Enhanced Autophagic Activity Improved the Root Growth and Nitrogen Utilization Ability of Apple Plants under Nitrogen Starvation
Abstract
:1. Introduction
2. Results
2.1. Overexpression of MdATG10 Alleviated the Growth Limitation of Apple Plants under Nitrogen Deficiency Stress
2.2. Overexpression of MdATG10 Alleviated the Damage on the Photosynthetic System in Apple Plants under Nitrogen Deficiency Stress
2.3. Apple Plants Overexpressing MdATG10 Maintained Better Growth and Activity of the Root System under Nitrogen-Deficiency Stress
2.4. Apple Plants Overexpressing MdATG10 Maintained a Better Nitrogen Retention Ability under Nitrogen Deficiency Stress
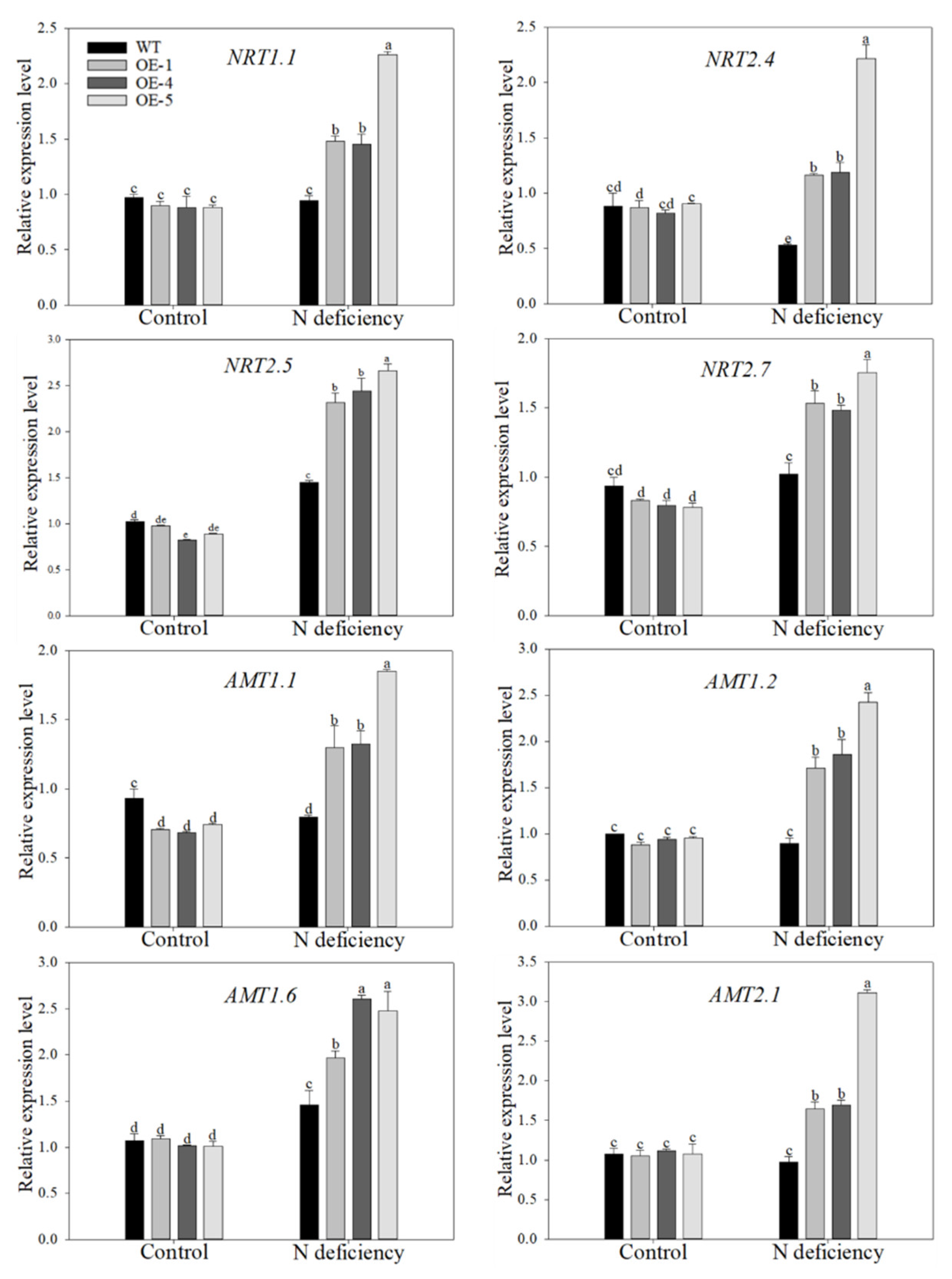
2.5. Overexpression of MdATG10 Promoted the Expression of Nitrogen Absorption Genes in Apple Leaves under Nitrogen Deficiency Stress
2.6. Overexpression of MdATG10 Intensified the Autophagic Activity in Apple Leaves under Nitrogen Deficiency Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatment
4.2. Growth Measurements
4.3. Evaluation of Photosynthetic Characteristics and Chlorophyll Fluorescence
4.4. Measurements of Root Architecture and Root Activity
4.5. Determination of Nitrogen Concentrations and Enzymatic Activities
4.6. RNA Extraction and qRT-PCR Analysis
4.7. Observation of Autophagosomes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Ding, G.D.; Yang, N.M.; White, P.J.; Ye, X.S.; Cai, H.M.; Lu, J.W.; Shi, L.; Xu, F.S. Comparative genome and transcriptome analysis unravels key factors of nitrogen use efficiency in Brassica napus L. Plant Cell Environ. 2020, 43, 712–731. [Google Scholar] [CrossRef]
- Leberecht, M.; Dannenmann, M.; Tejedor, J.; Simon, J.; Rennenberg, H.; Polle, A. Segregation of nitrogen use between ammonium and nitrate of ectomycorrhizas and beech trees. Plant Cell Environ. 2016, 39, 2691–2700. [Google Scholar] [CrossRef]
- Mu, X.H.; Chen, Q.W.; Chen, F.J.; Lixing, Y.X.; Mi, G.H. Within-leaf nitrogen allocation in Adaptation to low nitrogen supply in maize during grain-filling stage. Front. Plant Sci. 2016, 7, 699. [Google Scholar] [CrossRef]
- Mu, X.H.; Chen, Y.L. The physiological response of photosynthesis to nitrogen deficiency. Plant Physiol. Biochem. 2021, 158, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.F.; Sun, X.C.; Hu, C.X.; Tan, Q.L.; Zhao, X.H. Genotypic differences in nitrate uptake, translocation and assimilation of two Chinese cabbage cultivars Brassica campestris L. ssp Chinensis (L.). Plant Physiol. Biochem. 2013, 70, 14–20. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.N.; Gao, J.P.; Zhang, W.Z.; Yi, J.; Zhen, X.X.; Bi, C.Y.; He, D.W.; Liu, S.M.; Zhao, X.Y. Adaptation mechanism of roots to low and high nitrogen revealed by proteomic analysis. Rice 2021, 14, 1–14. [Google Scholar] [CrossRef]
- Gutierrez, R.A. Systems biology for enhanced plant nitrogen nutrition. Science 2012, 336, 1673–1675. [Google Scholar] [CrossRef]
- Evans, J.R.; Clarke, V.C. The nitrogen cost of photosynthesis. J. Exp. Bot. 2019, 70, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.L.; Song, H.X.; Liao, Q.; Yu, Y.; Jian, S.F.; Lepo, J.E.; Liu, Q.; Rong, X.M.; Tian, C.; Zeng, J.; et al. Nitrogen use efficiency is mediated by vacuolar nitrate sequestration capacity in roots of Brassica napus. Plant Physiol. 2016, 170, 1684–1698. [Google Scholar] [CrossRef] [Green Version]
- Qiu, K.; Li, Z.P.; Yang, Z.; Chen, J.Y.; Wu, S.X.; Zhu, X.Y.; Gao, S.; Gao, J.; Ren, G.D.; Kuai, B.K.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.B.; Li, C.; Fu, X.L.; Li, D.M.; Li, L.; Chen, X.D.; Wu, H.Y.; Cui, X.W.; Zhang, X.H.; Shen, H.Y.; et al. Effects of nitrate deficiency on nitrate assimilation and chlorophyll synthesis of detached apple leaves. Plant Physiol. Biochem. 2019, 142, 363–371. [Google Scholar] [CrossRef]
- Bassi, D.; Menossi, M.; Mattiello, L. Nitrogen supply influences photosynthesis establishment along the sugarcane leaf. Sci. Rep. 2018, 8, 2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Ju, C.X.; Buresh, R.J.; Wang, Z.Q.; Zhang, H.; Liu, L.J.; Yang, J.C.; Zhang, J.H. Root and shoot traits for rice varieties with higher grain yield and higher nitrogen use efficiency at lower nitrogen rates application. Field Crop. Res. 2015, 175, 47–55. [Google Scholar] [CrossRef]
- Huang, L.; Yu, L.J.; Zhang, X.; Fan, B.; Wang, F.Z.; Dai, Y.S.; Qi, H.; Zhou, Y.; Xie, L.J.; Xiao, S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019, 15, 407–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, W.; Beeckman, T.; Xu, G.H. Plant nitrogen nutrition: Sensing and signaling. Curr. Opin. Plant Biol. 2017, 39, 57–65. [Google Scholar] [CrossRef]
- Maeda, S.; Konishi, M.; Yanagisawa, S.; Omata, T. Nitrite transport activity of a novel HPP family protein conserved in cyanobacteria and chloroplasts. Plant Cell Physiol. 2014, 55, 1311–1324. [Google Scholar] [CrossRef] [Green Version]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018, 217, 35–53. [Google Scholar] [CrossRef] [Green Version]
- Coskun, D.; Britto, D.T.; Li, M.Y.; Becker, A.; Kronzucker, H.J. Rapid ammonia gas transport accounts for futile transmembrane cycling under NH3/NH4+ toxicity in plant roots. Plant Physiol. 2013, 163, 1859–1867. [Google Scholar] [CrossRef] [Green Version]
- McLoughlin, F.; Marshall, R.S.; Ding, X.X.; Chatt, E.C.; Kirkpatrick, L.D.; Augustine, R.C.; Li, F.Q.; Otegui, M.S.; Vierstra, R.D. Autophagy plays prominent roles in amino acid, nucleotide, and carbohydrate metabolism during fixed-carbon starvation in maize. Plant Cell 2020, 32, 2699–2724. [Google Scholar] [CrossRef]
- Yu, J.L.; Zhen, X.X.; Li, X.; Li, N.; Xu, F. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 2019, 10, 11. [Google Scholar] [CrossRef]
- Chen, Q.W.; Soulay, F.; Saudemont, B.; Elmayan, T.; Marmagne, A.; Masclaux-Daubresse, C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019, 60, 343–352. [Google Scholar] [CrossRef]
- Have, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar]
- Guiboileau, A.; Yoshimoto, K.; Soulay, F.; Bataille, M.P.; Avice, J.C.; Masclaux-Daubresse, C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012, 194, 732–740. [Google Scholar] [CrossRef]
- Kurusu, T.; Koyano, T.; Kitahata, N.; Kojima, M.; Hanamata, S.; Sakakibara, H.; Kuchitsu, K. Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 2017, 12, e1365211. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.Q.; Guo, Z.J.; Zhang, Z.J.; Jia, X.; Sun, Y.M.; Sun, X.; Wang, P.; Gong, X.Q.; Ma, F.W. The apple autophagy-related gene MdATG9 confers tolerance to low nitrogen in transgenic apple callus. Front. Plant Sci. 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Jia, X.; Huo, L.Q.; Che, R.M.; Gong, X.Q.; Wang, P.; Ma, F.W. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018, 41, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.R.; Suttangkakul, A.; Vierstra, R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008, 178, 1339–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, L.Q.; Guo, Z.J.; Jia, X.; Sun, X.; Wang, P.; Gong, X.Q.; Ma, F.W. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mugume, Y.; Basshama, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Guiboileau, A.; Avila-Ospina, L.; Yoshimoto, K.; Soulay, F.; Azzopardi, M.; Marmagne, A.; Lothier, J.; Masclaux-Daubresse, C. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 2013, 199, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Chen, M.; Zhong, L.; Liu, J.M.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; Guo, C.H.; Ma, Y.Z. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2015, 468, 800–806. [Google Scholar] [CrossRef]
- McLoughlin, F.; Augustine, R.C.; Marshall, R.S.; Li, F.Q.; Kirkpatrick, L.D.; Otegui, M.S.; Vierstra, R.D. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat. Plants 2018, 4, 1056–1070. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wei, Z.W.; Liang, D.; Zhou, S.S.; Li, Y.H.; Liu, C.H.; Ma, F.W. Enhanced salt resistance in apple plants overexpressing a Malus vacuolar Na+/H+ antiporter gene is associated with differences in stomatal behavior and photosynthesis. Plant Physiol. Biochem. 2013, 70, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Uribelarrea, M.; Crafts-Brandner, S.J.; Below, F.E. Physiological N response of field-grown maize hybrids (Zea mays L.) with divergent yield potential and grain protein concentration. Plant Soil 2009, 316, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Onodera, J.; Ohsumi, Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005, 280, 31582–31586. [Google Scholar] [CrossRef] [Green Version]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wiren, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzyakov, Y.; Xu, X.L. Competition between roots and microorganisms for nitrogen: Mechanisms and ecological relevance. New Phytol. 2013, 198, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Rentsch, D.; Schmidt, S.; Tegeder, M. Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 2007, 581, 2281–2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovacs, B.; Puskas-Preszner, A.; Huzsvai, L.; Levai, L.; Bodi, E. Effect of molybdenum treatment on molybdenum concentration and nitrate reduction in maize seedlings. Plant Physiol. Biochem. 2015, 96, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.L.; Li, M.J.; Zhou, K.; Sun, T.T.; Hu, L.Y.; Li, C.Y.; Ma, F.W. Uptake and metabolism of ammonium and nitrate in response to drought stress in Malus prunifolia. Plant Physiol. Biochem. 2018, 127, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Garnett, T.; Plett, D.; Conn, V.; Conn, S.; Rabie, H.; Rafalski, J.A.; Dhugga, K.; Tester, M.A.; Kaiser, B.N. Variation for N uptake system in maize: Genotypic response to N supply. Front. Plant Sci. 2015, 6, 936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.R.; Feng, H.M.; Tan, Y.W.; Xu, Y.L.; Miao, Q.S.; Xu, G.H. A putative 6-transmembrane nitrate transporter OsNRT1.1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 2016, 58, 590–599. [Google Scholar] [CrossRef] [Green Version]
- Ranathunge, K.; El-kereamy, A.; Gidda, S.; Bi, Y.M.; Rothstein, S.J. AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 2014, 65, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.N. Functional analysis of an arabidopsis T-DNA “knockout” of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol. 2002, 130, 1263–1275. [Google Scholar] [CrossRef]
- Ludewig, U.; Neuhäuser, B.; Dynowski, M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 2007, 581, 2301–2308. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Sun, X.; Jia, X.; Ma, F.W. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017, 256, 53–64. [Google Scholar] [CrossRef]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.Q.; Che, R.M.; Ma, F.W. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018, 16, 545–557. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.M.; Gao, T.T.; Zhang, Z.J.; Tan, K.X.; Li, C. The mitigation effects of exogenous dopamine on low nitrogen stress in Malus hupehensis. J. Integr. Agric. 2020, 19, 2709–2724. [Google Scholar] [CrossRef]
- Liang, B.W.; Gao, T.T.; Zhao, Q.; Ma, C.Q.; Chen, Q.; Wei, Z.W.; Li, C.Y.; Li, C.; Ma, F.W. Effects of exogenous dopamine on the uptake, transport, and resorption of apple ionome under moderate drought. Front. Plant Sci. 2018, 9, 755. [Google Scholar] [CrossRef] [PubMed]
- Perini, P.; Pasquali, G.; Margis-Pinheiro, M.; de Oliviera, P.R.; Revers, L. Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus × domestica Borkh.). Mol. Breed. 2014, 34, 829–842. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

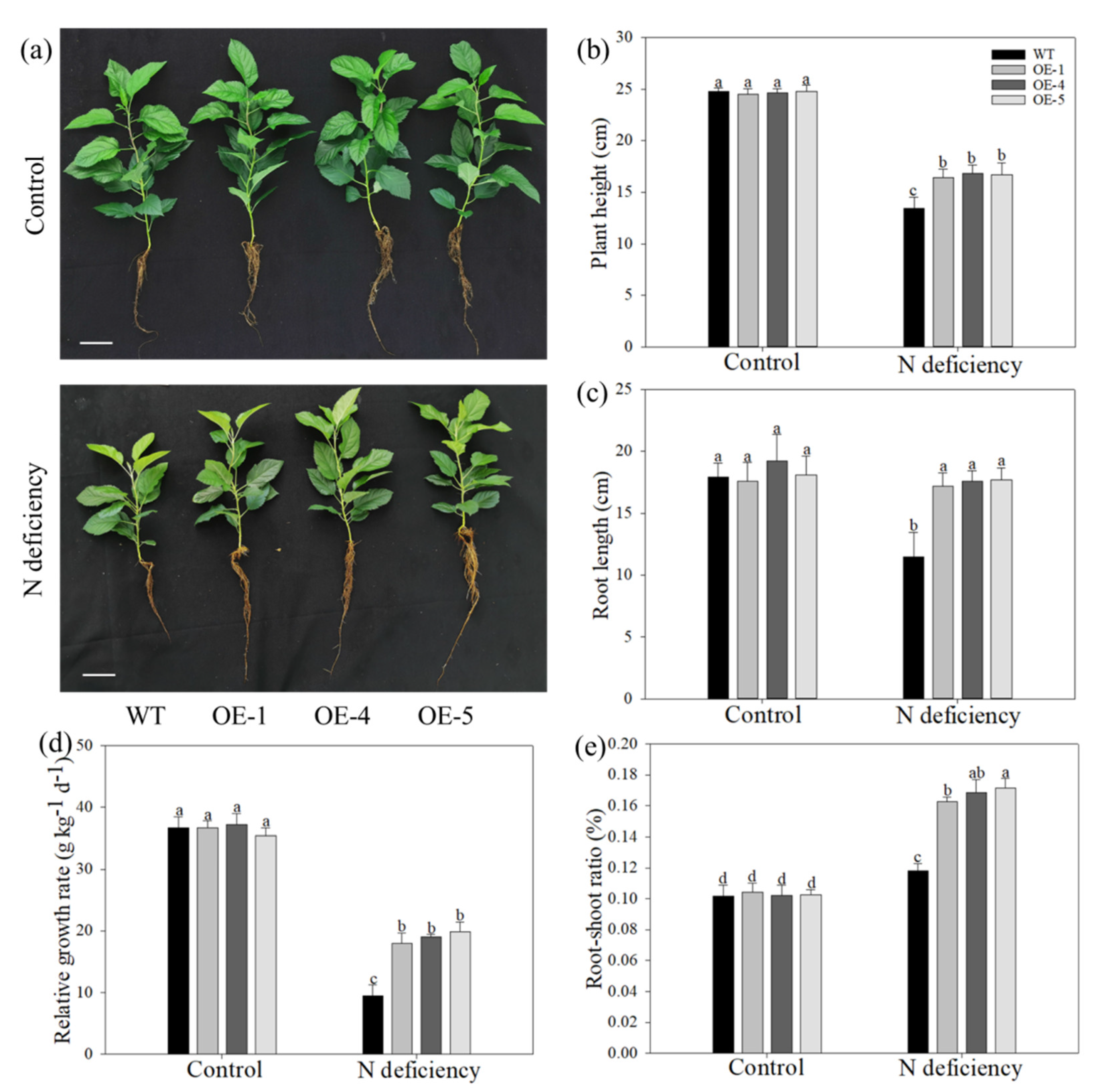

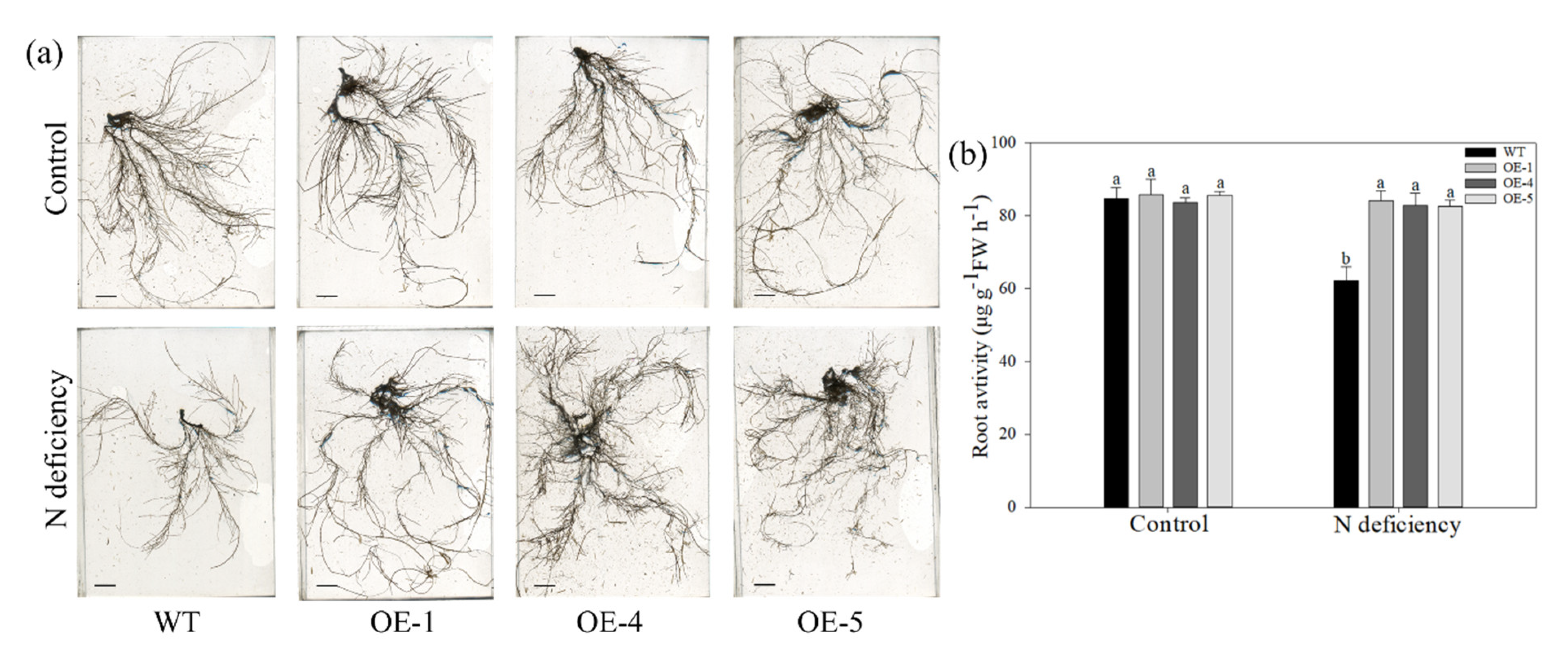
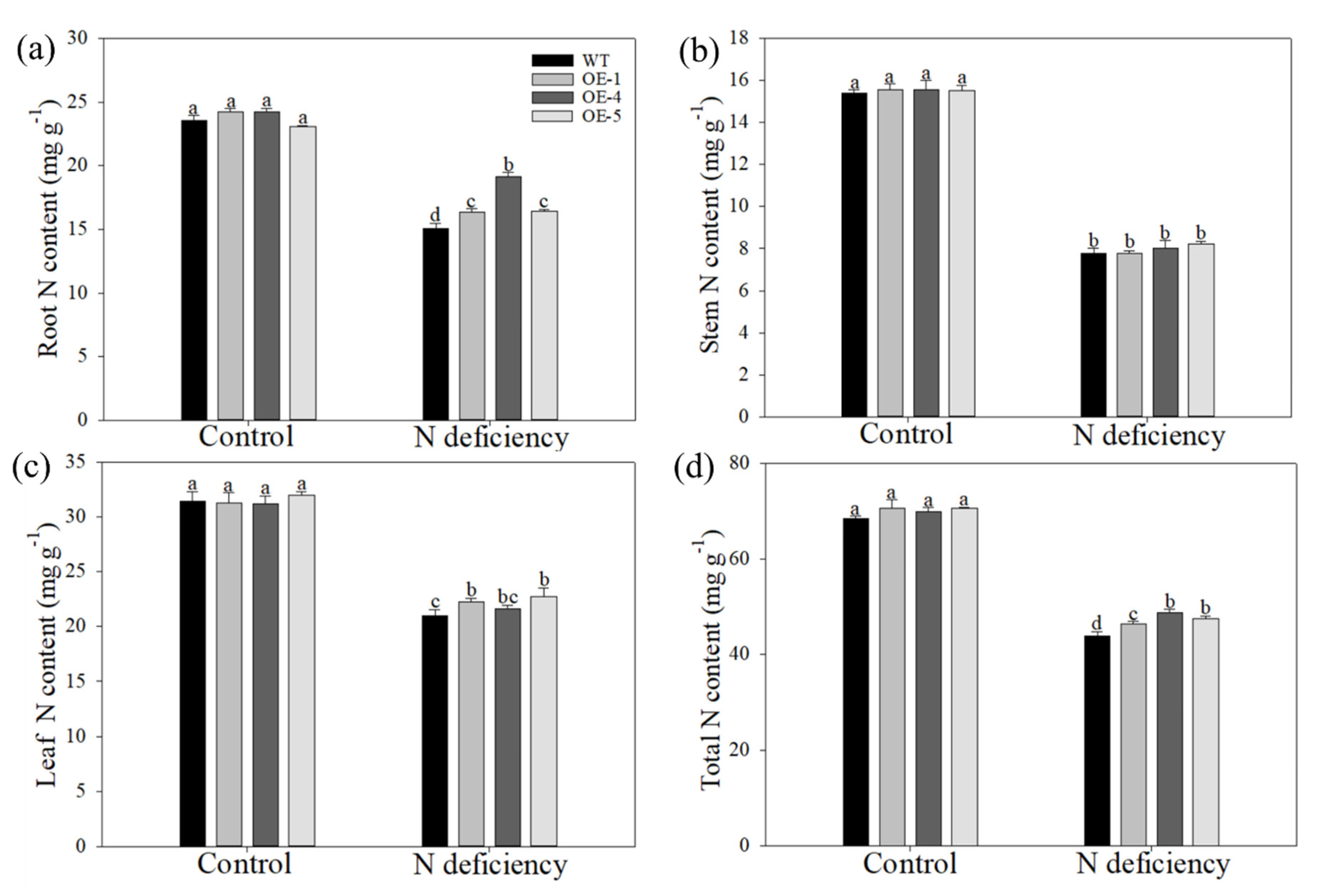
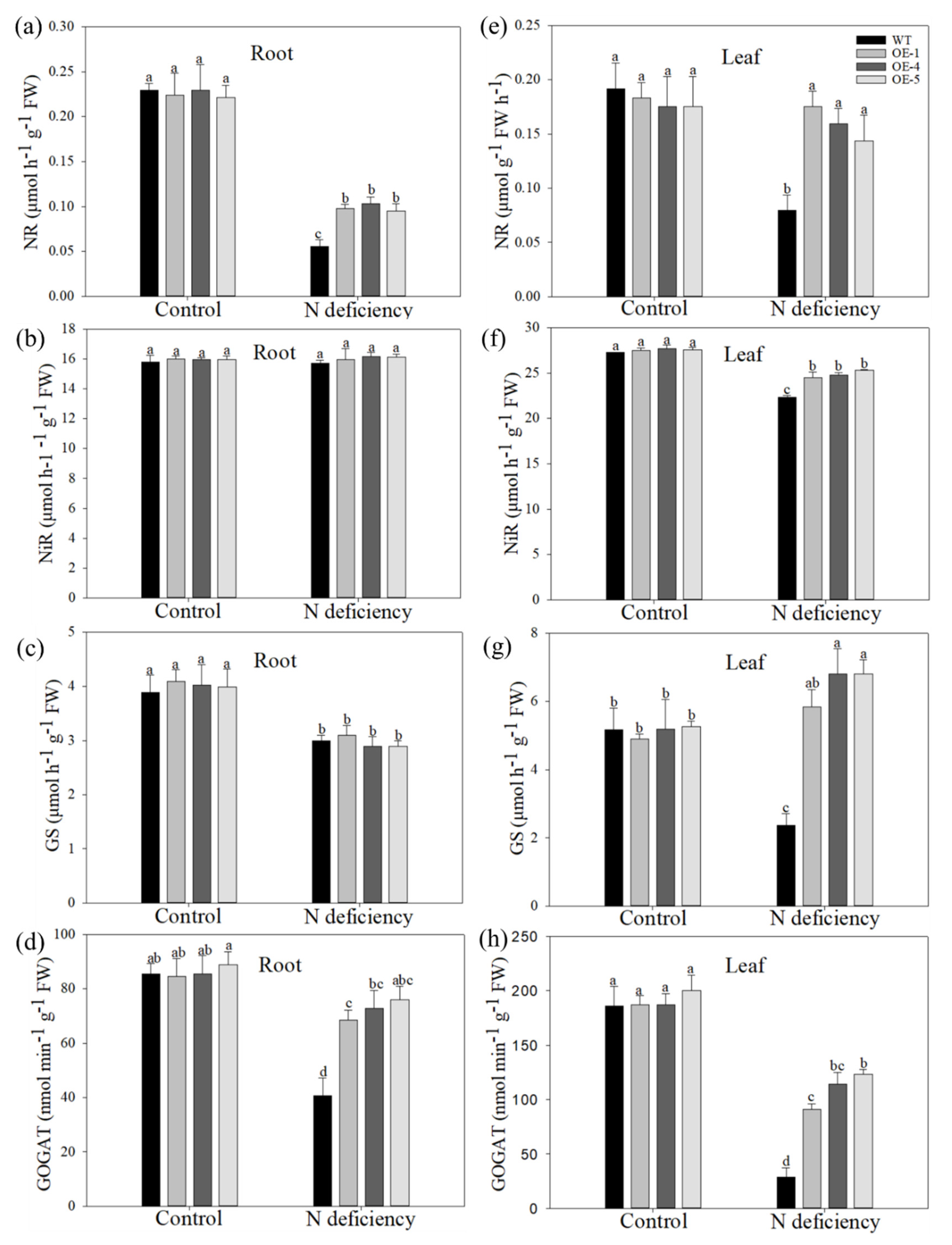

| Root FW (g plant−1) | Stem FW (g plant−1) | Leaf FW (g plant−1) | Total FW (g plant−1) | Root DW (g plant−1) | Stem DW (g plant−1) | Leaf DW (g plant−1) | Total DW (g plant−1) | |
|---|---|---|---|---|---|---|---|---|
| WT–CK | 1.063 ± 0.069 a | 1.429 ± 0.043 a | 3.675 ± 0.129 a | 6.168 ± 0.229 a | 0.206 ± 0.007 ab | 0.815 ± 0.027 a | 1.222 ± 0.093 a | 2.244 ± 0.118 a |
| OE–1–CK | 1.031 ± 0.109 a | 1.408 ± 0.029 a | 3.672 ± 0.075 a | 6.110 ± 0.054 a | 0.211 ± 0.006 ab | 0.803 ± 0.008 a | 1.223 ± 0.082 a | 2.237 ± 0.076 a |
| OE–4–CK | 1.081 ± 0.075 a | 1.436 ± 0.036 a | 3.634 ± 0.077 a | 6.151 ± 0.112 a | 0.207 ± 0.007 ab | 0.805 ± 0.005 a | 1.237 ± 0.113 a | 2.249 ± 0.118 a |
| OE–5–CK | 1.029 ± 0.071 a | 1.425 ± 0.036 a | 3.643 ± 0.058 a | 6.089 ± 0.128 a | 0.202 ±0.004 ab | 0.803 ± 0.004 a | 1.189 ± 0.062 a | 2.196 ± 0.062 a |
| WT–ND | 0.568 ± 0.076 b | 0.726 ± 0.029 c | 2.185 ± 0.069 c | 3.476 ± 0.059 c | 0.106 ± 0.009 d | 0.256 ± 0.009 c | 0.699 ± 0.045 c | 1.062 ± 0.049 c |
| OE–1–ND | 1.004 ± 0.037 a | 1.024 ± 0.101 b | 2.692 ± 0.065 b | 4.717 ± 0.065 b | 0.183 ± 0.005 c | 0.299 ± 0.009 b | 0.829 ± 0.012 bc | 1.312 ± 0.026 b |
| OE–4–ND | 1.122 ± 0.054 a | 1.065 ± 0.131 b | 2.739 ± 0.061 b | 4.927 ± 0.207 b | 0.197 ± 0.007 b | 0.303 ± 0.008 b | 0.866 ± 0.036 b | 1.367 ± 0.026 b |
| OE–5–ND | 1.097 ± 0.043 a | 1.069 ± 0.132 b | 2.938 ± 0.042 b | 4.905 ± 0.131 b | 0.201 ± 0.005 ab | 0.303 ± 0.008 b | 0.871 ± 0.026 b | 1.375 ± 0.029 b |
| Length (cm) | Surf Area (cm2) | Avg Diam (mm) | Root Volume (cm3) | Forks | |
|---|---|---|---|---|---|
| WT–CK | 1256.30 ± 42.30 ab | 93.85 ± 4.16 a | 0.24 ± 0.01 ab | 0.63 ± 0.02 ab | 26428 ± 3288.17 a |
| OE–1–CK | 1288.40 ± 26.28a | 93.88 ± 3.22 a | 0.28 ± 0.02 ab | 0.63 ± 0.01 a | 25322 ± 2156.41 a |
| OE–4–CK | 1296.20 ± 47.97 a | 93.08 ± 1.28 a | 0.28 ± 0.04 ab | 0.65 ± 0.12 ab | 26348 ± 2466.29 a |
| OE–5–CK | 1284.70 ± 53.31 a | 92.96 ± 2.45 a | 0.26 ± 0.02 ab | 0.65 ± 0.08 ab | 23746 ± 2466.29 a |
| WT–ND | 556.99 ± 26.72 d | 42.21 ± 2.25 c | 0.24 ± 0.01 b | 0.26 ± 0.02 c | 8692 ± 199.56 b |
| OE–1–ND | 927.00 ± 17.36 c | 78.33 ±8.01 b | 0.28 ± 0.03 ab | 0.55 ± 0.06 ab | 20846 ± 1929.27 a |
| OE–4–ND | 1160.50 ± 41.10 b | 102.81 ± 2.89 a | 0.25 ± 0.01 ab | 0.66 ± 0.01 ab | 22855 ± 1902.22 a |
| OE–5–ND | 1161.60 ± 34.84 b | 100.93 ± 3.28 a | 0.30 ± 0.03 a | 0.76 ± 0.04 a | 22432 ± 1805.93 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, L.; Guo, Z.; Wang, Q.; Cheng, L.; Jia, X.; Wang, P.; Gong, X.; Li, C.; Ma, F. Enhanced Autophagic Activity Improved the Root Growth and Nitrogen Utilization Ability of Apple Plants under Nitrogen Starvation. Int. J. Mol. Sci. 2021, 22, 8085. https://doi.org/10.3390/ijms22158085
Huo L, Guo Z, Wang Q, Cheng L, Jia X, Wang P, Gong X, Li C, Ma F. Enhanced Autophagic Activity Improved the Root Growth and Nitrogen Utilization Ability of Apple Plants under Nitrogen Starvation. International Journal of Molecular Sciences. 2021; 22(15):8085. https://doi.org/10.3390/ijms22158085
Chicago/Turabian StyleHuo, Liuqing, Zijian Guo, Qi Wang, Li Cheng, Xin Jia, Ping Wang, Xiaoqing Gong, Cuiying Li, and Fengwang Ma. 2021. "Enhanced Autophagic Activity Improved the Root Growth and Nitrogen Utilization Ability of Apple Plants under Nitrogen Starvation" International Journal of Molecular Sciences 22, no. 15: 8085. https://doi.org/10.3390/ijms22158085
APA StyleHuo, L., Guo, Z., Wang, Q., Cheng, L., Jia, X., Wang, P., Gong, X., Li, C., & Ma, F. (2021). Enhanced Autophagic Activity Improved the Root Growth and Nitrogen Utilization Ability of Apple Plants under Nitrogen Starvation. International Journal of Molecular Sciences, 22(15), 8085. https://doi.org/10.3390/ijms22158085






