VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture
Abstract
:1. Introduction
2. Results
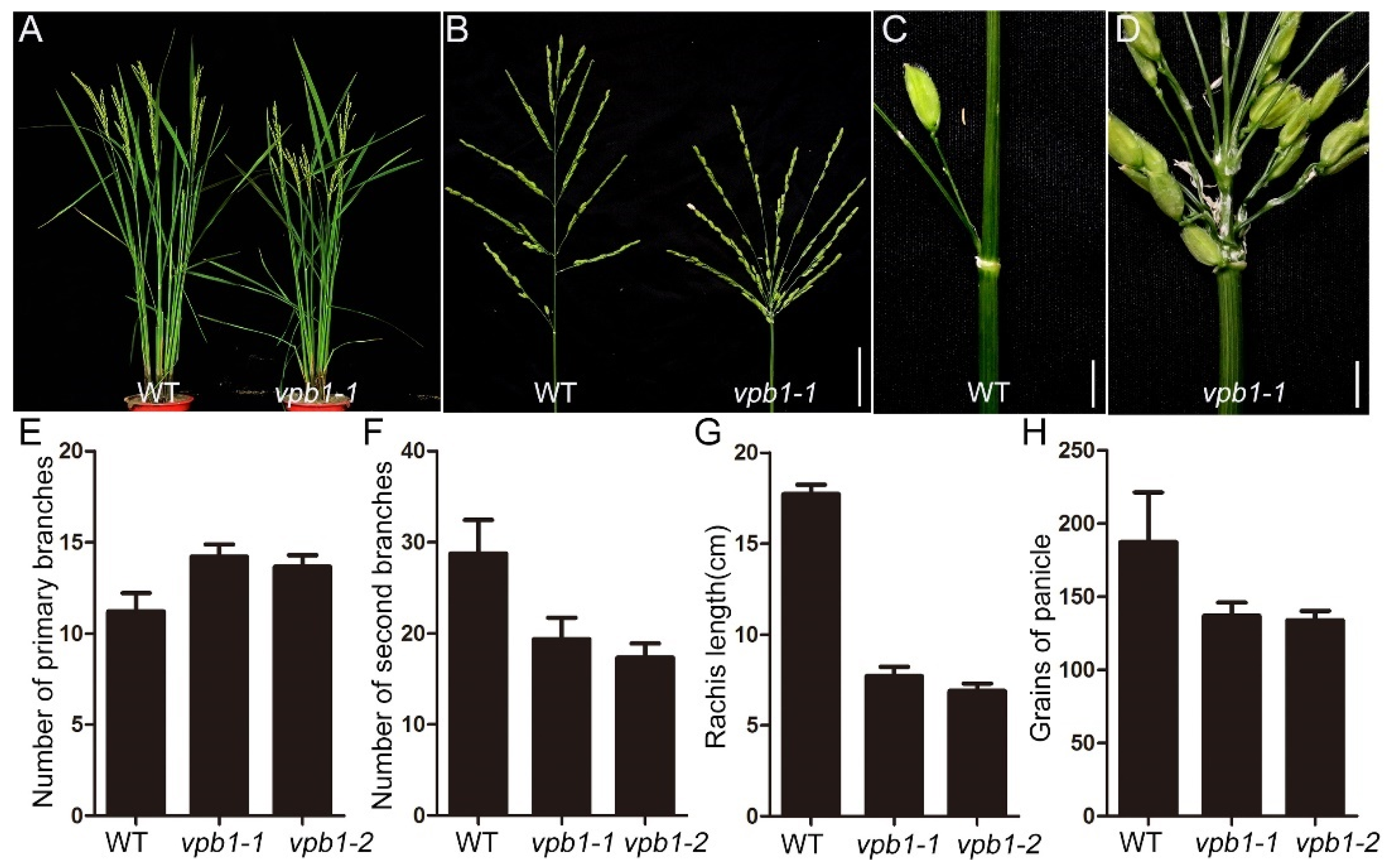
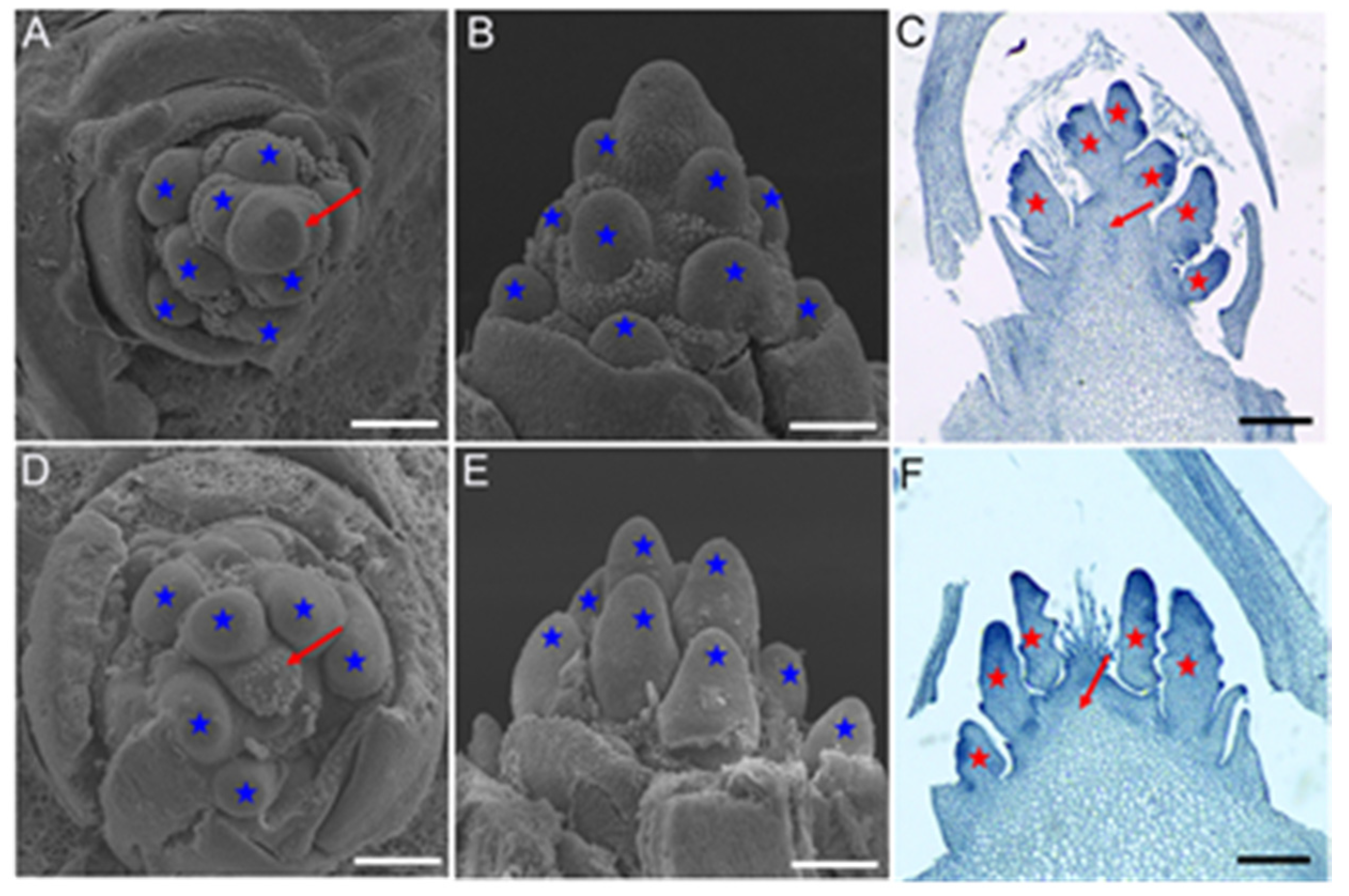
2.1. Inflorescence Phenotypes in vpb1 Mutant
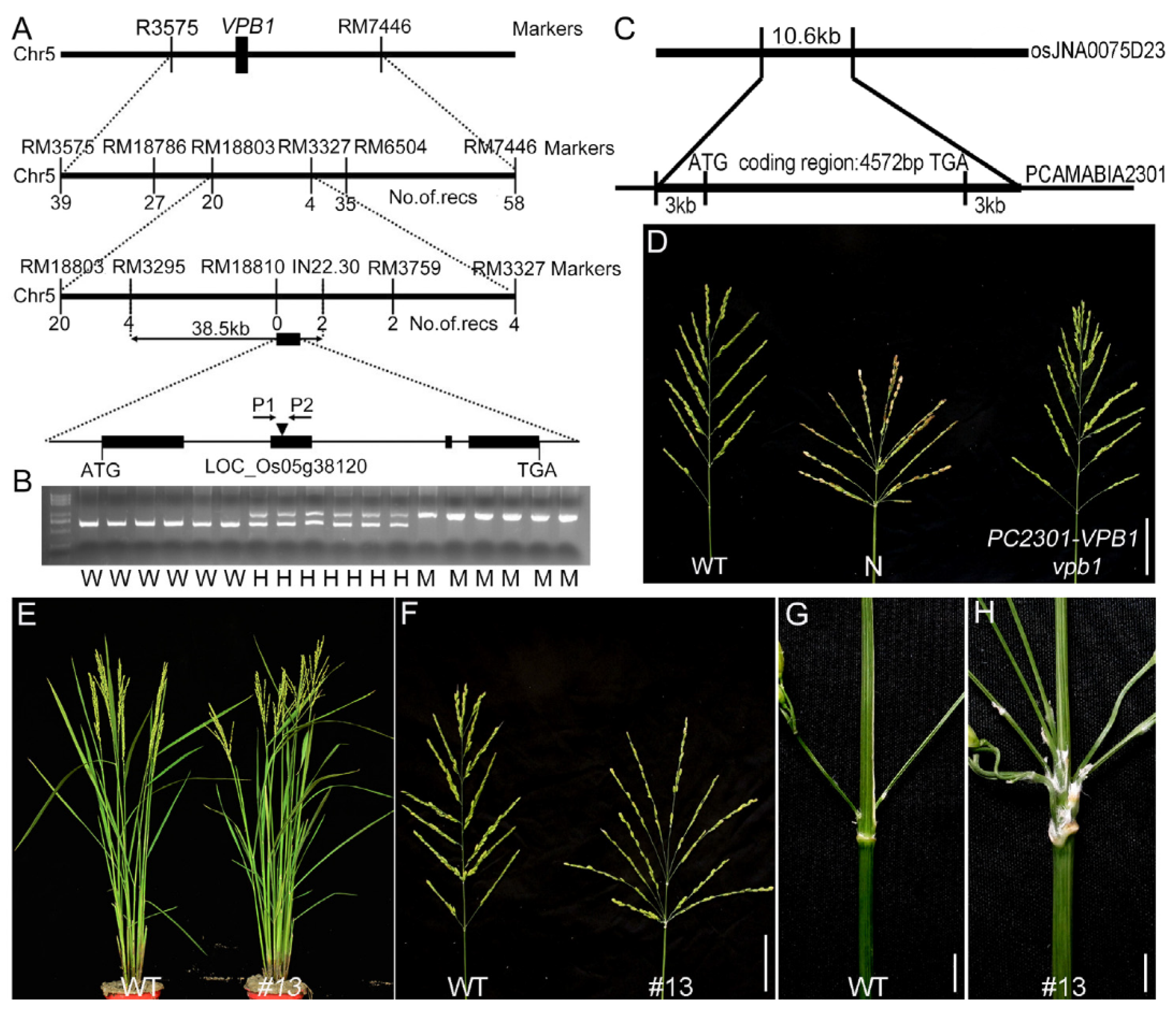
2.2. Map-Based Cloning of VPB1
2.3. VPB1 Encodes a BELL1-Type Transcription Factor
2.4. Expression Pattern of VPB1
2.5. Subcellular Localization and VPB1 Transcriptional Activity
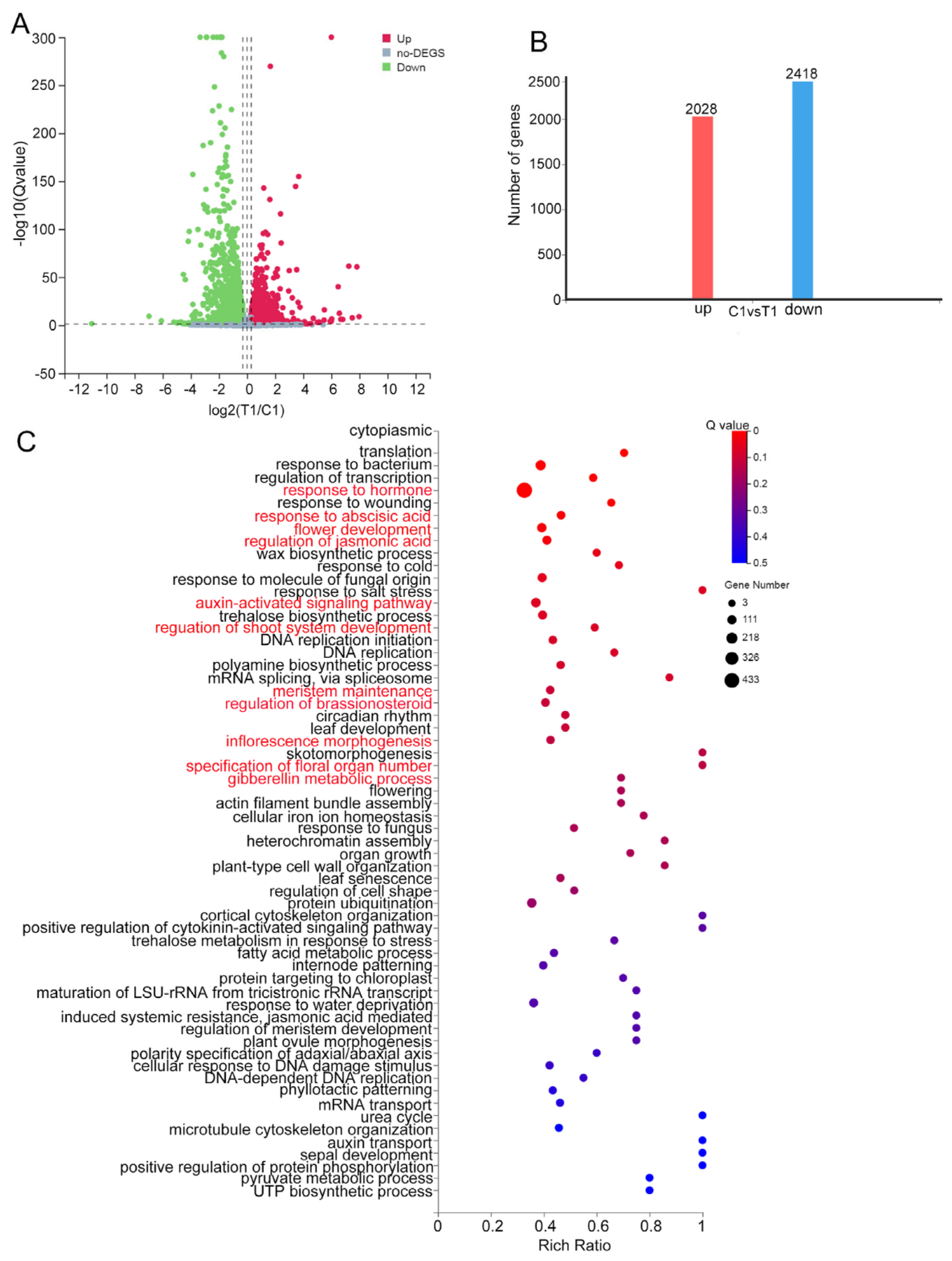
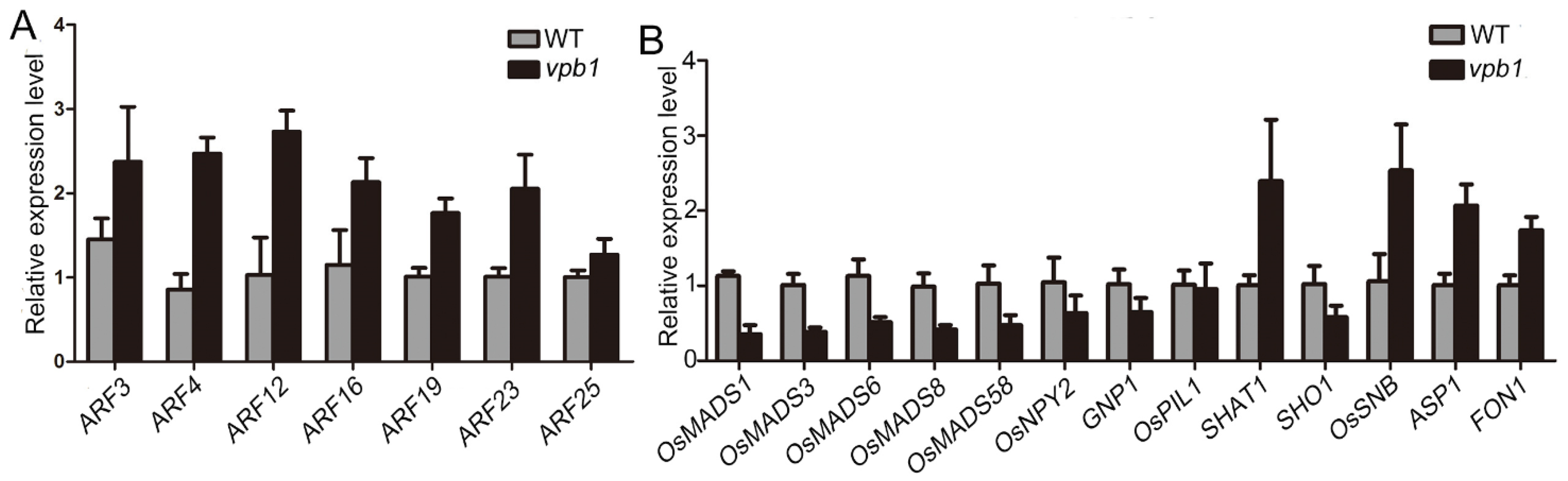
2.6. VPB1 Affects the Expression of Genes Involved in Inflorescence Development and Hormone Pathways
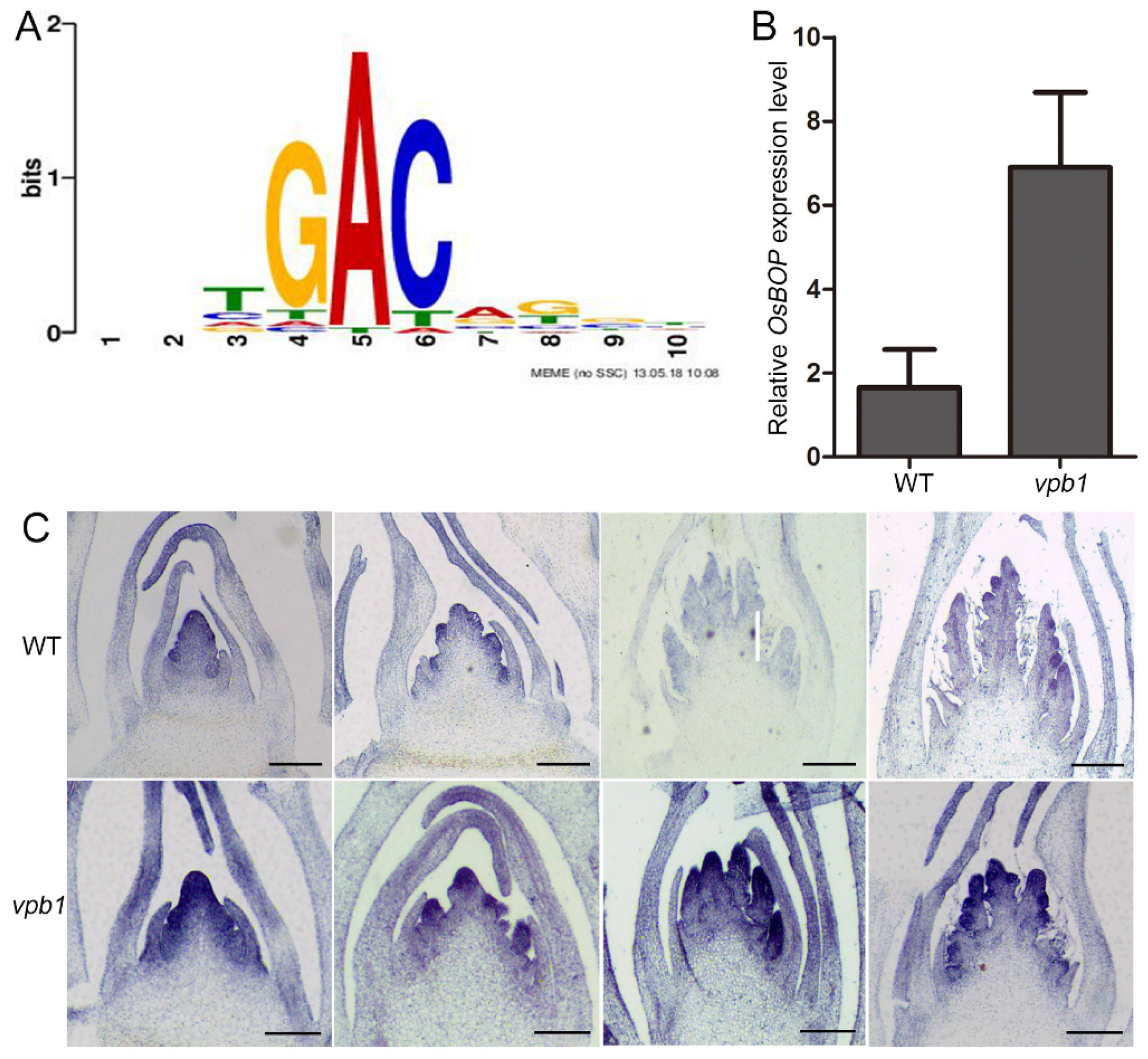
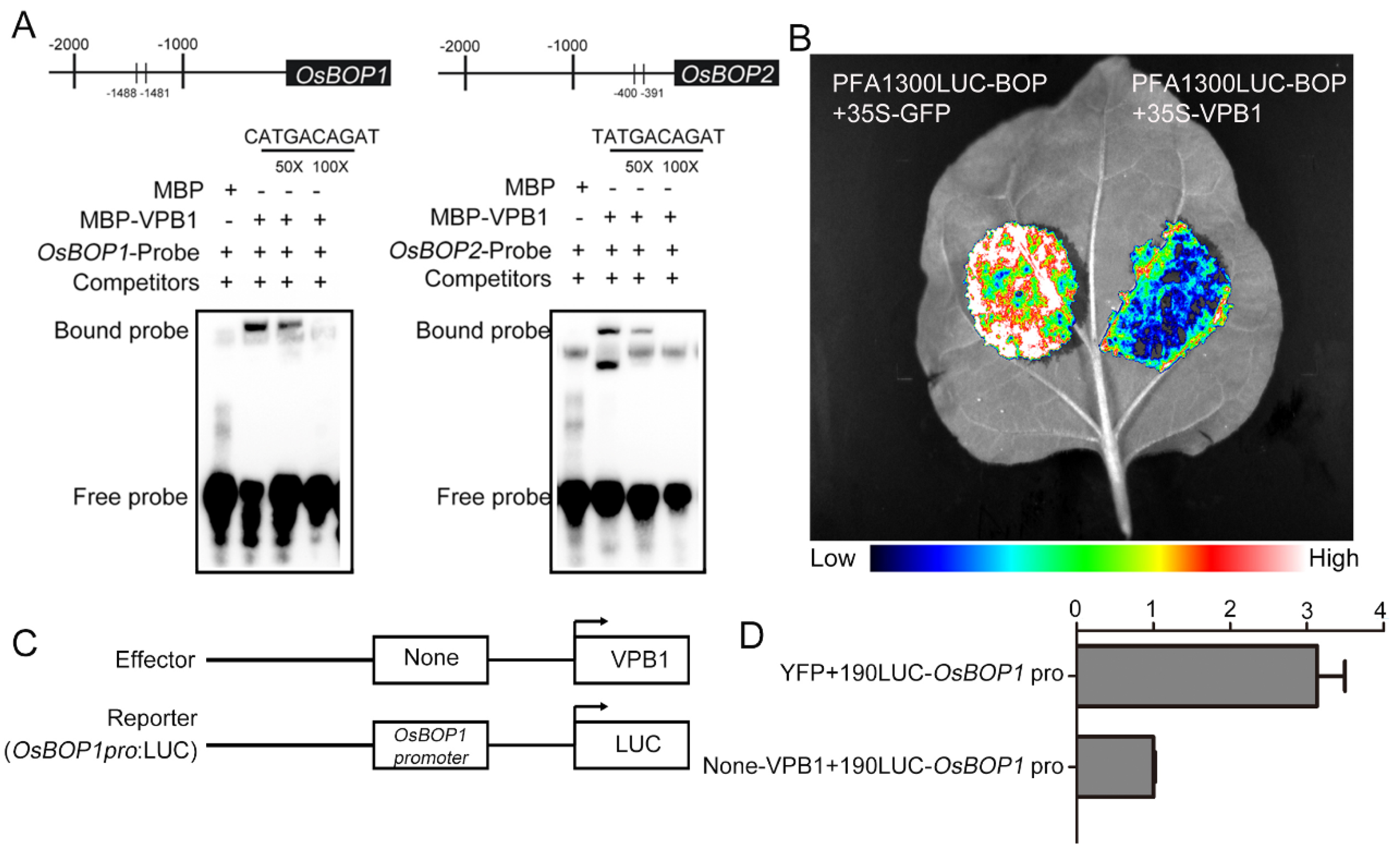
2.7. VPB1 Negatively Regulates OsBOP1 Expression
3. Discussion
3.1. VPB1 Regulates the Initiation and Arrangement of Primary Branch Meristems
3.2. VPB1 Belongs to a Functionally Conserved Gene Family
3.3. The VPB1 Gene Participates in a Complex Molecular Pathway to Regulate Panicle Development
3.4. VPB1 Regulates Inflorescence Development by Directly Binding to OsBOP1
3.5. The Role of BLH-KNOX Dimer Functions in Inflorescence Development
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Scanning Electron Microscopy
4.3. Histological Sectioning
4.4. Map-Based Cloning of VPB1
4.5. Plasmid Construction and Rice Transformation
4.6. Total RNA Isolation and qRT-PCR Analyses
4.7. In Situ Hybridization
4.8. Subcellular Localization
4.9. Transcriptional Activity Analysis
4.10. RNA-Seq Analysis
4.11. EMSA
4.12. The Transient Expression System in Tobacco
4.13. Accession Numbers
5. Conclusions and Future Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benlloch, R.; Berbel, A.; Serrano-Mislata, A.; Madueno, F. Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. 2007, 100, 659–676. [Google Scholar] [CrossRef]
- Gao, X.-Q.; Wang, N.; Wang, X.-L.; Zhang, X.S. Architecture of wheat inflorescence: Insights from rice. Trends Plant Sci. 2019, 24, 802–809. [Google Scholar] [CrossRef]
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef]
- Teo, Z.W.N.; Song, S.; Wang, Y.-Q.; Liu, J.; Yu, H. New insights into the regulation of inflorescence architecture. Trends Plant Sci. 2014, 19, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Bortiri, E.; Hake, S. Flowering and determinacy in maize. J. Exp. Bot. 2007, 58, 909–916. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Sunohara, H.; Nagato, Y. Developmental course of inflorescence and spikelet in rice. Breed. Sci. 2004, 54, 147–156. [Google Scholar] [CrossRef] [Green Version]
- Itoh, J.-I.; Nonomura, K.-I.; Ikeda, K.; Yamaki, S.; Inukai, Y.; Yamagishi, H.; Kitano, H.; Nagato, Y. Rice plant development: From zygote to spikelet. Plant Cell Physiol. 2005, 46, 23–47. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Zhang, Q. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 2010, 61, 421–442. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Kyozuka, J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 2009, 21, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, K.; Ito, M.; Nagasawa, N.; Kyozuka, J.; Nagato, Y. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 2007, 51, 1030–1040. [Google Scholar] [CrossRef]
- Ikeda-Kawakatsu, K.; Maekawa, M.; Izawa, T.; Itoh, J.-I.; Nagato, Y. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 2012, 69, 168–180. [Google Scholar] [CrossRef]
- Suzaki, T.; Sato, M.; Ashikari, M.; Miyoshi, M.; Nagato, Y.; Hirano, H.-Y. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 2004, 131, 5649–5657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T.; Toriba, T.; Fujimoto, M.; Tsutsumi, N.; Kitano, H.; Hirano, H.-Y. Conservation and diversification of meristem maintenance mechanism in oryza sativa: Function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006, 47, 1591–1602. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.; Yu, S.-I.; Park, J.-H.; Li, Y.; Han, J.-H.; Alavilli, H.; Cho, J.-I.; Kim, T.-H.; Jeon, J.-S.; Lee, B.-H. OsREL2, a rice TOPLESS homolog functions in axillary meristem development in rice inflorescence. Plant Biotechnol. Rep. 2012, 6, 213–224. [Google Scholar] [CrossRef]
- Yoshida, A.; Ohmori, Y.; Kitano, H.; Taguchi-Shiobara, F.; Hirano, H.-Y. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012, 70, 327–339. [Google Scholar] [CrossRef]
- Suzuki, C.; Tanaka, W.; Hirano, H.-Y. Transcriptional corepressor ASP1 and CLV-like signaling regulate meristem maintenance in Rice. Plant Physiol. 2019, 180, 1520–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.-Y. Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, A.; Sasao, M.; Yasuno, N.; Takagi, K.; Daimon, Y.; Chen, R.; Yamazaki, R.; Tokunaga, H.; Kitaguchi, Y.; Sato, Y.; et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc. Natl. Acad. Sci. USA 2013, 110, 767–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamant, O.; Pautot, V. Plant development: A TALE story. C.R. Biol. 2010, 333, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, A.M.; Etchells, J.P.; Canales, C.; Lagodienko, A.; Dickinson, H. VAAMANA—a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 2004, 328, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E.; Groover, A.T.; Fontana, J.R.; Martienssen, R.A. Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 2003, 130, 3941–3950. [Google Scholar] [CrossRef] [Green Version]
- Kanrar, S.; Bhattacharya, M.; Arthur, B.; Courtier, J.; Smith, H.M.S. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 2008, 54, 924–937. [Google Scholar] [CrossRef]
- Khan, M.; Ragni, L.; Tabb, P.; Salasini, B.C.; Chatfield, S.; Datla, R.; Lock, J.; Kuai, X.; Despres, C.; Proveniers, M.; et al. Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis. Plant Physiol. 2015, 169, 2166–2186. [Google Scholar] [CrossRef]
- Roeder, A.H.K.; Ferrándiz, C.; Yanofsky, M.F. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 2003, 13, 1630–1635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, H.M.S.; Campbell, B.C.; Hake, S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 2004, 14, 812–817. [Google Scholar] [CrossRef] [Green Version]
- Ung, N.; Lal, S.; Smith, H.M.S. The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical meristem in Arabidopsis. Plant Physiol. 2011, 156, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Patibanda, V.; Smith, H.M.S. A novel role of BELL1-like homeobox genes, PENNYWISE and POUND-FOOLISH, in floral patterning. Planta 2008, 229, 693–707. [Google Scholar] [CrossRef]
- Smith, H.M.S.; Hake, S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 2003, 15, 1717–1727. [Google Scholar] [CrossRef] [Green Version]
- Peaucelle, A.; Louvet, R.; Johansen, J.N.; Salsac, F.; Morin, H.; Fournet, F.; Belcram, K.; Gillet, F.; Hofte, H.; Laufs, P.; et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development 2011, 138, 4733–4741. [Google Scholar] [CrossRef] [Green Version]
- Kanrar, S.; Onguka, O.; Smith, H.M.S. Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 2006, 224, 1163–1173. [Google Scholar] [CrossRef]
- Rutjens, B.; Bao, D.; van Eck-Stouten, E.; Brand, M.; Smeekens, S.; Proveniers, M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009, 58, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Bencivenga, S.; Serrano-Mislata, A.; Bush, M.; Fox, S.; Sablowski, R. Control of oriented tissue growth through repression of organ boundary genes promotes stem morphogenesis. Dev Cell. 2016, 39, 198–208. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Abraham-Juarez, M.-J.; Maeno, A.; Dong, Z.; Aromdee, D.; Meeley, R.; Shiroishi, T.; Nonomura, K.-i.; Hake, S. KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell 2017, 29, 1105–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.; Cho, L.-H.; Kim, S.L.; Choi, H.; Koh, H.-J.; An, G. The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 2014, 79, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.-H.; Antt, H.W.; Koh, H.-J.; An, G. KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 2017, 174, 312–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Tanaka, W.; Toriba, T.; Suzuki, C.; Maeno, A.; Tsuda, K.; Shiroishi, T.; Kurata, T.; Sakamoto, T.; Murai, M.; et al. BELL1-like homeobox genes regulate inflorescence architecture and meristem maintenance in rice. Plant J. 2019, 98, 465–478. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.G. CRISPR/Cas9-based multiplex genome editing in monocot and dicot plants. Curr. Protoc. Mol. Biol 2016, 115, 31–36. [Google Scholar] [CrossRef]
- Gómez-Mena, C.; Sablowski, R. ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 2008, 20, 2059–2072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Liu, Y.; He, H.; Liu, Y.; Qi, J.; Zhang, X.; Li, Y.; Mao, Y.; Zhou, S.; Zheng, X.; et al. A molecular framework underlying the compound leaf pattern of Medicago truncatula. Nat. Plants. 2020, 6, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Hu, Y.; Liang, W.; Yin, C.; Yang, X.; Ping, B.; Li, A.; Jia, R.; Chen, M.; Luo, Z.; Cai, Q.; et al. Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 2015, 8, 1366–1384. [Google Scholar] [CrossRef]
- Itoh, J.-I.; Kitano, H.; Matsuoka, M.; Nagato, Y. SHOOT ORGANIZATION genes regulate shoot apical meristem organization and the pattern of leaf primordium initiation in rice. Plant Cell 2000, 12, 2161–2174. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, Y.; Mi, X.-F.; Shan, J.-X.; Li, X.-M.; Xu, J.-L.; Lin, H.-X. The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 2016, 12, e1006386. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yan, L.; Ge, C.; Yao, X.-F.; Han, X.; Wang, R.; Xiong, L.; Jiang, L.; Liu, C.-M.; Zhao, Y. PINOID is required for formation of the stigma and style in rice. Plant Physiol. 2019, 180, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Ma, X.; Zhao, S.; Tang, Y.; Liu, F.; Gu, P.; Fu, Y.; Zhu, Z.; Cai, H.; Sun, C.; et al. The APETALA2-like transcription factor SUPERNUMERARY BRACT controls rice seed shattering and seed size. Plant Cell 2019, 31, 17–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrés, F.; Romera-Branchat, M.; Martínez-Gallegos, R.; Patel, V.; Schneeberger, K.; Jang, S.; Altmüller, J.; Nürnberg, P.; Coupland, G. Floral induction in Arabidopsis thaliana by FLOWERING LOCUS T requires direct repression of BLADE-ON-PETIOLE genes by homeodomain protein PENNYWISE. Plant Physiol. 2015, 169, 2187–2199. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-N.; Lee, T.-Y.; Hung, Y.-C.; Li, G.-Z.; Tseng, K.-C.; Liu, Y.-H.; Kuo, P.-L.; Zheng, H.-Q.; Chang, W.-C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, 1155–1163. [Google Scholar] [CrossRef] [Green Version]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [Green Version]
- Simonini, S.; Stephenson, P.; Østergaard, L. A molecular framework controlling style morphology in Brassicaceae. Development 2018, 145, dev158105. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Fu, Y.; Zhao, S.; Gu, P.; Zhu, Z.; Sun, C.; Tan, L. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 2015, 14, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Xu, H.; Hepworth, S.R. BLADE-ON-PETIOLE genes: Setting boundaries in development and defense. Plant Sci. 2014, 215, 157–171. [Google Scholar] [CrossRef]
- Jost, M.; Taketa, S.; Mascher, M.; Himmelbach, A.; Yuo, T.; Shahinnia, F.; Rutten, T.; Druka, A.; Schmutzer, T.; Steuernagel, B.; et al. A homolog of Blade-On-Petiole 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 2016, 171, 1113–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Park, S.J.; van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [Green Version]
- Toriba, T.; Tokunaga, H.; Shiga, T.; Nie, F.; Naramoto, S.; Honda, E.; Tanaka, K.; Taji, T.; Itoh, J.-I.; Kyozuka, J. BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 2019, 10, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Ragni, L.; Belles-Boix, E.; Günl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Li, X.; Yuan, W.; Chen, G.; Kilian, A.; Li, J.; Xu, C.; Li, X.; Zhou, D.-X.; Wang, S.; et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003, 35, 418–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Sun, S.; Jin, J.; Fu, D.; Yang, X.; Weng, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 15504–15509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.-J.; Song, Q.-X.; Chen, H.-W.; Zou, H.-F.; Wei, W.; Kang, X.-S.; Ma, B.; Zhang, W.-K.; Zhang, J.-S.; Chen, S.-Y. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 2010, 232, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Bart, R.; Chern, M.; Park, C.J.; Bartley, L.; Ronald, P.C. A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2006, 2, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, 122–129. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Fu, D.; Xu, T.; Wu, C. VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture. Int. J. Mol. Sci. 2021, 22, 7909. https://doi.org/10.3390/ijms22157909
Li M, Fu D, Xu T, Wu C. VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture. International Journal of Molecular Sciences. 2021; 22(15):7909. https://doi.org/10.3390/ijms22157909
Chicago/Turabian StyleLi, Mu, Debao Fu, Tingting Xu, and Changyin Wu. 2021. "VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture" International Journal of Molecular Sciences 22, no. 15: 7909. https://doi.org/10.3390/ijms22157909
APA StyleLi, M., Fu, D., Xu, T., & Wu, C. (2021). VPB1 Encoding BELL-like Homeodomain Protein Is Involved in Rice Panicle Architecture. International Journal of Molecular Sciences, 22(15), 7909. https://doi.org/10.3390/ijms22157909





