P.P.; Wolbink, G.J. Clinical response to adalimumab: Relationship to anti-adalimumab antibodies and serum adalimumab concentrations in Rheumatoid Arthritis. Ann. Rheum. Dis. 2007, 66, 921–926. [Google Scholar] [CrossRef]
- Dixon, W.G.; Hyrich, K.L.; Watson, K.D.; Lunt, M.; Galloway, J.; Ustianowski, A.; BSRBR Control Centre Consortium; Symmons, D.P.M. PDrug-specific risk of tuberculosis in patients with Rheumatoid Arthritis treated with anti-TNF therapy: Results from the British Society for Rheumatology Biologics Register (BSRBR). Ann. Rheum. Dis. 2010, 69, 522–528. [Google Scholar] [CrossRef]
- Niechcial, A.; Butter, M.; Manz, S.; Obialo, N.; Babler, K.; van der Lely, L.; Lang, S.; Gottier, C.; McCole, D.F.; Scharl, M.; et al. Presence of PTPN2 SNP rs1893217 enhances the anti-inflamatory effect of spermidine. Inflamm. Bowel Dis. 2020, 26, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ni, L.; Yang, T.; Mao, P.; Huang, X.; Luo, Y.; Jiang, Z.; Hu, L.; Zhao, Y.; Fu, Z.; et al. Preventive and therapeutic spermidine treatment attenuated acute colitis in mice. J. Agric. Food Chem. 2021, 69, 1864–1876. [Google Scholar] [CrossRef] [PubMed]
- Scharl, M.; Mwinyi, J.; Fischbeck, A.; Leucht, K.; Eloranta, J.J.; Arikkat, J.; Rogler, G. Crohn’s disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm. Bowel Dis. 2012, 18, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Vang, T.; Congia, M.; Macis, M.D.; Musuemci, L.; Orru, V.; Zavattari, P.; Nika, K.; Tautz, L.; Taskén, K.; Cucca, F.; et al. Autoiumme-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 2005, 37, 1317–1319. [Google Scholar] [CrossRef]
- Serrano, A.; Marquez, A.; Mackie, S.L.; Carmona, F.D.; Solans, R.; Miranda-Filloy, J.A.; Hernández-Rodríguez, J.; Cid, M.C.; Castañeda, S.; Morado, I.; et al. Identification of the PTPN22 functional variant R620W as susceptibility genetic factor for giant cell arteritis. Ann. Rheum. Dis. 2013, 72, 1882–1886. [Google Scholar] [CrossRef]
- Spalinger, M.R.; Lang, S.; Weber, A.; Frei, P.; Fried, M.; Rogler, G.; Scharl, M. Loss of protein tyrosine phosphatase nonreceptor type 22 regulates interferon-gamma-induced signaling in human monocytes. Gastroenterology 2013, 144, 978–988.e10. [Google Scholar] [CrossRef]
- Roda, G.; Chien Ng, S.; Kotze, P.G.; Argollo, M.; Panaccione, R.; Spinelli, A.; Kaser, A.; Peyrin-Biroulet, L.; Danese, S. Crohn’s disease. Nat. Rev. Dis. Primers 2020, 6, 22. [Google Scholar] [CrossRef]
- Brennan, F.M.; McInnes, I.B. Evidence that cytokines play a role in Rheumatoid Arthritis. J. Clin. Investig. 2008, 118, 3537–3545. [Google Scholar] [CrossRef] [PubMed]
- Cope, A.P. T cells in Rheumatoid Arthritis. Arthritis Res. Ther. 2008, 10, S1. [Google Scholar] [CrossRef] [PubMed]
- Koetz, K.; Bryl, E.; Spickschen, K.; O’Fallon, W.M.; Goronzy, J.J.; Weyand, C.M. T cell homeostasis in patients with Rheumatoid Arthritis. Proc. Natl. Acad. Sci. USA 2000, 97, 9203–9208. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, J.R.; Wilson, C.B. Regulation of interferon-gamma during innate and adaptive immune. Adv. Immunol. 2007, 96, 41–101. [Google Scholar]
- Bajnok, A.; Ivanova, M.; Rigó, J.; Toldi, G. The distribution of activation markers and selectins on peripheral T lymphocytes in preeclampsia. Mediat. Inflamm. 2017, 2017, 8045161. [Google Scholar] [CrossRef]
- Hussain, T.; Tan, B.; Ren, W.; Rahu, N.; Dad, R.; Husain Kalhoro, D.; Yin, Y. Polyamines: Therapeutic perspectives in oxidative stress and inflammatory diseases. Amino Acids 2017, 49, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in aging and disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef]
- Madeo, F.; Bauer, M.A.; Carmona-Gutierrez, D.; Kroemer, G. Spermidine: A physiological autophagy inducer acting as an anti-aging vitamin in humans? Autophagy 2018, 15, 165–168. [Google Scholar] [CrossRef]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Weiss, T.S.; Herfarth, H.; Obermeier, F.; Ouart, J.; Vogl, D.; Schölmerich, J.; Jauch, K.-W.; Rogler, G. Intracellular polyamine levels of intestinal epithelial cells in inflammatory bowel disease. Inflamm. Bowel Dis. 2004, 10, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kang, S.C. Natural polyamine inhibits mouse skin inflammation and macrophage activation. Inflamm. Res. 2013, 62, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Park, H.Y. Anti-inflammatory effects of spermidine in lipopolysaccharide-stimulated BV2 microglial cells. J. Biomed. Sci. 2012, 19, 31. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, T.; Qin, W.; Huang, B.; Chen, W.; Li, S.; Li, J. Upregulated PTPN2 induced by inflammatory response or oxidative stress stimulates the progression of thyroid cancer. Biochem. Biophys. Res. Commun. 2020, 522, 21–25. [Google Scholar] [CrossRef]
- Stewart, T.M.; Dunston, T.T.; Woster, P.M.; Casero, R.A. Polyamine catabolism and oxidative damage. J. Biol. Chem. 2018, 293, 18736–18745. [Google Scholar] [CrossRef] [PubMed]


| Gene | RefSNP | Mutation | Location | Mean Allele Frequency | Mutation Phenotype | Reference |
|---|---|---|---|---|---|---|
| PTPN2 | rs478582 | T → C | Intron 3 | C = 0.3158 | High susceptibility to RA, TID, MS, and Celiac disease | [16] |
| PTPN22 | rs2476601 | G → A | R620W | A = 0.0588 | High susceptibility to CD, RA, TID, MS, SLE, and Celiac disease | [20] |
| Gene | Component | Sequence (5′ → 3′) |
|---|---|---|
| PTPN2 | sgRNA | AUUAUACUACGUCAAUUCAC +scaffold |
| Donor DNA | CTACCTCAAGTAAAAAATGCATTTTAGTTTCCTGTGAATT GACGCAGTATAATAGCTCATAGTTACATTAATCTGCATAT (80 bases) | |
| Donor DNA reverse complement | ATATGCAGATTAATGTAACTATGAGCTATTATACTGCGTC AATTCACAGGAAACTAAAATGCATTTTTTACTTGAGGTAG (80 bases) | |
| Forward PCR Primer | CAGGGCTGTCTTTCCCCCTA (20 bases) | |
| Reverse PCR Primer | GCAAAGTGTCCACCTTTGAT (20 bases) | |
| PTPN22 | sgRNA | AAUGAUUCAGGUGUCCGUAC +scaffold |
| Donor DNA | AGCTTCCTCAACCACAATAAATGATTCAGGTGTCC TACAGGAAGTGGAGGGGGGATTTCATCATCTATCC (70 bases) | |
| Donor DNA reverse complement | GGATAGATGATGAAATCCCCCCTCCACTTCCTGTA GGACACCTGAATCATTTATTGTGGTTGAGGAAGCT (70 bases) | |
| Forward PCR Primer | CGCCCAGCCCTACTTTTGAG (20 bases) | |
| Reverse PCR Primer | CCATGCCCATCCCACACTTT (20 bases) |
| Gene | Forward Primer Sequence (5′ → 3′) | Reverse Primer Sequence (5′ → 3′) |
|---|---|---|
| GAPDH | 5′-CTTTTGCAGACCACAGTCCATG-3′ (22 bases) | 5′-TTTTCTAGACGGCAGGTCAGG-3′ (21 bases) |
| PTPN2 | 5′-CTAGAGGGTTAGCGAGCCTCA-3′ (21 bases) | 5′- TCATGTGGGAATGATTTTTGGTCAC-3′ (25 bases) |
| PTPN22 | 5′-TAGTTTTATTTGCAGGTGTACTTGCAG-3′ (27 bases) | 5′-TGGTCAAGATGCTGCCTAACATT-3′ (23 bases) |
| Cell Group | [IFN-γ] ± SD (pg/mL) | [TNF-α] ± SD (pg/mL) |
|---|---|---|
| WT | 0.029 ± 0.003 | 0.293 ± 0.042 |
| PTPN2 (TT → CC) | 0.060 ± 0.004 * | 0.507 ± 0.014 * |
| PTPN22 (GG → AA) | 0.054 ± 0.002 * | 0.408 ± 0.039 * |
| Treatment | Spermidine Concentration (μM) | IFN-γ ± SD (pg/mL) | TNF-α ± SD (pg/mL) |
|---|---|---|---|
| WT | - | 0.029 ± 0.003 | 0.293 ± 0.042 |
| WT+PHA | - | 0.283 ± 0.026 | 0.785 ± 0.012 |
| 10 | 0.124 ± 0.005 * | 0.359 ± 0.012 * | |
| 20 | 0.172 ± 0.010 * | 0.353 ± 0.022 * | |
| PTPN2 | - | 0.060 ± 0.004 | 0.408 ± 0.039 |
| PTPN2+PHA | - | 0.359 ± 0.020 | 4.87 ± 0.292 |
| 10 | 0.115 ± 0.010 * | 0.469 ± 0.015 * | |
| 20 | 0.164 ± 0.006 * | 0.484 ± 0.016 * | |
| PTPN22 | - | 0.054 ± 0.002 | 0.507 ± 0.014 |
| PTPN22+PHA | - | 0.237 ± 0.009 | 10.8 ± 0.318 |
| 10 | 0.108 ± 0.010 * | 0.609 ± 0.021 * | |
| 20 | 0.130 ± 0.010 * | 0.408 ± 0.039 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, A.M.; Qasem, A.; Naser, S.A. Modulation of PTPN2/22 Function by Spermidine in CRISPR-Cas9-Edited T-Cells Associated with Crohn’s Disease and Rheumatoid Arthritis. Int. J. Mol. Sci. 2021, 22, 8883. https://doi.org/10.3390/ijms22168883
Shaw AM, Qasem A, Naser SA. Modulation of PTPN2/22 Function by Spermidine in CRISPR-Cas9-Edited T-Cells Associated with Crohn’s Disease and Rheumatoid Arthritis. International Journal of Molecular Sciences. 2021; 22(16):8883. https://doi.org/10.3390/ijms22168883
Chicago/Turabian StyleShaw, Ameera M., Ahmad Qasem, and Saleh A. Naser. 2021. "Modulation of PTPN2/22 Function by Spermidine in CRISPR-Cas9-Edited T-Cells Associated with Crohn’s Disease and Rheumatoid Arthritis" International Journal of Molecular Sciences 22, no. 16: 8883. https://doi.org/10.3390/ijms22168883
APA StyleShaw, A. M., Qasem, A., & Naser, S. A. (2021). Modulation of PTPN2/22 Function by Spermidine in CRISPR-Cas9-Edited T-Cells Associated with Crohn’s Disease and Rheumatoid Arthritis. International Journal of Molecular Sciences, 22(16), 8883. https://doi.org/10.3390/ijms22168883







