Atorvastatin Attenuates Programmed Death Ligand-1 (PD-L1) Induction in Human Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Results
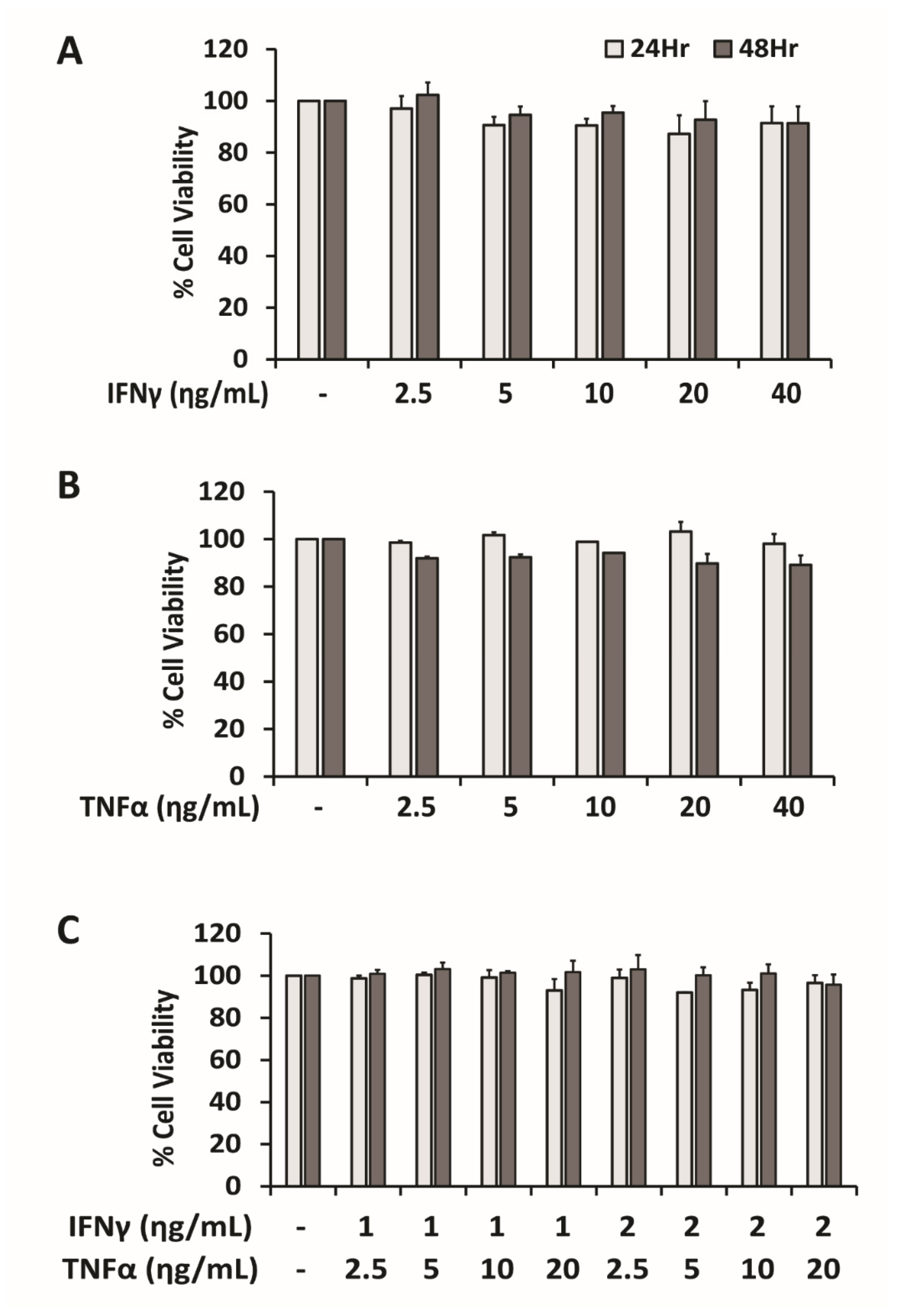
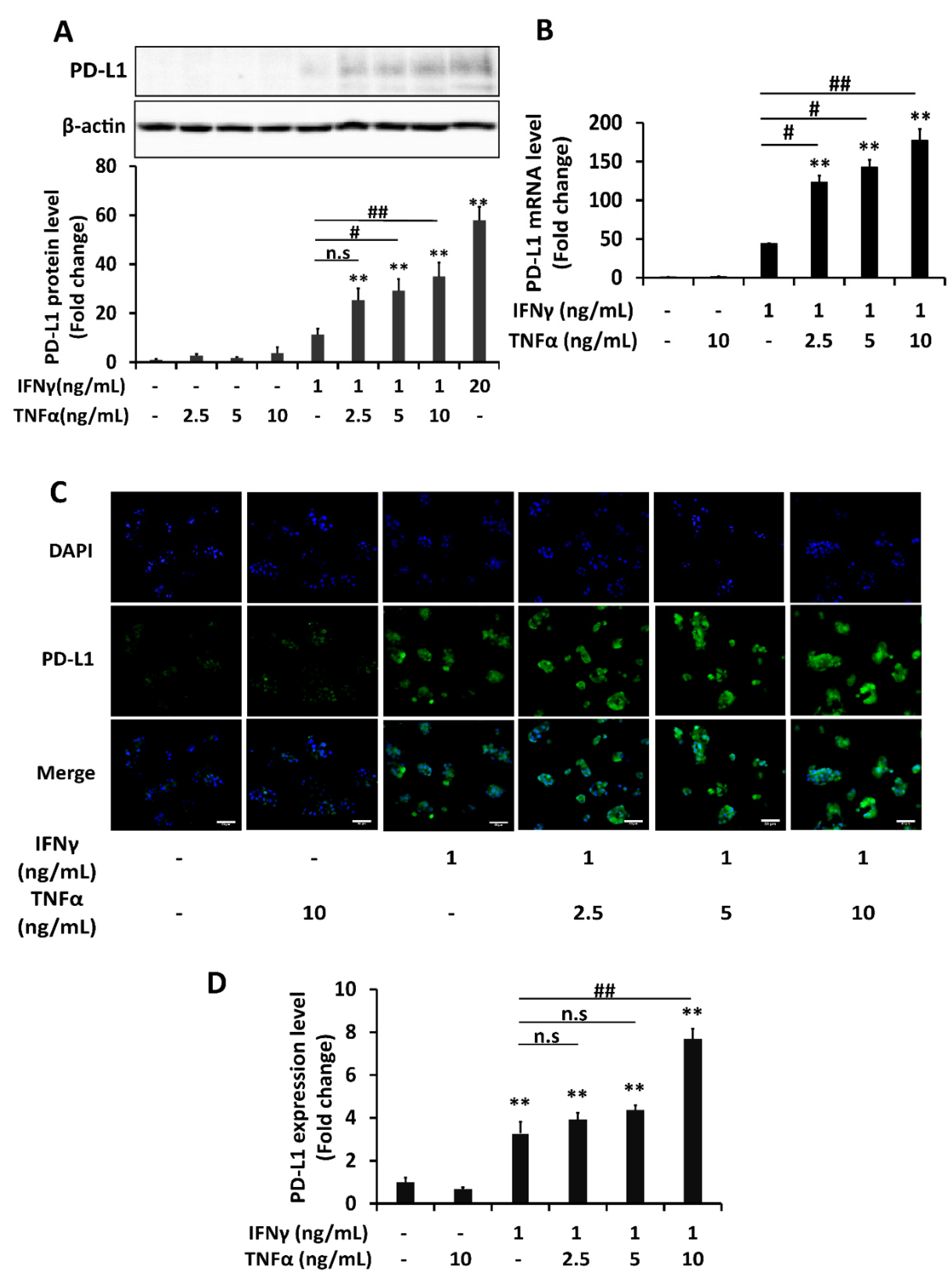
2.1. PD-L1 Expression Is Synergistically Upregulated by Combination of IFNγ and TNFα in HepG2 Cells
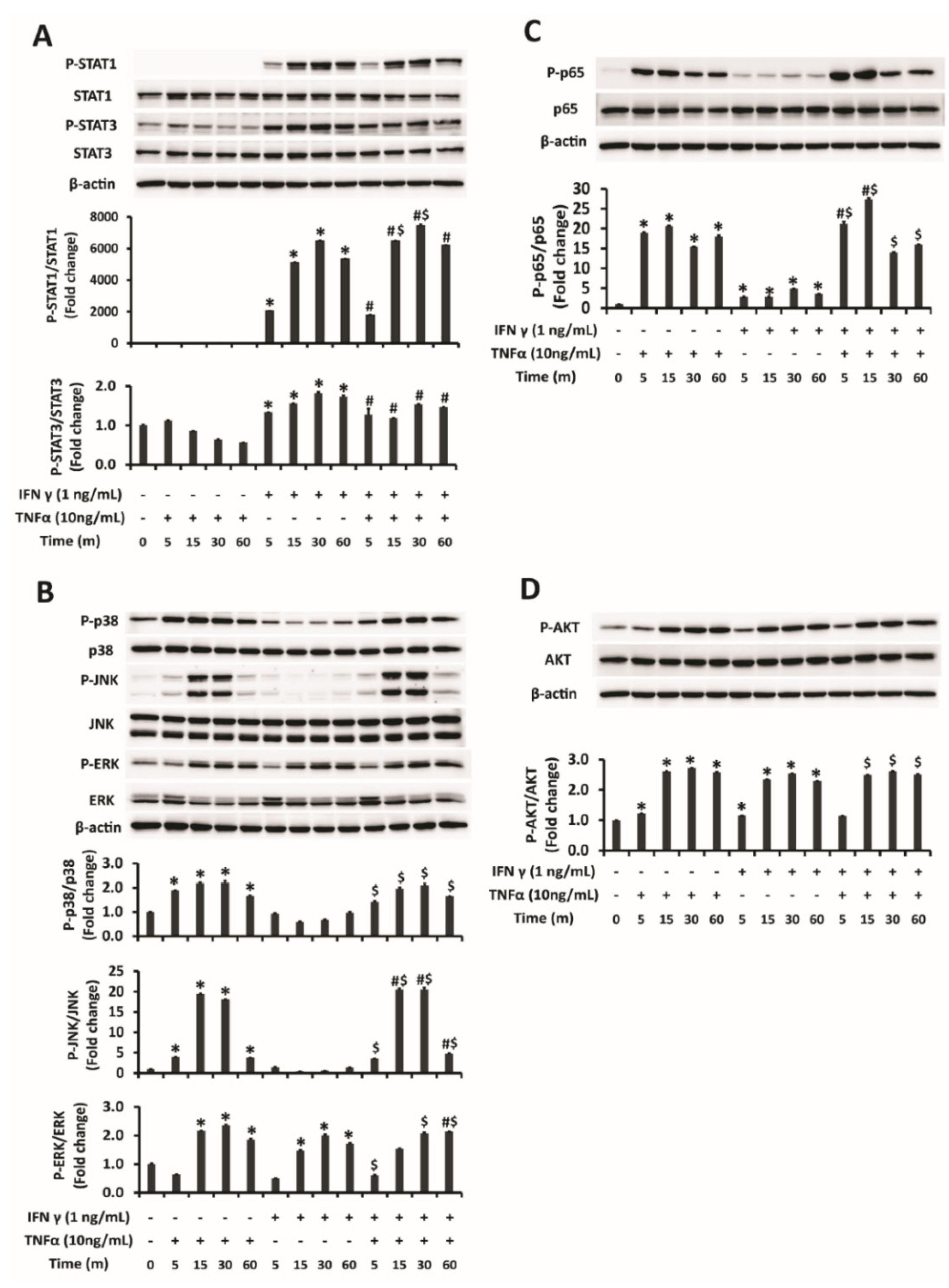
2.2. Activation of JNK of MAPK Pathway and NFĸB in Addition to STAT1 Is Responsible for Augmented Upregulation of PD-L1 by Combination Treatment of IFNγ and TNFα
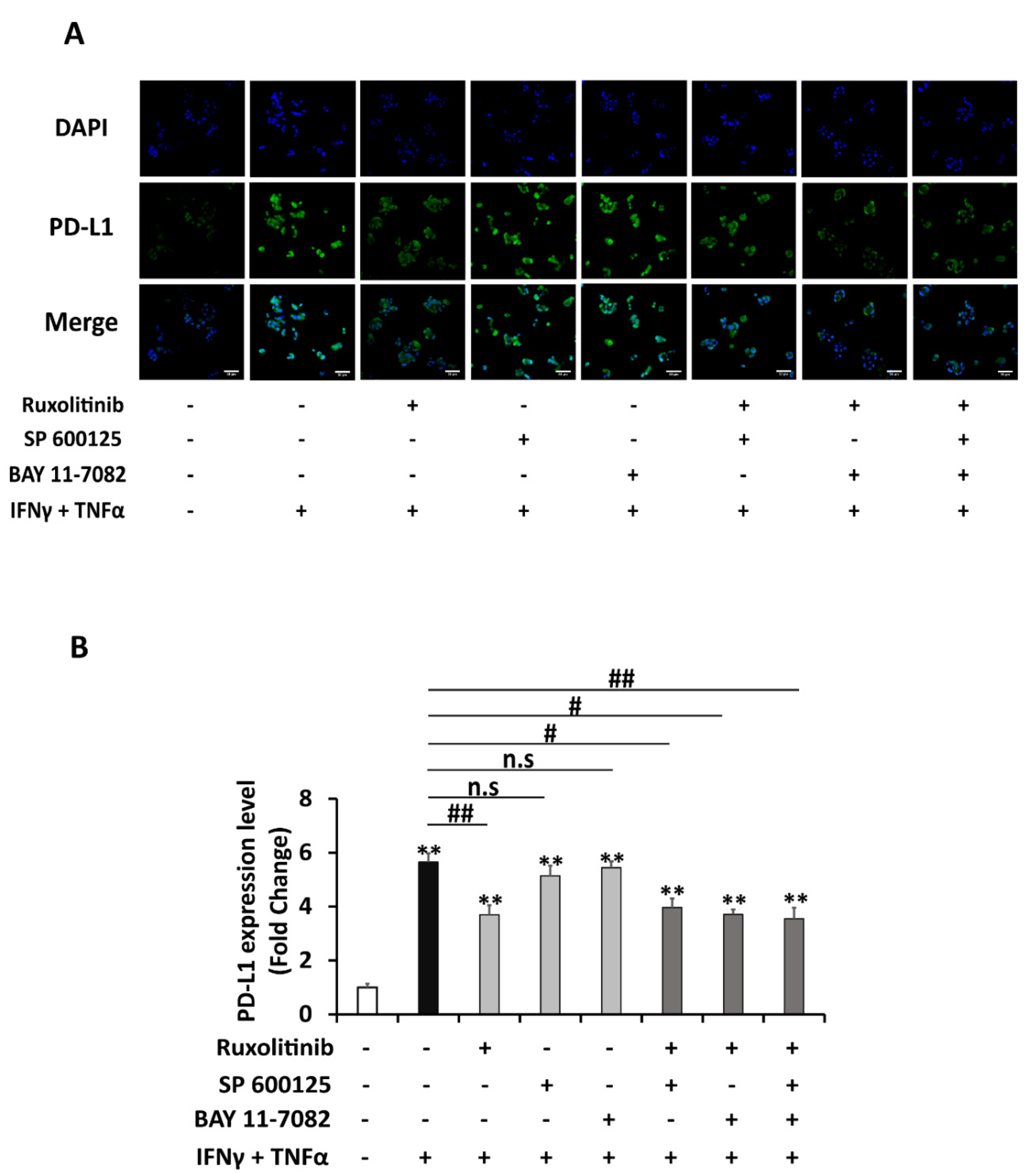
2.3. STAT1 Signaling Is Mainly Responsible for Enhanced Upregulation of PD-L1 by Combination Treatment of IFNγ and TNFα
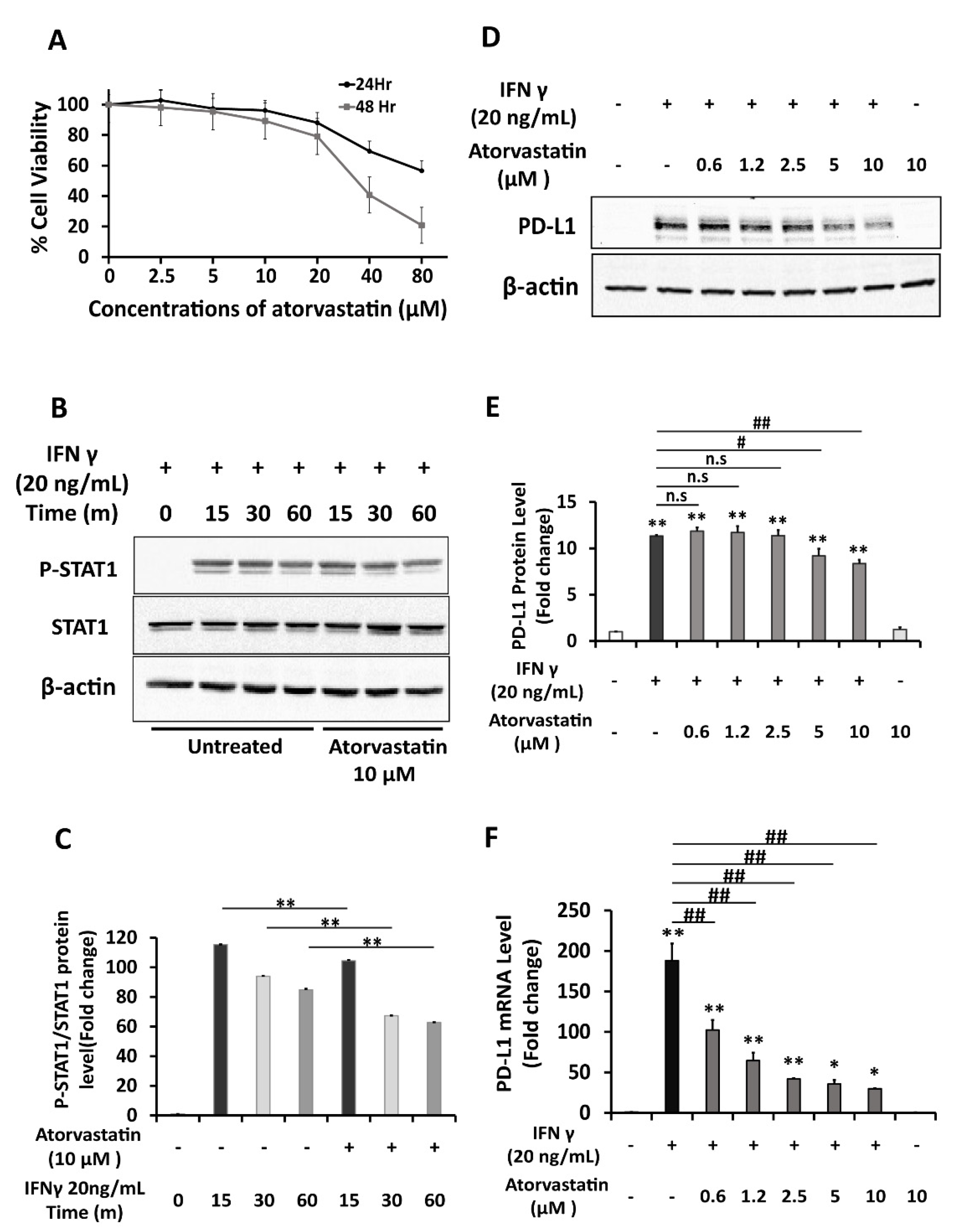
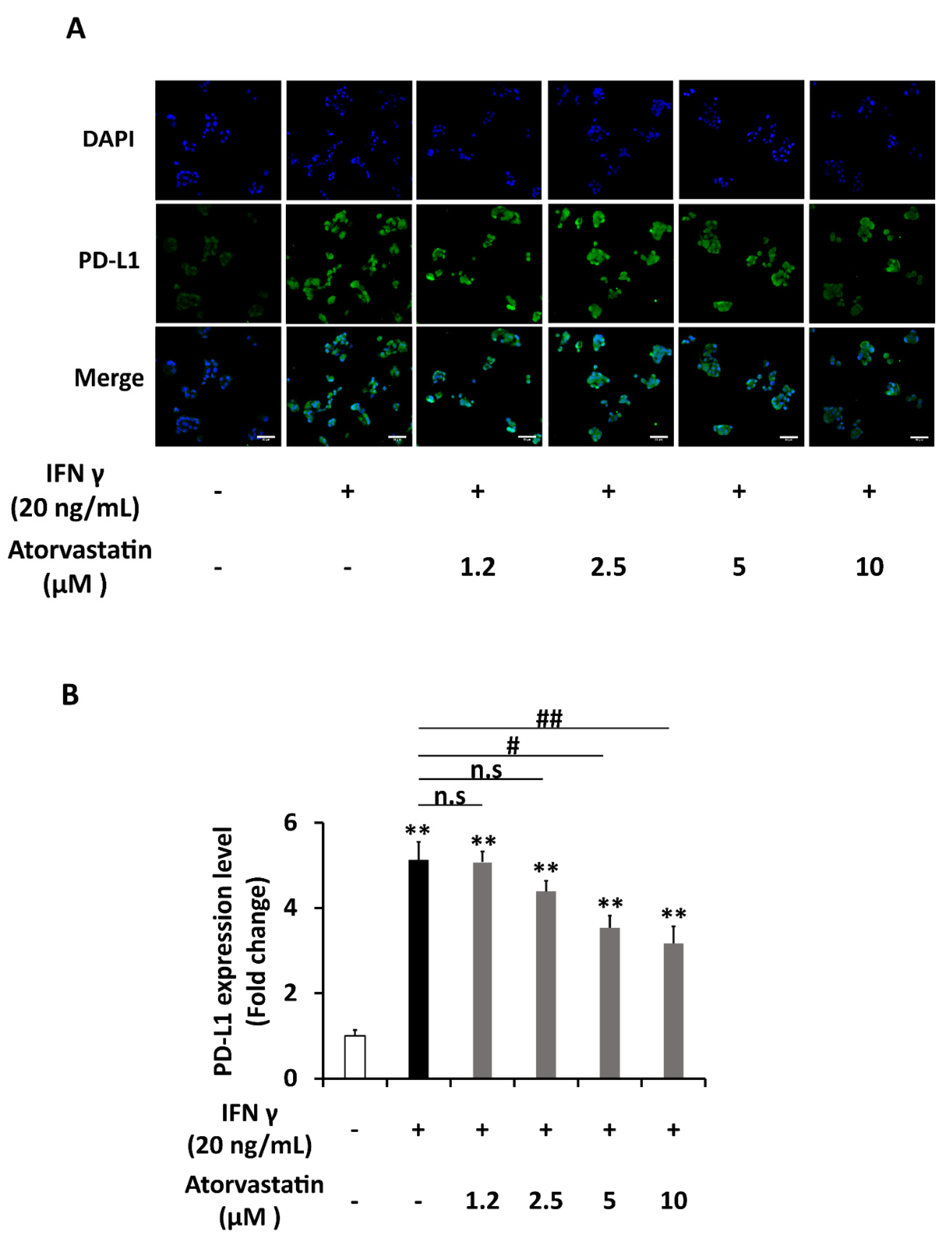
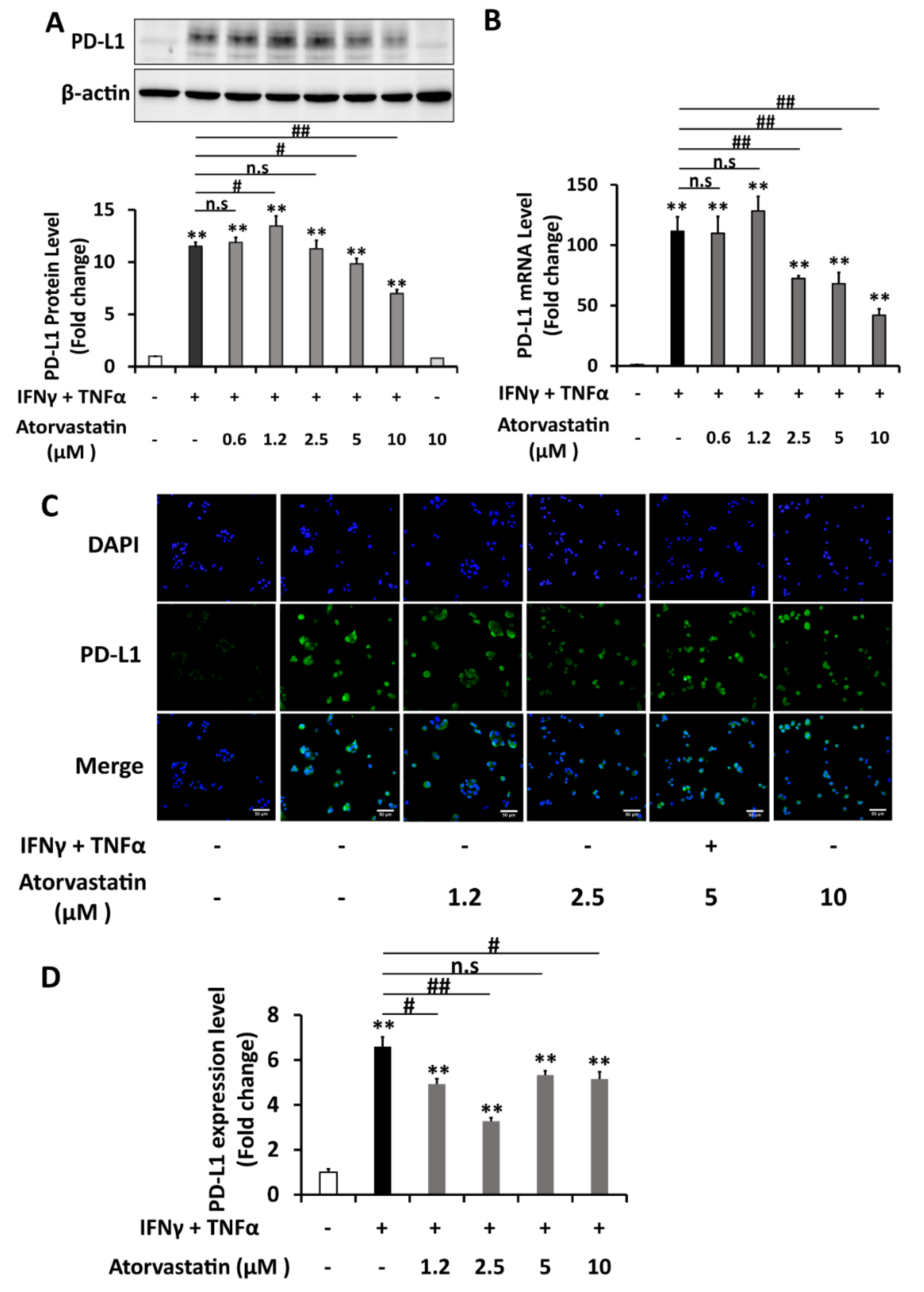
2.4. Atorvastatin Inhibits Induction of PD-L1 Expression in HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Cytotoxic Assay
4.4. Real-Time RT-PCR
| human PD-L1 forward primer | 5′ AAATGGAACCTGGCGAAAGC-3′ |
| human PD-L1 reverse primer | 5′ GATGAGCCCCTCAGGCATTT-3′ |
| human GAPDH forward primer | 5′TGGTATCGTGGAAGGACTCATGAC-3′ |
| human GAPDH reverse primer | 5′ATGCCACTCAGCTTCCCGTTCAGC-3′ |
4.5. Western Blotting Assay
4.6. Immunocytochemistry
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Johnston, M.P.; Khakoo, S.I. Immunotherapy for hepatocellular carcinoma: Current and future. World J. Gastroenterol. 2019, 25, 2977–2989. [Google Scholar] [CrossRef]
- Chen, H.J.; Hu, M.; Xu, F.G.; Xu, H.J.; She, J.J.; Xia, H.P. Understanding the inflammation-cancer transformation in the development of primary liver cancer. Hepatoma. Res. 2018, 4, 29. [Google Scholar] [CrossRef]
- Fu, J.; Xu, D.; Liu, Z.; Shi, M.; Zhao, P.; Fu, B.; Zhang, Z.; Yang, H.; Zhang, H.; Zhou, C.; et al. Increased regulatory T cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology 2007, 132, 2328–2339. [Google Scholar] [CrossRef]
- Arasanz, H.; Gato-Canas, M.; Zuazo, M.; Ibanez-Vea, M.; Breckpot, K.; Kochan, G.; Escors, D. PD1 signal transduction pathways in T cells. Oncotarget 2017, 8, 51936–51945. [Google Scholar] [CrossRef] [PubMed]
- Kythreotou, A.; Siddique, A.; Mauri, F.A.; Bower, M.; Pinato, D.J. Pd-L1. J. Clin. Pathol. 2018, 71, 189–194. [Google Scholar] [CrossRef]
- Solinas, A.; Calvisi, D.F. Programmed death ligand 1 expression in hepatocellular carcinoma: A prognostic marker and therapeutic target for liver cancer? Hepatology 2016, 64, 1847–1849. [Google Scholar] [CrossRef]
- Abiko, K.; Matsumura, N.; Hamanishi, J.; Horikawa, N.; Murakami, R.; Yamaguchi, K.; Yoshioka, Y.; Baba, T.; Konishi, I.; Mandai, M. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br. J. Cancer 2015, 112, 1501–1509. [Google Scholar] [CrossRef]
- Xie, Q.K.; Zhao, Y.J.; Pan, T.; Lyu, N.; Mu, L.W.; Li, S.L.; Shi, M.D.; Zhang, Z.F.; Zhou, P.H.; Zhao, M. Programmed death ligand 1 as an indicator of pre-existing adaptive immune responses in human hepatocellular carcinoma. Oncoimmunology 2016, 5, e1181252. [Google Scholar] [CrossRef]
- Schoop, R.; Wahl, P.; Le Hir, M.; Heemann, U.; Wang, M.; Wuthrich, R.P. Suppressed T-cell activation by IFN-gamma-induced expression of PD-L1 on renal tubular epithelial cells. Nephrol. Dial. Transpl. 2004, 19, 2713–2720. [Google Scholar] [CrossRef]
- Lim, S.O.; Li, C.W.; Xia, W.; Cha, J.H.; Chan, L.C.; Wu, Y.; Chang, S.S.; Lin, W.C.; Hsu, J.M.; Hsu, Y.H.; et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell 2016, 30, 925–939. [Google Scholar] [CrossRef]
- Hartley, G.; Regan, D.; Guth, A.; Dow, S. Regulation of PD-L1 expression on murine tumor-associated monocytes and macrophages by locally produced TNF-alpha. Cancer Immunol. Immunother. 2017, 66, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, J.; Zhang, N.; Zhuang, M.; Zong, Z.; Zou, J.; Li, G.; Wang, X.; Zhou, H.; Zhang, L.; et al. Cross-talk between TNF-alpha and IFN-gamma signaling in induction of B7-H1 expression in hepatocellular carcinoma cells. Cancer Immunol. Immunother. 2018, 67, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Stancu, C.; Sima, A. Statins: Mechanism of action and effects. J. Cell Mol. Med. 2001, 5, 378–387. [Google Scholar] [CrossRef]
- Yi, C.; Song, Z.; Wan, M.; Chen, Y.; Cheng, X. Statins intake and risk of liver cancer: A dose-response meta analysis of prospective cohort studies. Medicine 2017, 96, e7435. [Google Scholar] [CrossRef] [PubMed]
- Sarrabayrouse, G.; Pich, C.; Teiti, I.; Tilkin-Mariame, A.F. Regulatory properties of statins and rho gtpases prenylation inhibitiors to stimulate melanoma immunogenicity and promote anti-melanoma immune response. Int. J. Cancer 2017, 140, 747–755. [Google Scholar] [CrossRef]
- Greenwood, J.; Steinman, L.; Zamvil, S.S. Statin therapy and autoimmune disease: From protein prenylation to immunomodulation. Nat. Rev. Immunol. 2006, 6, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Han, D.; Kilaru, B.K.; Franek, B.S.; Niewold, T.B.; Reder, A.T. Inhibition of interferon-beta responses in multiple sclerosis immune cells associated with high-dose statins. Arch. Neurol. 2012, 69, 1303–1309. [Google Scholar] [CrossRef]
- Chung, H.K.; Lee, I.K.; Kang, H.; Suh, J.M.; Kim, H.; Park, K.C.; Kim, D.W.; Kim, Y.K.; Ro, H.K.; Shong, M. Statin inhibits interferon-gamma-induced expression of intercellular adhesion molecule-1 (ICAM-1) in vascular endothelial and smooth muscle cells. Exp. Mol. Med. 2002, 34, 451–461. [Google Scholar] [CrossRef][Green Version]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, J.; Deng, X.; Xiong, F.; Ge, J.; Xiang, B.; Wu, X.; Ma, J.; Zhou, M.; Li, X.; et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 2019, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Jiang, W.; Tian, D.; Guo, Q.; Shen, Z. Icotinib inhibits the proliferation of hepatocellular carcinoma cells in vitro and in vivo dependently on EGFR activation and PDL1 expression. Onco Targets Ther. 2018, 11, 8227–8237. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Mamessier, E.; Adelaide, J.; Chaffanet, M.; Ali, H.R.; Viens, P.; Caldas, C.; Birnbaum, D.; Bertucci, F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 2015, 6, 5449–5464. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Meng, L.; Liu, K.; Ji, B. Targeting the PD-L1/DNMT1 axis in acquired resistance to sorafenib in human hepatocellular carcinoma. Oncol. Rep. 2017, 38, 899–907. [Google Scholar] [CrossRef]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Gough, D.J.; Levy, D.E.; Johnstone, R.W.; Clarke, C.J. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008, 19, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Concha-Benavente, F.; Srivastava, R.M.; Trivedi, S.; Lei, Y.; Chandran, U.; Seethala, R.R.; Freeman, G.J.; Ferris, R.L. Identification of the Cell-Intrinsic and -Extrinsic Pathways Downstream of EGFR and IFNgamma That Induce PD-L1 Expression in Head and Neck Cancer. Cancer Res. 2016, 76, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Mimura, K.; Teh, J.L.; Okayama, H.; Shiraishi, K.; Kua, L.F.; Koh, V.; Smoot, D.T.; Ashktorab, H.; Oike, T.; Suzuki, Y.; et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018, 109, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.Y.; Zhou, R.; Hu, G.; Li, X.; Splinter, P.L.; O’Hara, S.P.; LaRusso, N.F.; Soukup, G.A.; Dong, H.; Chen, X.M. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J. Immunol. 2009, 182, 1325–1333. [Google Scholar] [CrossRef]
- Zhang, X.; Zeng, Y.; Qu, Q.; Zhu, J.; Liu, Z.; Ning, W.; Zeng, H.; Zhang, N.; Du, W.; Chen, C.; et al. PD-L1 induced by IFN-gamma from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int. J. Clin. Oncol. 2017, 22, 1026–1033. [Google Scholar] [CrossRef]
- Lee, S.K.; Seo, S.H.; Kim, B.S.; Kim, C.D.; Lee, J.H.; Kang, J.S.; Maeng, P.J.; Lim, J.S. IFN-gamma regulates the expression of B7-H1 in dermal fibroblast cells. J. Dermatol. Sci. 2005, 40, 95–103. [Google Scholar] [CrossRef]
- Liu, J.; Hamrouni, A.; Wolowiec, D.; Coiteux, V.; Kuliczkowski, K.; Hetuin, D.; Saudemont, A.; Quesnel, B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood 2007, 110, 296–304. [Google Scholar] [CrossRef]
- Kondo, A.; Yamashita, T.; Tamura, H.; Zhao, W.; Tsuji, T.; Shimizu, M.; Shinya, E.; Takahashi, H.; Tamada, K.; Chen, L.; et al. Interferon-gamma and tumor necrosis factor-alpha induce an immunoinhibitory molecule, B7-H1, via nuclear factor-kappaB activation in blasts in myelodysplastic syndromes. Blood 2010, 116, 1124–1131. [Google Scholar] [CrossRef]
- Guerra, B.; Recio, C.; Aranda-Tavio, H.; Guerra-Rodriguez, M.; Garcia-Castellano, J.M.; Fernandez-Perez, L. The Mevalonate Pathway, a Metabolic Target in Cancer Therapy. Front. Oncol. 2021, 11, 626971. [Google Scholar] [CrossRef]
- Omori, M.; Okuma, Y.; Hakozaki, T.; Hosomi, Y. Statins improve survival in patients previously treated with nivolumab for advanced non-small cell lung cancer: An observational study. Mol. Clin. Oncol. 2019, 10, 137–143. [Google Scholar] [CrossRef]
- Okoye, I.; Namdar, A.; Xu, L.; Crux, N.; Elahi, S. Atorvastatin downregulates co-inhibitory receptor expression by targeting Ras-activated mTOR signalling. Oncotarget 2017, 8, 98215–98232. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Qin, H.; Benveniste, E.N. Simvastatin inhibits IFN-gamma-induced CD40 gene expression by suppressing STAT-1alpha. J. Leukoc. Biol. 2007, 82, 436–447. [Google Scholar] [CrossRef]
- Ma, X.; Bi, E.; Lu, Y.; Su, P.; Huang, C.; Liu, L.; Wang, Q.; Yang, M.; Kalady, M.F.; Qian, J.; et al. Cholesterol Induces CD8(+) T Cell Exhaustion in the Tumor Microenvironment. Cell Metab. 2019, 30, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, M.; Guemiri, R.; Druillennec, S.; Girault, I.; Malka-Mahieu, H.; Shen, S.; Allard, D.; Martineau, S.; Welsch, C.; Agoussi, S.; et al. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat. Med. 2018, 24, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, T.S.; Hu-Lieskovan, S.; Ribas, A. Mechanisms of Resistance to PD-1 and PD-L1 Blockade. Cancer J. 2018, 24, 47–53. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shwe, T.H.; Pothacharoen, P.; Phitak, T.; Wudtiwai, B.; Kongtawelert, P. Atorvastatin Attenuates Programmed Death Ligand-1 (PD-L1) Induction in Human Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 8755. https://doi.org/10.3390/ijms22168755
Shwe TH, Pothacharoen P, Phitak T, Wudtiwai B, Kongtawelert P. Atorvastatin Attenuates Programmed Death Ligand-1 (PD-L1) Induction in Human Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences. 2021; 22(16):8755. https://doi.org/10.3390/ijms22168755
Chicago/Turabian StyleShwe, Thuzar Hla, Peraphan Pothacharoen, Thanyaluck Phitak, Benjawan Wudtiwai, and Prachya Kongtawelert. 2021. "Atorvastatin Attenuates Programmed Death Ligand-1 (PD-L1) Induction in Human Hepatocellular Carcinoma Cells" International Journal of Molecular Sciences 22, no. 16: 8755. https://doi.org/10.3390/ijms22168755
APA StyleShwe, T. H., Pothacharoen, P., Phitak, T., Wudtiwai, B., & Kongtawelert, P. (2021). Atorvastatin Attenuates Programmed Death Ligand-1 (PD-L1) Induction in Human Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences, 22(16), 8755. https://doi.org/10.3390/ijms22168755






