Seeing Keratinocyte Proteins through the Looking Glass of Intrinsic Disorder
Abstract
:1. Introduction
1.1. Protein Intrinsic Disorder and Keratinocyte Biology
1.2. Epidermal Specialization and Keratinocyte-Related Protein Intrinsic Disorder
- Proteins of infecting bacterial and viral pathogens, the latter notably highlighting HPV oncoproteins;
- The dermal extracellular matrix protein elastin;
- Subdomains of familiar keratinocyte proteins, e.g., EGF receptor C-terminus and keratin N- and C-termini, for mediating protein–protein interactions in signaling and structural assembly, respectively;
- Specializations of non-human skin proteins for bio-reflectance or protection via skin-associated toxins in other organisms.
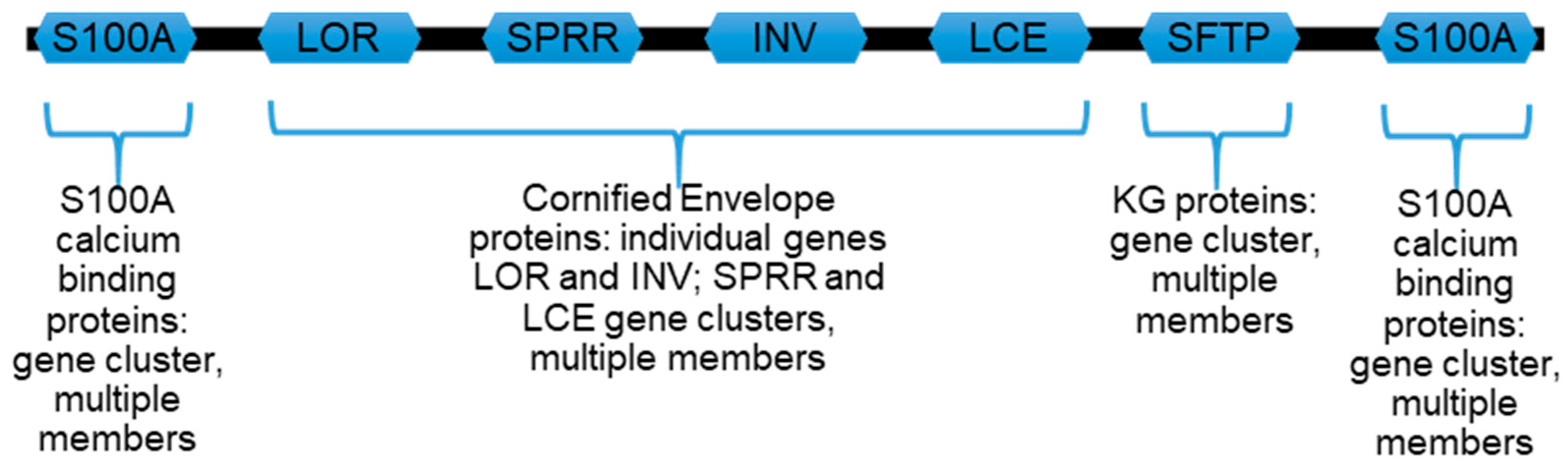
1.3. Proteins Encoded by Genes of the Human EDC Are Enriched for ID Traits
2. Bioinformatic Evaluations of Keratinocyte-Specific Proteins from the EDC Locus
2.1. Assessing Protein Intrinsic Disorder Encoded in the EDC
2.1.1. S100A Proteins
2.1.2. Loricrin, Involucrin, SPRR, and LCE
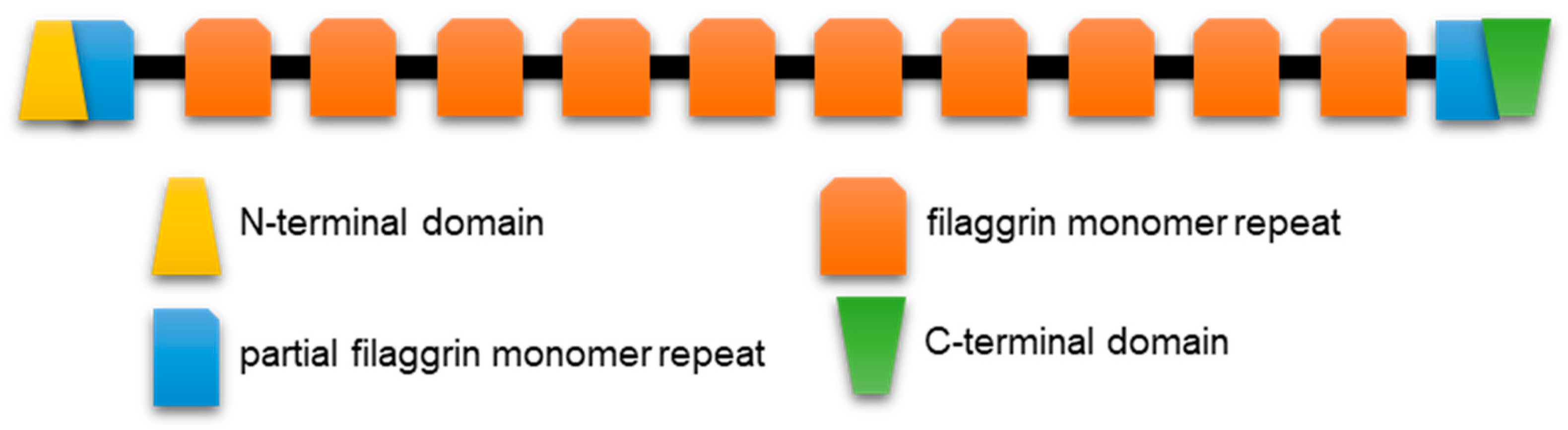
2.1.3. Profilaggrin and Related S100 Fused-Type Proteins (SFTPs)
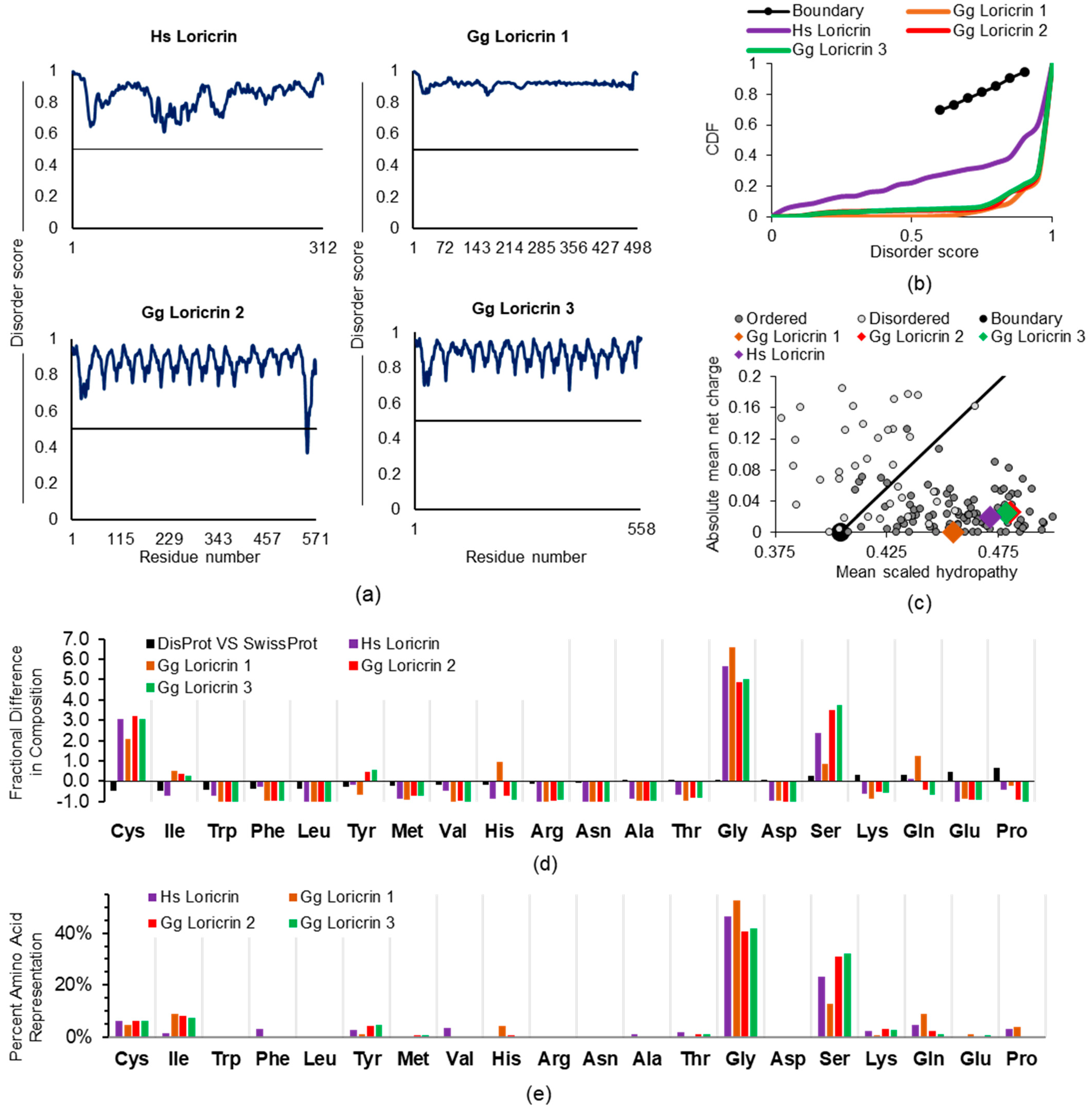
2.2. Examining Intrinsic Disorder in the EDC Proteins of Non-Human Species
2.2.1. General EDC Protein Considerations across Species
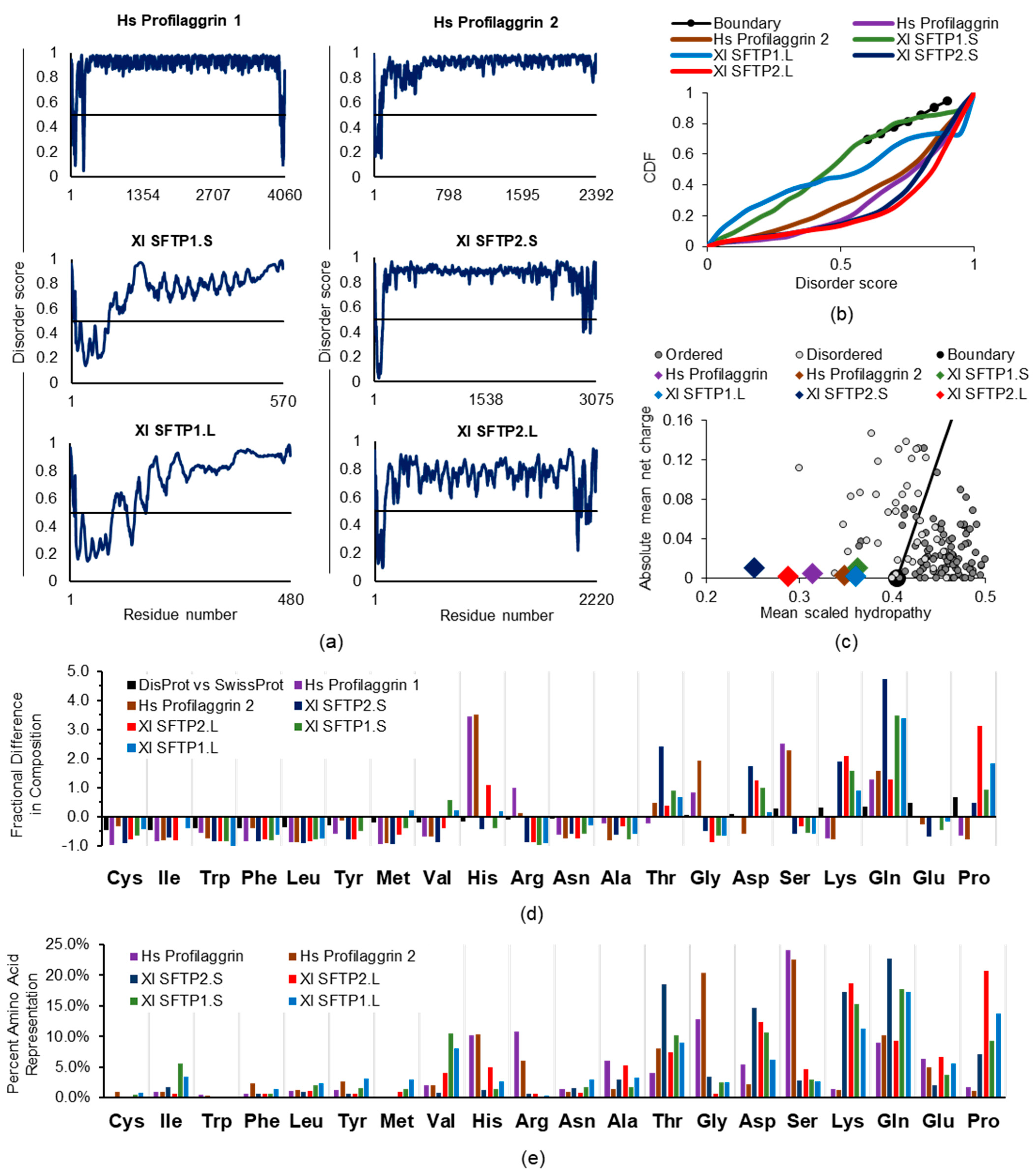
2.2.2. SFTP Disorder: Frog Versus Human Sequence Considerations
2.2.3. SFTP Disorder and Liquid–Liquid Phase Separation: Frog Versus Human Sequence Evaluations
2.2.4. SFTP Liquid–Liquid Phase Separation: Further In Silico Assessments
3. Methodology
3.1. Literature Inquiry
3.2. Bioinformatics Assessments
4. Conclusions
4.1. Protein ID in the Keratinocyte Proteome Facilitates Cell Function
4.2. Investigations of Protein ID Synergize with Keratinocyte Biology
- Despite the few reports to date, the importance of IDPs in cutaneous biology can be expected to be the rule, not the exception. Intrinsic disorder is likely integral to the conformation and function not only of numerous endogenous keratinocyte proteins but also in therapeutically counteracting biofilm proteins of skin bacteria, e.g., Staphylococcus epidermidis, and oncogenic proteins of keratinocyte-tropic papilloma virus [25,29].
- Keratinocyte IDPs and proteins with extensive IDR, especially in upper epidermal strata, may be particularly favored in these superficial cells. Typical IDP qualities of being minimally affected (i.e., denatured) by harsh conditions or possessing “conformational plasticity” [9] could lend to understanding the resiliency of these cells as they are subjected to varying surface environmental assaults.
- The ID derived from numerous repeats enriched for disorder-promoting amino acids within keratinocyte-specific proteins may advantageously contribute to increased mammalian SFTP proteolytic sensitivity, efficiently yielding antimicrobial and hydrative peptides, respectively, from hornerin and filaggrin [5,8], and might be harnessed for clinical benefit by purposefully regulating the breakdown.
- Learning from human profilaggrin protein liquid–liquid phase separation for KG, it is worth conducting future direct biophysical experimentation with other keratinocyte-manifested structures such as cornified envelopes in the context of proteinaceous membraneless organelles (PMLO). This is especially appropriate in light of CE involucrin and loricrin self–self-protein interactions and their repeating amino acid motifs [74], which are biomolecular condensate- and PMLO-germane characteristics [75]. Strengthening this rationale is loricrin’s very favorable, pro-granule scoring in the LARKS and catGRANULE analysis. Investigation of keratinocyte sheath-like CE could add to the granule, speckle, and droplet categories of PMLO.
- Approximately 30 years of intense and elegant structural biology investigations of epithelial proteins such as keratins ([106] for review) have yielded a tremendous understanding regarding their function in tissue health and multiple disease states. We must now also face the fact that about one half of all eukaryotic proteins reside in a “dark proteome” [107] where conformational studies are often thwarted because of ID. As with Alice, we must reconsider what we think we know. It stands to reason that many keratinocyte-specific and -expressed proteins under current investigation, along with those of more established structure–function relationships, might be newly and revealingly viewed through the looking glass of ID. In this regard, keratinocyte normal physiology and pathophysiology can also be greatly affected by more broadly expressed IDPs, such as TNIP1, a repressor of inflammatory signaling, as we recently reported [53,108]. Additionally, there are the new lessons that the unique proteins of epidermal keratinocytes might provide to the IDP field. An integrated approach recently reviewed by Fuxreiter and colleagues [109] for membraneless organelles, protein–protein interaction, and phase separation in neurodegenerative disorders provides a roadmap for combining and applying ID computational and biophysical methodologies to other cell types. Such investigations of keratinocyte proteins could be a sea change moment for conformational understanding and subsequent translational cutaneous health benefits.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Csizmok, V.; Follis, A.V.; Kriwacki, R.W.; Forman-Kay, J.D. Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling. Chem. Rev. 2016, 116, 6424–6462. [Google Scholar] [CrossRef]
- Uversky, V.N. New technologies to analyse protein function: An intrinsic disorder perspective. F1000Research 2020, 9, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog. Mol. Biol. Transl. Sci. 2019, 166, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rice, G.; Rompolas, P. Advances in resolving the heterogeneity and dynamics of keratinocyte differentiation. Curr. Opin. Cell Biol. 2020, 67, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Latendorf, T.; Gerstel, U.; Wu, Z.; Bartels, J.; Becker, A.; Tholey, A.; Schröder, J.-M. Cationic Intrinsically Disordered Antimicrobial Peptides (CIDAMPs) Represent a New Paradigm of Innate Defense with a Potential for Novel Anti-Infectives. Sci. Rep. 2019, 9, 3331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuusa, J.; Koski, M.K.; Ruskamo, S.; Tasanen, K. The intracellular domain of BP180/collagen XVII is intrinsically disordered and partially folds in an anionic membrane lipid-mimicking environment. Amino Acids 2020, 52, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiroz, F.G.; Fiore, V.F.; Levorse, J.; Polak, L.; Wong, E.; Pasolli, H.A.; Fuchs, E. Liquid-liquid phase separation drives skin barrier formation. Science 2020, 367, eaax9554. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; McLean, W.I. One Remarkable Molecule: Filaggrin. J. Investig. Dermatol. 2012, 132, 751–762. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Paradoxes and wonders of intrinsic disorder: Stability of instability. Intrinsically Disord. Proteins 2017, 5, e1327757. [Google Scholar] [CrossRef] [Green Version]
- Shamilov, R.; Staid, M.J.; Aneskievich, B.J. In Silico and In Vitro Considerations of Keratinocyte Nuclear Receptor Protein Structural Order for Improving Experimental Analysis. Methods Mol. Biol. 2019, 2109, 93–111. [Google Scholar] [CrossRef]
- Levenson, R.; Bracken, C.; Sharma, C.; Santos, J.; Arata, C.; Malady, B.; Morse, D.E. Calibration between trigger and color: Neutralization of a genetically encoded coulombic switch and dynamic arrest precisely tune reflectin assembly. J. Biol. Chem. 2019, 294, 16804–16815. [Google Scholar] [CrossRef] [Green Version]
- Kurvits, L.; Reimann, E.; Kadastik-Eerme, L.; Truu, L.; Kingo, K.; Erm, T.; Koks, S.; Taba, P.; Planken, A. Serum Amyloid Alpha Is Downregulated in Peripheral Tissues of Parkinson’s Disease Patients. Front. Neurosci. 2019, 13, 13. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, K.; Sako, Y. Single-Molecule Förster Resonance Energy Transfer Measurement Reveals the Dynamic Partially Ordered Structure of the Epidermal Growth Factor Receptor C-Tail Domain. J. Phys. Chem. B 2018, 123, 571–581. [Google Scholar] [CrossRef]
- Moens, M.A.; Pérez-Tris, J.; Cortey, M.; Benitez, L. Identification of two novel CRESS DNA viruses associated with an Avipoxvirus lesion of a blue-and-gray Tanager ( Thraupis episcopus ). Infect. Genet. Evol. 2018, 60, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, A.; Deka, G.; Prabhavathi, M.; Savithri, H.; Murthy, M.; Raja, A. Structural and biophysical characterization of Rv3716c, a hypothetical protein from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2018, 495, 982–987. [Google Scholar] [CrossRef]
- Singh, I.; Singh, S.; Verma, V.; Uversky, V.N.; Chandra, R. In silico evaluation of the resistance of the T790M variant of epidermal growth factor receptor kinase to cancer drug Erlotinib. J. Biomol. Struct. Dyn. 2017, 36, 4209–4219. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, S.; Pomès, R. The liquid structure of elastin. eLife 2017, 6, e26526. [Google Scholar] [CrossRef]
- Yarawsky, A.; English, L.R.; Whitten, S.T.; Herr, A.B. The Proline/Glycine-Rich Region of the Biofilm Adhesion Protein Aap Forms an Extended Stalk that Resists Compaction. J. Mol. Biol. 2017, 429, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Keppel, T.R.; Sarpong, K.; Murray, E.M.; Monsey, J.; Zhu, J.; Bose, R. Biophysical Evidence for Intrinsic Disorder in the C-terminal Tails of the Epidermal Growth Factor Receptor (EGFR) and HER3 Receptor Tyrosine Kinases. J. Biol. Chem. 2017, 292, 597–610. [Google Scholar] [CrossRef] [Green Version]
- Muiznieks, L.D.; Miao, M.; Sitarz, E.E.; Keeley, F.W. Contribution of domain 30 of tropoelastin to elastic fiber formation and material elasticity. Biopolymers 2016, 105, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Levenson, R.; Bracken, C.; Bush, N.; Morse, D.E. Cyclable Condensation and Hierarchical Assembly of Metastable Reflectin Proteins, the Drivers of Tunable Biophotonics. J. Biol. Chem. 2016, 291, 4058–4068. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Merillat, S.A.; Vincent, M.; Huber, A.; Basrur, V.; Mangelberger, D.; Zeng, L.; Elenitoba-Johnson, K.; Miller, R.A.; Irani, D.N.; et al. Loss of the Ubiquitin-conjugating Enzyme UBE2W Results in Susceptibility to Early Postnatal Lethality and Defects in Skin, Immune, and Male Reproductive Systems. J. Biol. Chem. 2016, 291, 3030–3042. [Google Scholar] [CrossRef] [Green Version]
- Bray, D.; Walsh, T.R.; Noro, M.G.; Notman, R. Complete Structure of an Epithelial Keratin Dimer: Implications for Intermediate Filament Assembly. PLoS ONE 2015, 10, e0132706. [Google Scholar] [CrossRef] [Green Version]
- Kornreich, M.; Avinery, R.; Malka-Gibor, E.; Laser-Azogui, A.; Beck, R. Order and disorder in intermediate filament proteins. FEBS Lett. 2015, 589, 2464–2476. [Google Scholar] [CrossRef] [Green Version]
- Whelan, F.; Potts, J.R. Two repetitive, biofilm-forming proteins from Staphylococci: From disorder to extension. Biochem. Soc. Trans. 2015, 43, 861–866. [Google Scholar] [CrossRef]
- Mukherjee, S.; Panda, A.; Ghosh, T.C. Elucidating evolutionary features and functional implications of orphan genes in Leishmania major. Infect. Genet. Evol. 2015, 32, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Kwan, A.H.; Stokes, P.H.; Mackay, J.P.; Cubeddu, L.; Matthews, J.M. The Structure of an LIM-Only Protein 4 (LMO4) and Deformed Epidermal Autoregulatory Factor-1 (DEAF1) Complex Reveals a Common Mode of Binding to LMO4. PLoS ONE 2014, 9, e109108. [Google Scholar] [CrossRef]
- Richer, B.C.; Seeger, K. The hinge region of type VII collagen is intrinsically disordered. Matrix Biol. 2014, 36, 77–83. [Google Scholar] [CrossRef]
- Xue, B.; Ganti, K.; Rabionet, A.; Banks, L.; Uversky, V. Disordered Interactome of Human Papillomavirus. Curr. Pharm. Des. 2014, 20, 1274–1292. [Google Scholar] [CrossRef]
- Yates, C.M.; Sternberg, M. The Effects of Non-Synonymous Single Nucleotide Polymorphisms (nsSNPs) on Protein–Protein Interactions. J. Mol. Biol. 2013, 425, 3949–3963. [Google Scholar] [CrossRef] [PubMed]
- Scorciapino, M.A.; Manzo, G.; Rinaldi, A.C.; Sanna, R.; Casu, M.; Pantic, J.M.; Lukic, M.L.; Conlon, J.M. Conformational Analysis of the Frog Skin Peptide, Plasticin-L1, and Its Effects on Production of Proinflammatory Cytokines by Macrophages. Biochemistry 2013, 52, 7231–7241. [Google Scholar] [CrossRef] [PubMed]
- Akinshina, A.; Jambon-Puillet, E.; Warren, P.B.; Noro, M.G. Self-consistent field theory for the interactions between keratin intermediate filaments. BMC Biophys. 2013, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Graham, L.D.; Glattauer, V.; Li, N.; Tyler, M.J.; Ramshaw, J.A. The adhesive skin exudate of Notaden bennetti frogs (Anura: Limnodynastidae) has similarities to the prey capture glue of Euperipatoides sp. velvet worms (Onychophora: Peripatopsidae). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2013, 165, 250–259. [Google Scholar] [CrossRef]
- Lewitzky, M.; Simister, P.C.; Feller, S.M. Beyond ‘Furballs’ and ‘Dumpling Soups’-Towards a Molecular Architecture of Signaling Complexes and Networks. FEBS Lett. 2012, 586, 2740–2750. [Google Scholar] [CrossRef] [Green Version]
- Shan, Y.; Eastwood, M.P.; Zhang, X.; Kim, E.T.; Arkhipov, A.; Dror, R.O.; Jumper, J.; Kuriyan, J.; Shaw, D.E. Oncogenic Mutations Counteract Intrinsic Disorder in the EGFR Kinase and Promote Receptor Dimerization. Cell 2012, 149, 860–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauscher, S.; Pomès, R. Structural Disorder and Protein Elasticity. Adv. Exp. Med. Biol. 2012, 725, 159–183. [Google Scholar] [CrossRef]
- Lehoux, M.; Fradet-Turcotte, A.; Lussier-Price, M.; Omichinski, J.G.; Archambault, J. Inhibition of Human Papillomavirus DNA Replication by an E1-Derived p80/UAF1-Binding Peptide. J. Virol. 2012, 86, 3486–3500. [Google Scholar] [CrossRef] [Green Version]
- Majczak, G.; Lilla, S.; Garay-Malpartida, M.; Markovic, J.; Medrano, F.; De Nucci, G.; E Belizário, J. Prediction and biochemical characterization of intrinsic disorder in the structure of proteolysis-inducing factor/dermcidin. Genet. Mol. Res. 2007, 6, 1000–1011. [Google Scholar]
- Uversky, V.N.; Roman, A.; Oldfield, A.C.J.; Dunker†, A.K. Protein Intrinsic Disorder and Human Papillomaviruses: Increased Amount of Disorder in E6 and E7 Oncoproteins from High Risk HPVs. J. Proteome Res. 2006, 5, 1829–1842. [Google Scholar] [CrossRef]
- Oh, I.; Strong, C.D.G. The Molecular Revolution in Cutaneous Biology: EDC and Locus Control. J. Investig. Dermatol. 2017, 137, e101–e104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, B.; Tilli, C.M.; Hardman, M.; Avilion, A.A.; MacLeod, M.C.; Ashcroft, G.S.; Byrne, C. Late Cornified Envelope Family in Differentiating Epithelia—Response to Calcium and Ultraviolet Irradiation. J. Investig. Dermatol. 2005, 124, 1062–1070. [Google Scholar] [CrossRef]
- Kypriotou, M.; Huber, M.; Hohl, D. The human epidermal differentiation complex: Cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp. Dermatol. 2012, 21, 643–649. [Google Scholar] [CrossRef]
- Strasser, B.; Mlitz, V.; Hermann, M.; Rice, R.H.; Eigenheer, R.A.; Alibardi, L.; Tschachler, E.; Eckhart, L. Evolutionary Origin and Diversification of Epidermal Barrier Proteins in Amniotes. Mol. Biol. Evol. 2014, 31, 3194–3205. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, Z.A.; Strong, C.D.G. Recent Positive Selection in Genes of the Mammalian Epidermal Differentiation Complex Locus. Front. Genet. 2017, 7, 227. [Google Scholar] [CrossRef] [Green Version]
- Mlitz, V.; Strasser, B.; Jaeger, K.; Hermann, M.; Ghannadan, M.; Buchberger, M.; Alibardi, L.; Tschachler, E.; Eckhart, L. Trichohyalin-Like Proteins Have Evolutionarily Conserved Roles in the Morphogenesis of Skin Appendages. J. Investig. Dermatol. 2014, 134, 2685–2692. [Google Scholar] [CrossRef] [Green Version]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 2016, 14, 1. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. et Biophys. Acta (BBA)-Bioenery 2021, 1868, 118876. [Google Scholar] [CrossRef]
- Ryan, V.; Fawzi, N.L. Physiological, Pathological, and Targetable Membraneless Organelles in Neurons. Trends Neurosci. 2019, 42, 693–708. [Google Scholar] [CrossRef]
- Bratek-Skicki, A.; Pancsa, R.; Mészáros, B.; Van Lindt, J.; Tompa, P. A guide to regulation of the formation of biomolecular condensates. FEBS J. 2020, 287, 1924–1935. [Google Scholar] [CrossRef] [Green Version]
- Kosik, K.S.; Han, S. Tau Condensates. Adv. Exp. Med. Biol. 2019, 1184, 327–339. [Google Scholar] [CrossRef]
- Babinchak, W.; Surewicz, W.K. Liquid–Liquid Phase Separation and Its Mechanistic Role in Pathological Protein Aggregation. J. Mol. Biol. 2020, 432, 1910–1925. [Google Scholar] [CrossRef]
- Fuxreiter, M.; Vendruscolo, M. Generic nature of the condensed states of proteins. Nat. Cell Biol. 2021, 23, 587–594. [Google Scholar] [CrossRef]
- Shamilov, R.; Vinogradova, O.; Aneskievich, B.J. The Anti-Inflammatory Protein TNIP1 Is Intrinsically Disordered with Structural Flexibility Contributed by Its AHD1-UBAN Domain. Biomolecules 2020, 10, 1531. [Google Scholar] [CrossRef]
- Wolf, R.; Ruzicka, T.; Yuspa, S.H. Novel S100A7 (psoriasin)/S100A15 (koebnerisin) subfamily: Highly homologous but distinct in regulation and function. Amino Acids 2010, 41, 789–796. [Google Scholar] [CrossRef]
- Leśniak, W.; Graczyk-Jarzynka, A. The S100 proteins in epidermis: Topology and function. Biochim. et Biophys. Acta (BBA)-Gen. Subj. 2015, 1850, 2563–2572. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. et Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [Green Version]
- Mlitz, V.; Hussain, T.; Tschachler, E.; Eckhart, L. Filaggrin has evolved from an “S100 fused-type protein” (SFTP) gene present in a common ancestor of amphibians and mammals. Exp. Dermatol. 2017, 26, 955–957. [Google Scholar] [CrossRef] [Green Version]
- Candi, E.; Melino, G.; Mei, G.; Tarcsa, E.; Chung, S.-I.; Marekov, L.N.; Steinert, P.M. Biochemical, Structural, and Transglutaminase Substrate Properties Of Human Loricrin, the Major Epidermal Cornified Cell Envelope Protein. J. Biol. Chem. 1995, 270, 26382–26390. [Google Scholar] [CrossRef] [Green Version]
- Hohl, D.; Mehrel, T.; Lichti, U.; Turner, M.L.; Roop, D.R.; Steinert, P.M. Characterization of human loricrin. Structure and function of a new class of epidermal cell envelope proteins. J. Biol. Chem. 1991, 266, 6626–6636. [Google Scholar] [CrossRef]
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef]
- Holthaus, K.B.; Alibardi, L.; Tschachler, E.; Eckhart, L. Identification of epidermal differentiation genes of the tuatara provides insights into the early evolution of lepidosaurian skin. Sci. Rep. 2020, 10, 12844. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.-L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. Pac. Symp. Biocomput. 2011, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Ishitsuka, Y.; Roop, D.R. Loricrin: Past, Present, and Future. Int. J. Mol. Sci. 2020, 21, 2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacic, V.; Uversky, V.N.; Dunker, A.K.; Lonardi, S. Composition Profiler: A tool for discovery and visualization of amino acid composition differences. BMC Bioinform. 2007, 8, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. The intrinsic disorder alphabet. III. Dual personality of serine. Intrinsically Disord. Proteins 2015, 3, e1027032. [Google Scholar] [CrossRef] [Green Version]
- Steven, A.; Bisher, M.; Roop, D.; Steinert, P. Biosynthetic pathways of filaggrin and loricrin—two major proteins expressed by terminally differentiated epidermal keratinocytes. J. Struct. Biol. 1990, 104, 150–162. [Google Scholar] [CrossRef]
- Ruff, K.M.; Roberts, S.; Chilkoti, A.; Pappu, R.V. Advances in Understanding Stimulus-Responsive Phase Behavior of Intrinsically Disordered Protein Polymers. J. Mol. Biol. 2018, 430, 4619–4635. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernon, R.M.; Forman-Kay, J.D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019, 58, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.P.; Sawaya, M.R.; Boyer, D.R.; Goldschmidt, L.; Rodriguez, J.A.; Cascio, D.; Chong, L.; Gonen, T.; Eisenberg, D.S. Atomic Structures of Low-Complexity Protein Segments Reveal Kinked Beta Sheets that Assemble Networks. Science 2018, 359, 698–701. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, B.; Gotor, N.L.; Dhar, R.; Cirillo, D.; Baldrighi, M.; Tartaglia, G.G.; Lehner, B. A Concentration-Dependent Liquid Phase Separation Can Cause Toxicity upon Increased Protein Expression. Cell Rep. 2016, 16, 222–231. [Google Scholar] [CrossRef] [Green Version]
- Michibata, H.; Chiba, H.; Wakimoto, K.; Seishima, M.; Kawasaki, S.; Okubo, K.; Mitsui, H.; Torii, H.; Imai, Y. Identification and Characterization of a Novel Component of the Cornified Envelope, Cornifelin. Biochem. Biophys. Res. Commun. 2004, 318, 803–813. [Google Scholar] [CrossRef]
- Wagner, T.; Beer, L.; Gschwandtner, M.; Eckhart, L.; Kalinina, P.; Laggner, M.; Ellinger, A.; Gruber, R.; Kuchler, U.; Golabi, B.; et al. The Differentiation-Associated Keratinocyte Protein Cornifelin Contributes to Cell-Cell Adhesion of Epidermal and Mucosal Keratinocytes. J. Investig. Dermatol. 2019, 139, 2292–2301.e9. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Oddi, S.; Terrinoni, A.; Paradisi, A.; Ranalli, M.; Finazzi-Agró, A.; Melino, G. Transglutaminase 5 Cross-links Loricrin, Involucrin, and Small Proline-rich Proteins in Vitro. J. Biol. Chem. 2001, 276, 35014–35023. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Sandilands, A.; Sutherland, C.; Irvine, A.; McLean, W.H.I. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294. [Google Scholar] [CrossRef] [Green Version]
- McLean, W. Filaggrin failure-from ichthyosis vulgaris to atopic eczema and beyond. Br. J. Dermatol. 2016, 175, 4–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, J.; Hsu, C.-Y.; Haftek, M.; Nachat, R.; de Koning, H.D.; Gardinal-Galera, I.; Hitomi, K.; Balica, S.; Jean-Decoster, C.; Schmitt, A.-M.; et al. Hornerin is a component of the epidermal cornified cell envelopes. FASEB J. 2011, 25, 1567–1576. [Google Scholar] [CrossRef]
- Kuechle, M.K.; Thulin, C.D.; Presland, R.; Dale, B.A. Profilaggrin Requires both Linker and Filaggrin Peptide Sequences to Form Granules: Implications for Profilaggrin Processing In Vivo11This work was presented in part at the Society of Investigative Dermatology meeting in 1997. J. Investig. Dermatol. 1999, 112, 843–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brettmann, E.; Strong, C.D.G. Recent evolution of the human skin barrier. Exp. Dermatol. 2018, 27, 859–866. [Google Scholar] [CrossRef]
- Strasser, B.; Mlitz, V.; Fischer, H.; Tschachler, E.; Eckhart, L. Comparative genomics reveals conservation of filaggrin and loss of caspase-14 in dolphins. Exp. Dermatol. 2015, 24, 365–369. [Google Scholar] [CrossRef] [Green Version]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef] [Green Version]
- Quiroz, F.G.; Chilkoti, A. Sequence heuristics to encode phase behaviour in intrinsically disordered protein polymers. Nat. Mater. 2015, 14, 1164–1171. [Google Scholar] [CrossRef]
- Martin, E.W.; Holehouse, A.S. Intrinsically disordered protein regions and phase separation: Sequence determinants of assembly or lack thereof. Emerg. Top. Life Sci. 2020, 4, 307–329. [Google Scholar] [CrossRef]
- Fisher, R.S.; Elbaum-Garfinkle, S. Tunable multiphase dynamics of arginine and lysine liquid condensates. Nat. Commun. 2020, 11, 4628. [Google Scholar] [CrossRef]
- Alibardi, L. Ultrastructural localization of histidine-labelled interkeratin matrix during keratinization of amphibian epidermis. Acta Histochem. 2003, 105, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Theillet, F.-X.; Kalmar, L.; Tompa, P.; Han, K.-H.; Selenko, P.; Dunker, A.K.; Daughdrill, G.W.; Uversky, V.N. The alphabet of intrinsic disorder. Intrinsically Disord. Proteins 2013, 1, e24360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of Intrinsic Disorder in Protein Domains and Families: I. A Database of Conserved Predicted Disordered Regions. J. Proteome Res. 2006, 5, 879–887. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Oldfield, C.J.; Yan, W.; Shen, B.; Dunker, A.K. Intrinsically Disordered Domains: Sequence Disorder Function Relationships. Protein Sci. 2019, 28, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Pappu, R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA 2013, 110, 13392–13397. [Google Scholar] [CrossRef] [Green Version]
- Holehouse, A.S.; Das, R.; Ahad, J.N.; Richardson, M.O.; Pappu, R.V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys. J. 2017, 112, 16–21. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.T.; Mooney, C.; Lopez, C.; Pollastri, G.; Shields, D.C. Prediction of polyproline II secondary structure propensity in proteins. R. Soc. Open Sci. 2020, 7, 191239. [Google Scholar] [CrossRef] [PubMed]
- Vandelli, A.; Monti, M.; Milanetti, E.; Armaos, A.; Rupert, J.; Zacco, E.; Bechara, E.; Delli Ponti, R.; Tartaglia, G.G. Structural Analysis of SARS-CoV-2 Genome and Predictions of the Human Interactome. Nucleic Acids Res. 2020, 48, 11270–11283. [Google Scholar] [CrossRef] [PubMed]
- Harami, G.M.; Kovács, Z.J.; Pancsa, R.; Pálinkás, J.; Baráth, V.; Tárnok, K.; Málnási-Csizmadia, A.; Kovács, M. Phase separation by ssDNA binding protein controlled via protein−protein and protein−DNA interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 26206–26217. [Google Scholar] [CrossRef] [PubMed]
- Gotor, N.L.; Armaos, A.; Calloni, G.; Burgas, M.T.; Vabulas, R.M.; De Groot, N.S.; Tartaglia, G.G. RNA-binding and prion domains: The Yin and Yang of phase separation. Nucleic Acids Res. 2020, 48, 9491–9504. [Google Scholar] [CrossRef] [PubMed]
- Ambadipudi, S.; Biernat, J.; Riedel, D.; Mandelkow, E.; Zweckstetter, M. Liquid–liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nat. Commun. 2017, 8, 275. [Google Scholar] [CrossRef]
- Bellay, J.; Han, S.; Michaut, M.; Kim, T.; Costanzo, M.; Andrews, B.J.; Boone, C.; Bader, G.D.; Myers, C.L.; Kim, P.M. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 2011, 12, R14. [Google Scholar] [CrossRef]
- Pauwels, K.; Lebrun, P.; Tompa, P. To be Disordered or Not to be Disordered: Is that Still a Question for Proteins in the Cell? Cell Mol. Life Sci. 2017, 74, 3185–3204. [Google Scholar] [CrossRef]
- Farahi, N.; Lazar, T.; Wodak, S.; Tompa, P.; Pancsa, R. Integration of Data from Liquid–Liquid Phase Separation Databases Highlights Concentration and Dosage Sensitivity of LLPS Drivers. Int. J. Mol. Sci. 2021, 22, 3017. [Google Scholar] [CrossRef]
- Itakura, A.K.; Futia, R.A.; Jarosz, D.F. It Pays To Be in Phase. Biochemistry 2018, 57, 2520–2529. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [Google Scholar] [CrossRef]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K. Predicting intrinsic disorder from amino acid sequence. Proteins Struct. Funct. Bioinform. 2003, 53, 566–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence Complexity of Disordered Protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Uversky, V.N.; Finkelstein, A.V. Life in Phases: Intra-and Inter-Molecular Phase Transitions in Protein Solutions. Biomolecules 2019, 9, 842. [Google Scholar] [CrossRef] [Green Version]
- McSwiggen, D.T.; Mir, M.; Darzacq, X.; Tjian, R. Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 2019, 33, 1619–1634. [Google Scholar] [CrossRef]
- Jacob, J.T.; Coulombe, P.A.; Kwan, R.; Omary, B. Types I and II Keratin Intermediate Filaments. Cold Spring Harb. Perspect. Biol. 2018, 10, a018275. [Google Scholar] [CrossRef] [Green Version]
- Hatos, A.; Hajdu-Soltész, B.; Monzon, A.M.; Palopoli, N.; Álvarez, L.; Aykac-Fas, B.; Bassot, C.; I Benítez, G.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2019, 48, D269–D276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamilov, R.; Ackley, T.W.; Aneskievich, B.J. Enhanced Wound Healing-and Inflammasome-Associated Gene Expression in TNFAIP3-Interacting Protein 1-(TNIP1-) Deficient HaCaT Keratinocytes Parallels Reduced Reepithelialization. Mediat. Inflamm. 2020, 2020, 5919150. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; van den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| 1st Author | Journal Citation | Proteins Reported as IDPs or with IDR |
|---|---|---|
| Tuusa, J. | [6] | hemidesmosome protein BP180 |
| Garcia Quiroz, F. | [7] | filaggrin in keratohyalin granules |
| Shamilov, R. | [10] | nuclear receptor N-terminal AF-1 |
| Levenson, R. | [11] | squid skin light-reflecting proteins |
| Latendorf, T. | [5] | hornerin antimicrobial peptides |
| Kurvits, L. | [12] | serum amyloid in skin biopsies |
| Okamoto, K. | [13] | C-terminus of EGF-R |
| Moens, M. | [14] | avian skin viral protein |
| Gopalan, A. | [15] | TB proteins in skin lesions |
| Uversky, V. | [9] | biofilms of pathogenic Staphylococcus epidermidis |
| Singh, I. | [16] | C-terminus of EGF-R |
| Rauscher, S. | [17] | skin ECM protein elastin |
| Yarawsky, A. | [18] | biofilms of pathogenic Staphylococcus epidermidis |
| Keppel, T. | [19] | C-terminus of EGF-R |
| Muiznieks, L. | [20] | skin ECM protein elastin |
| Levenson, R. | [21] | squid skin light-reflecting proteins |
| Wang, B. | [22] | ubiquitin-conjugating enzyme |
| Bray, D. | [23] | keratin N- and C-termini |
| Kornreich, M. | [24] | keratin N- and C-termini |
| Whelan, F. | [25] | biofilms of pathogenic Staphylococcus epidermidis |
| Mukherjee, S. | [26] | proteins of cutaneous pathogen Leishmania major |
| Joseph, S. | [27] | DNA-binding protein |
| Richer, B. | [28] | skin ECM collagen subdomain |
| Xue, B. | [29] | HPV oncoproteins |
| Yates, C. | [30] | C-terminus of EGF-R |
| Scorciapino, M. | [31] | frog skin antibacterial peptide |
| Akinshina, A. | [32] | keratin N- and C-termini |
| Graham, L. | [33] | frog skin-secreted adhesive protein |
| Lewitzky, M. | [34] | C-terminus of EGF-R |
| Shan, Y. | [35] | C-terminus of EGF-R |
| Rauscher, S. | [36] | skin ECM protein elastin |
| Lehoux, M. | [37] | HPV oncoproteins |
| Majczak, G. | [38] | antimicrobial dermicidin protein |
| Uversky, V. | [39] | HPV oncoproteins |
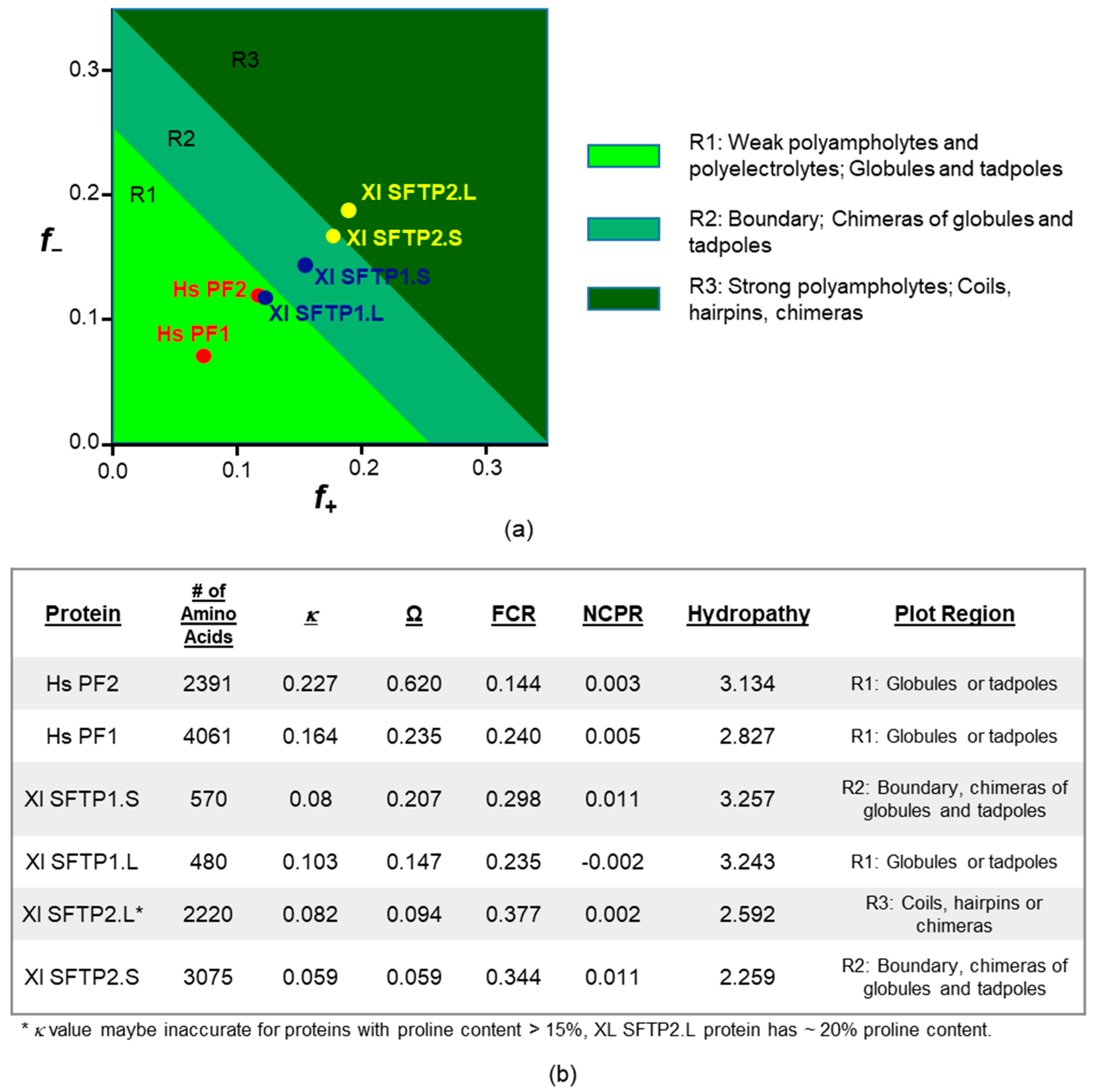
| Trait | Hs PF1 | Hs PF2 | Xl SFTP2.S | Xl SFTP2.L | Xl SFTP1.S | Xl SFTP1.L |
|---|---|---|---|---|---|---|
| Sequence source | UniProt P20930 | UniProt Q5D862 | GenBank: OCT66701.1 | GenBank: OCT69537.1 [57] | [57] | GenBank: XP_018087213.1 |
| # AA | 4061 | 2391 | 3075 | 2220 | 570 | 480 |
| MW | 435 kDa | 248 kDa | 349 kDa | 249 kDa | 64 kDa | 54 kDa |
| Theoretical pI | 9.24 | 8.45 | 8.77 | 8.03 | 8.65 | 6.90 |
| ARG bias | 0.885 | 0.824 | 0.031 | 0.033 | 0.012 | 0.036 |
| ARG % | 10.8 | 6.1% | 0.60 | 0.60 | 0.2% | 0.4% |
| LYS % | 1.40 | 1.3% | 17.20 | 18.30 | 15.3% | 11.2% |
| HIS % | 10.20 | 10.3 | 1.30 | 4.80 | 1.4% | 2.7% |
| PONDR-FIT score w/N-term | 0.895 | 0.896 | 0.852 | 0.740 | 0.722 | 0.705 |
| Aliphatic index | 19.76 | 15.97 | 15.62 | 26.91 | 62.39 | 49.65 |
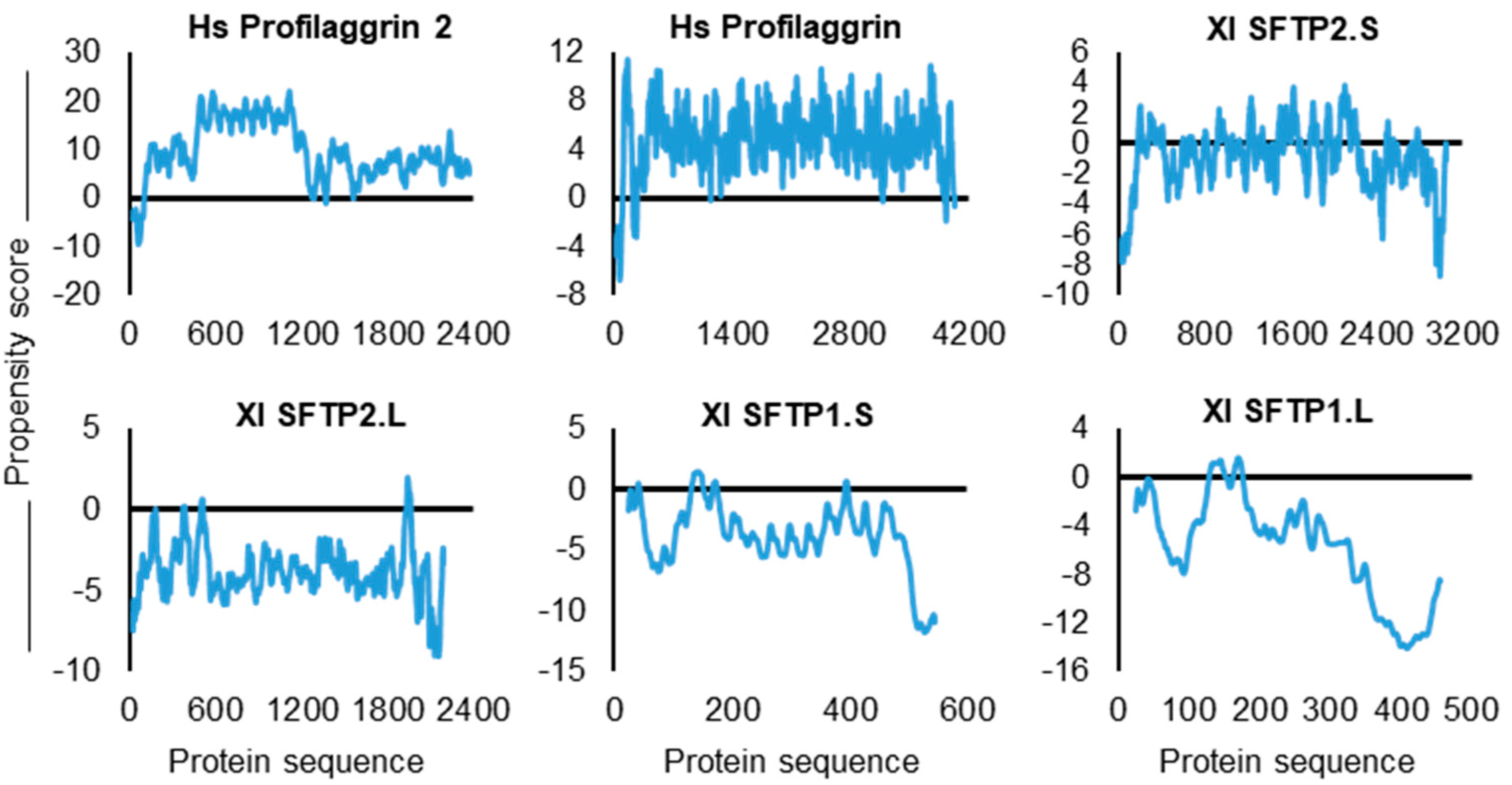
| SFTP | Propensity Score | Gly | Arg | Phe | SUM: Gly, Arg, Phe |
|---|---|---|---|---|---|
| Hs PF2 | 4.418 | 20.30% | 6.10% | 2.30% | 28.70% |
| Hs PF1 | 3.553 | 12.80% | 10.80% | 0.60% | 24.20% |
| Xl SFTP2.S | 2.019 | 3.50% | 0.60% | 0.60% | 4.70% |
| Xl SFTP2.L | 1.179 | 0.80% | 0.60% | 0.90% | 2.30% |
| Xl SFTP1.S | 0.561 | 2.50% | 0.20% | 0.70% | 3.40% |
| Xl SFTP1.L | 0.063 | 2.50% | 0.40% | 1.50% | 4.40% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shamilov, R.; Robinson, V.L.; Aneskievich, B.J. Seeing Keratinocyte Proteins through the Looking Glass of Intrinsic Disorder. Int. J. Mol. Sci. 2021, 22, 7912. https://doi.org/10.3390/ijms22157912
Shamilov R, Robinson VL, Aneskievich BJ. Seeing Keratinocyte Proteins through the Looking Glass of Intrinsic Disorder. International Journal of Molecular Sciences. 2021; 22(15):7912. https://doi.org/10.3390/ijms22157912
Chicago/Turabian StyleShamilov, Rambon, Victoria L. Robinson, and Brian J. Aneskievich. 2021. "Seeing Keratinocyte Proteins through the Looking Glass of Intrinsic Disorder" International Journal of Molecular Sciences 22, no. 15: 7912. https://doi.org/10.3390/ijms22157912
APA StyleShamilov, R., Robinson, V. L., & Aneskievich, B. J. (2021). Seeing Keratinocyte Proteins through the Looking Glass of Intrinsic Disorder. International Journal of Molecular Sciences, 22(15), 7912. https://doi.org/10.3390/ijms22157912






