The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation
Abstract
1. Introduction
2. Literature Search Strategy
3. Fras-Related Extracellular Matrix 1 (FREM1)
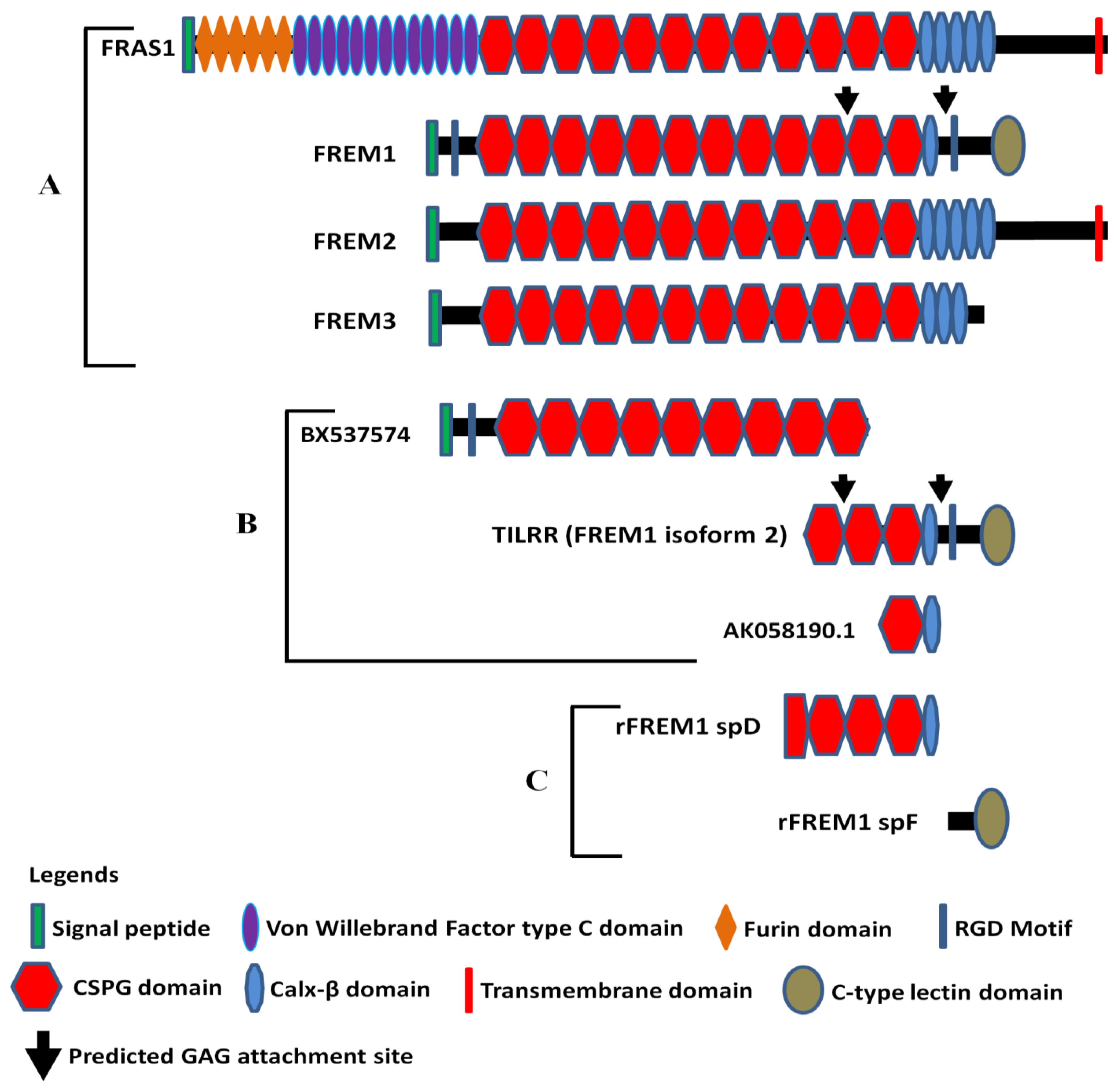
3.1. Molecular Structure of FREM1 and Its Splice Variants
3.2. FREM1 Expression, Localization, and Interactions
3.3. FREM1 and Immune Cell Infiltration
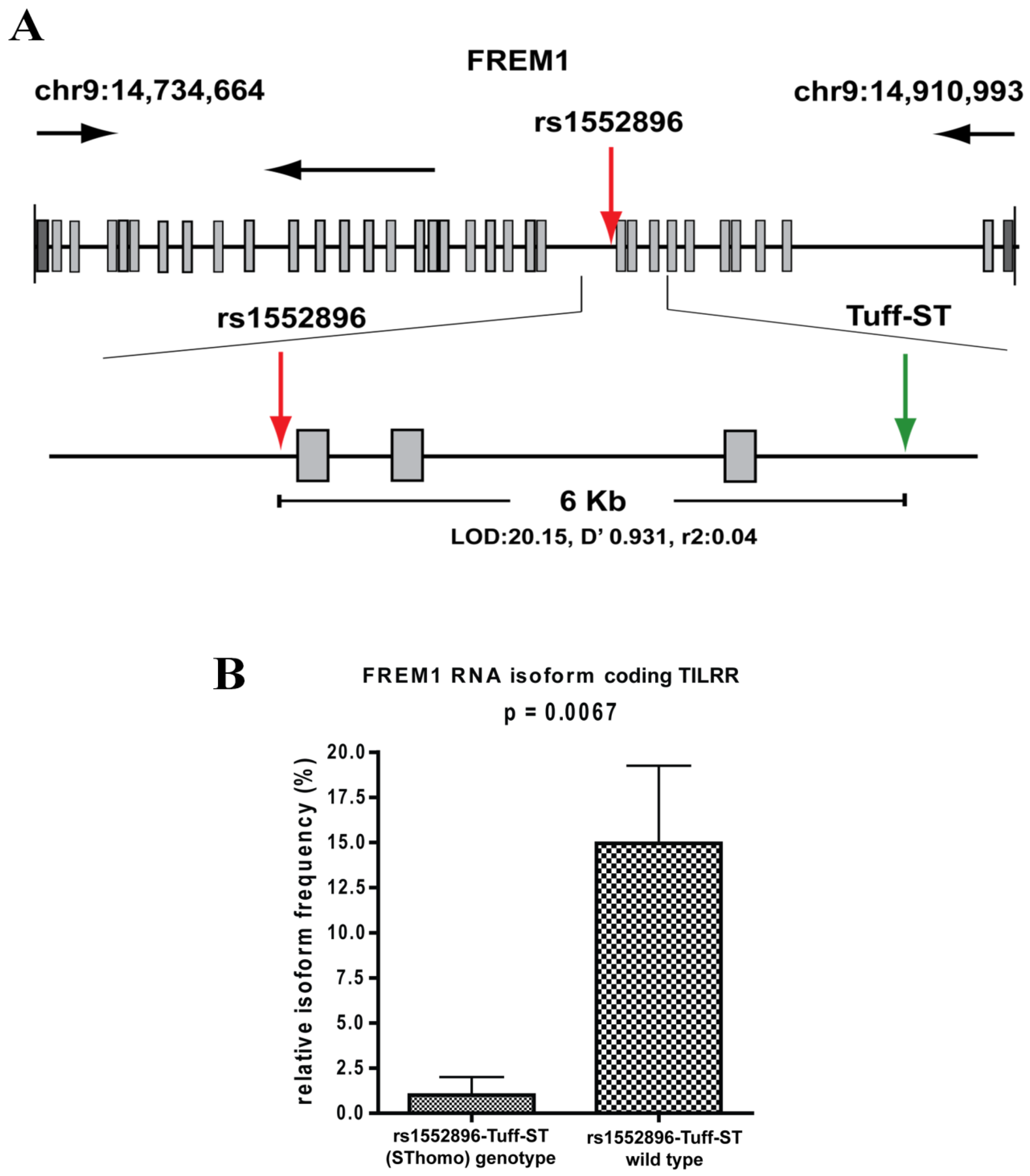
3.4. FREM1 Polymorphism and HIV-1 Resistance
3.5. Monoclonal Antibodies to Study FREM1 and Its Splice Variant TILRR
4. Toll-like Interleukin -1 Receptor Regulator (TILRR)
4.1. Characteristics of TILRR
4.2. TILRR Expression
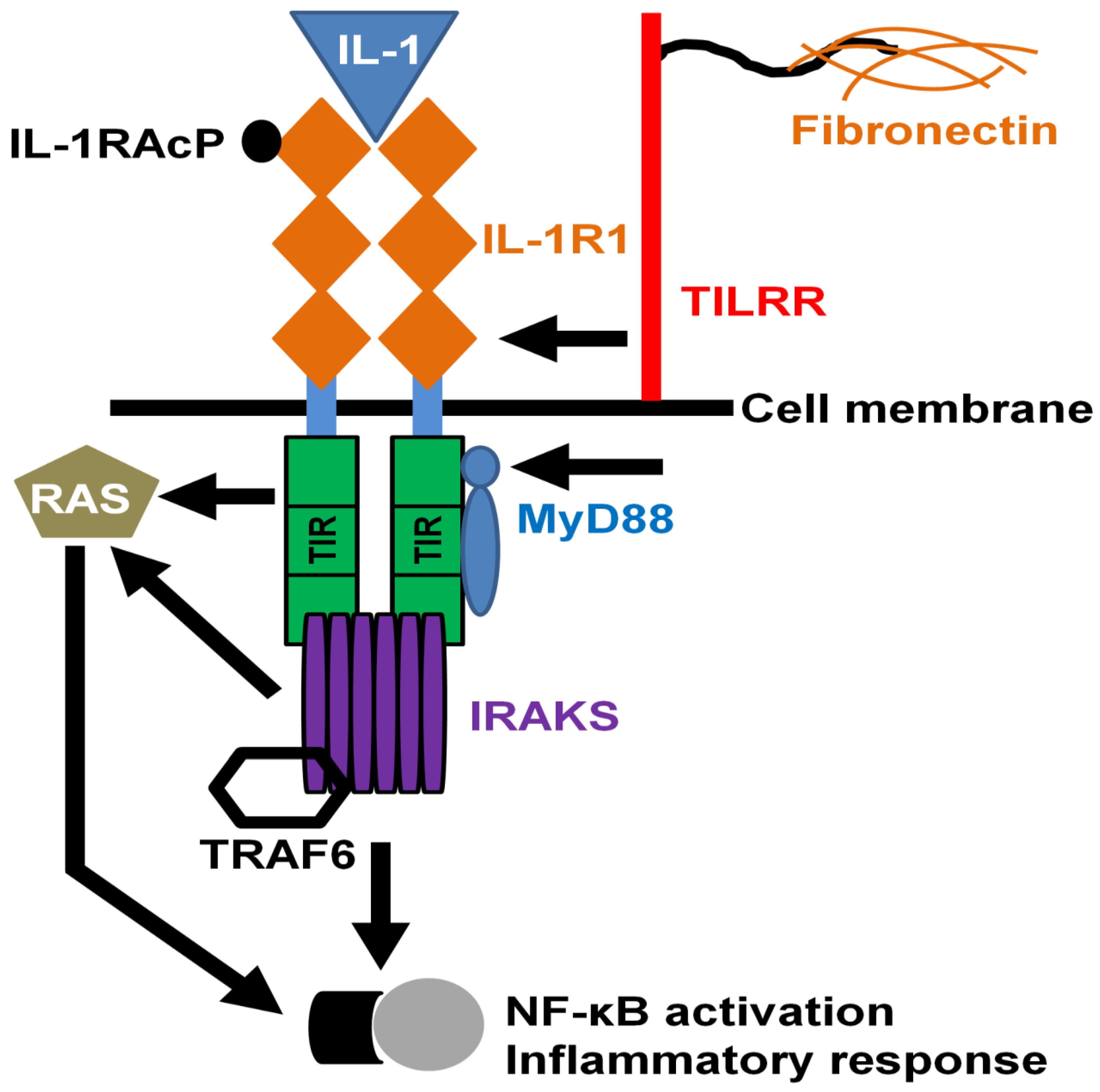
4.3. TILRR in Innate Signal Transduction and Inflammation
4.4. TILRR and Production of Inflammatory Mediators
4.5. TILRR and Migration of HIV-1 Target Cells
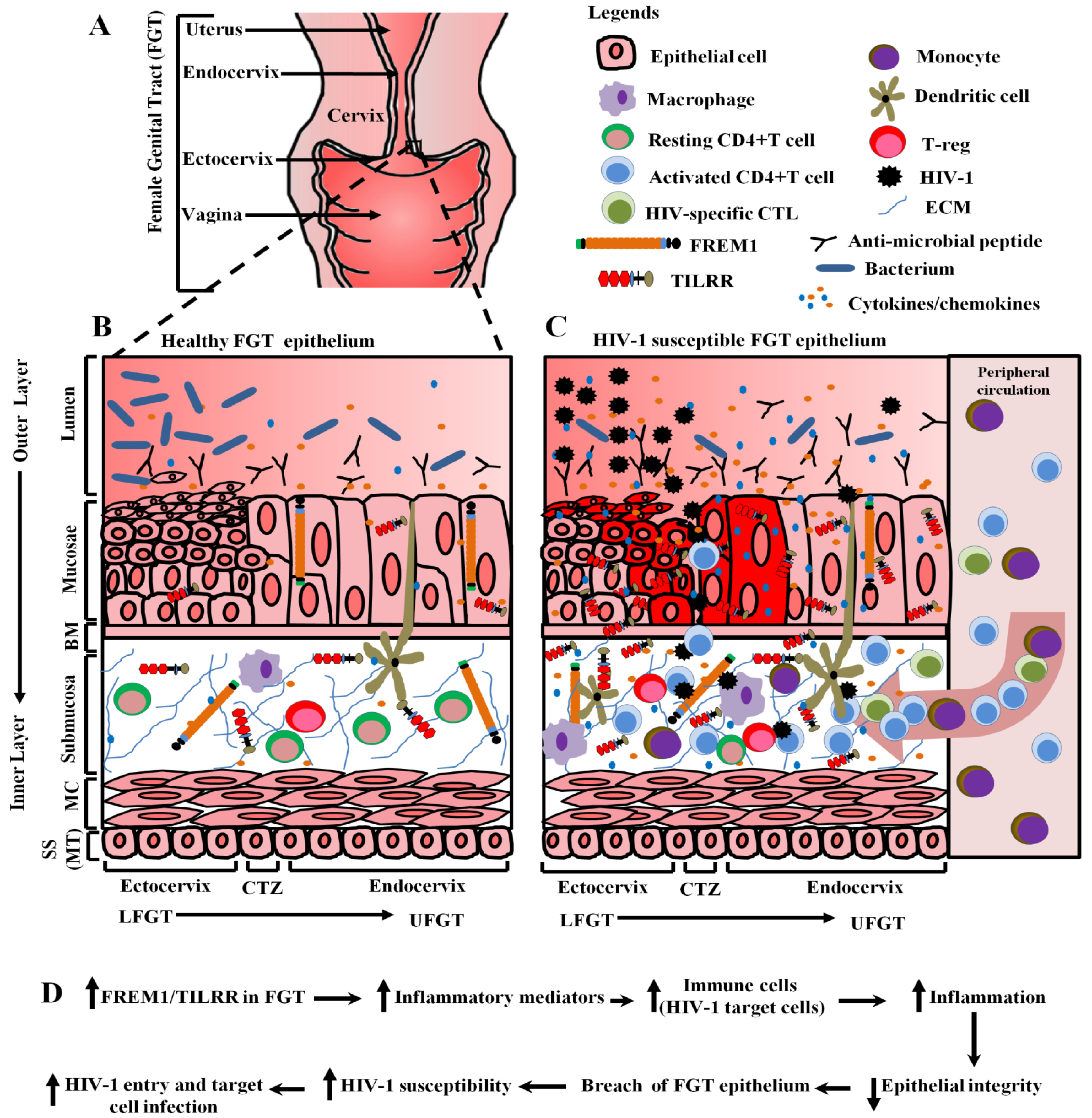
4.6. Potential Role of FREM1 and Its Isoform TILRR in Vaginal HIV-1 Acquisition
5. Conclusions and Future Direction
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Short, K.; Wiradjaja, F.; Smyth, I. Let‘s stick together: The role of the Fras1 and Frem proteins in epidermal adhesion. IUBMB Life 2007, 59, 427–435. [Google Scholar] [CrossRef]
- Kiyozumi, D.; Sugimoto, N.; Sekiguchi, K. Breakdown of the reciprocal stabilization of QBRICK/Frem1, Fras1, and Frem2 at the basement membrane provokes Fraser syndrome-like defects. Proc. Natl. Acad. Sci. USA 2006, 103, 11981–11986. [Google Scholar] [CrossRef]
- Smyth, I.; Du, X.; Taylor, M.S.; Justice, M.J.; Beutler, B.; Jackson, I.J. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proc. Natl. Acad. Sci. USA 2004, 101, 13560–13565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shephard, F.; Kim, H.B.; Palmer, I.R.; McHarg, S.; Fowler, G.J.; O’Neill, L.A.; Kiss-Toth, E.; Qwarnstrom, E.E. TILRR, a novel IL-1RI co-receptor, potentiates MyD88 recruitment to control Ras-dependent amplification of NF-kappaB. J. Biol. Chem. 2010, 285, 7222–7232. [Google Scholar] [CrossRef] [PubMed]
- Chacon-Camacho, O.F.; Zenker, M.; Schanze, D.; Ledesma-Gil, J.; Zenteno, J.C. Novel FREM1 mutations in a patient with MOTA syndrome: Clinical findings, mutation update and review of FREM1-related disorders literature. Eur. J. Med. Genet. 2017, 60, 190–194. [Google Scholar] [CrossRef]
- Zhang, X.; Pino, G.M.; Shephard, F.; Kiss-Toth, E.; Qwarnstrom, E.E. Distinct control of MyD88 adapter-dependent and Akt kinase-regulated responses by the interleukin (IL)-1RI co-receptor, TILRR. J. Biol. Chem. 2012, 287, 12348–12352. [Google Scholar] [CrossRef]
- Lacap, P.A.; Huntington, J.D.; Luo, M.; Nagelkerke, N.J.; Bielawny, T.; Kimani, J.; Wachihi, C.; Ngugi, E.N.; Plummer, F.A. Associations of human leukocyte antigen DRB with resistance or susceptibility to HIV-1 infection in the Pumwani Sex Worker Cohort. AIDS 2008, 22, 1029–1038. [Google Scholar] [CrossRef]
- Price, H.; Lacap, P.; Tuff, J.; Wachihi, C.; Kimani, J.; Ball, T.B.; Luo, M.; Plummer, F.A. A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS 2010, 24, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Hardie, R.A.; Knight, E.; Bruneau, B.; Semeniuk, C.; Gill, K.; Nagelkerke, N.; Kimani, J.; Wachihi, C.; Ngugi, E.; Luo, M.; et al. A common human leucocyte antigen-DP genotype is associated with resistance to HIV-1 infection in Kenyan sex workers. AIDS 2008, 22, 2038–2042. [Google Scholar] [CrossRef]
- Hardie, R.A.; Luo, M.; Bruneau, B.; Knight, E.; Nagelkerke, N.J.; Kimani, J.; Wachihi, C.; Ngugi, E.N.; Plummer, F.A. Human leukocyte antigen-DQ alleles and haplotypes and their associations with resistance and susceptibility to HIV-1 infection. AIDS 2008, 22, 807–816. [Google Scholar] [CrossRef]
- Simonsen, J.N.; Plummer, F.A.; Ngugi, E.N.; Black, C.; Kreiss, J.K.; Gakinya, M.N.; Waiyaki, P.; D’Costa, L.J.; Ndinya-Achola, J.O.; Piot, P.; et al. HIV infection among lower socioeconomic strata prostitutes in Nairobi. AIDS 1990, 4, 139–144. [Google Scholar] [CrossRef]
- Kreiss, J.K.; Koech, D.; Plummer, F.A.; Holmes, K.K.; Lightfoote, M.; Piot, P.; Ronald, A.R.; Ndinya-Achola, J.O.; D’Costa, L.J.; Roberts, P.; et al. AIDS virus infection in Nairobi prostitutes. Spread of the epidemic to East Africa. N. Engl. J. Med. 1986, 314, 414–418. [Google Scholar] [CrossRef]
- Plummer, F.A.; Simonsen, J.N.; Cameron, D.W.; Ndinya-Achola, J.O.; Kreiss, J.K.; Gakinya, M.N.; Waiyaki, P.; Cheang, M.; Piot, P.; Ronald, A.R.; et al. Cofactors in male-female sexual transmission of human immunodeficiency virus type 1. J. Infect. Dis. 1991, 163, 233–239. [Google Scholar] [CrossRef]
- Fowke, K.R.; Nagelkerke, N.J.; Kimani, J.; Simonsen, J.N.; Anzala, A.O.; Bwayo, J.J.; MacDonald, K.S.; Ngugi, E.N.; Plummer, F.A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 1996, 348, 1347–1351. [Google Scholar] [CrossRef]
- Luo, M.; Sainsbury, J.; Tuff, J.; Lacap, P.A.; Yuan, X.Y.; Hirbod, T.; Kimani, J.; Wachihi, C.; Ramdahin, S.; Bielawny, T.; et al. A genetic polymorphism of FREM1 is associated with resistance against HIV infection in the Pumwani sex worker cohort. J. Virol. 2012, 86, 11899–11905. [Google Scholar] [CrossRef] [PubMed]
- Kashem, M.A.; Li, H.; Toledo, N.P.; Omange, R.W.; Liang, B.; Liu, L.R.; Li, L.; Yang, X.; Yuan, X.-Y.; Kindrachuk, J.; et al. Toll-like Interleukin 1 Receptor Regulator Is an Important Modulator of Inflammation Responsive Genes. Front. Immunol. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Fichorova, R.N.; Tucker, L.D.; Anderson, D.J. The molecular basis of nonoxynol-9-induced vaginal inflammation and its possible relevance to human immunodeficiency virus type 1 transmission. J. Infect. Dis. 2001, 184, 418–428. [Google Scholar] [CrossRef] [PubMed]
- Dabee, S.; Barnabas, S.L.; Lennard, K.S.; Jaumdally, S.Z.; Gamieldien, H.; Balle, C.; Happel, A.U.; Murugan, B.D.; Williamson, A.L.; Mkhize, N.; et al. Defining characteristics of genital health in South African adolescent girls and young women at high risk for HIV infection. PLoS ONE 2019, 14, e0213975. [Google Scholar] [CrossRef] [PubMed]
- Cohen, C.R.; Moscicki, A.B.; Scott, M.E.; Ma, Y.; Shiboski, S.; Bukusi, E.; Daud, I.; Rebbapragada, A.; Brown, J.; Kaul, R. Increased levels of immune activation in the genital tract of healthy young women from sub-Saharan Africa. AIDS 2010, 24, 2069–2074. [Google Scholar] [CrossRef]
- Kaul, R.; Prodger, J.; Joag, V.; Shannon, B.; Yegorov, S.; Galiwango, R.; McKinnon, L. Inflammation and HIV Transmission in Sub-Saharan Africa. Curr. HIV/AIDS Rep. 2015, 12, 216–222. [Google Scholar] [CrossRef]
- Pan, X.; Baldauf, H.M.; Keppler, O.T.; Fackler, O.T. Restrictions to HIV-1 replication in resting CD4+ T lymphocytes. Cell Res. 2013, 23, 876–885. [Google Scholar] [CrossRef]
- Biancotto, A.; Iglehart, S.J.; Vanpouille, C.; Condack, C.E.; Lisco, A.; Ruecker, E.; Hirsch, I.; Margolis, L.B.; Grivel, J.C. HIV-1 induced activation of CD4+ T cells creates new targets for HIV-1 infection in human lymphoid tissue ex vivo. Blood 2008, 111, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Wietgrefe, S.W.; Li, Q.; Shore, M.D.; Duan, L.; Reilly, C.; Lifson, J.D.; Haase, A.T. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. USA 2004, 101, 5640–5645. [Google Scholar] [CrossRef] [PubMed]
- Passmore, J.A.; Jaspan, H.B.; Masson, L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr. Opin. HIV AIDS 2016, 11, 156–162. [Google Scholar] [CrossRef]
- Masson, L.; Passmore, J.A.; Liebenberg, L.J.; Werner, L.; Baxter, C.; Arnold, K.B.; Williamson, C.; Little, F.; Mansoor, L.E.; Naranbhai, V.; et al. Genital inflammation and the risk of HIV acquisition in women. Clin. Infect. Dis. 2015, 61, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Kiyozumi, D.; Osada, A.; Sugimoto, N.; Weber, C.N.; Ono, Y.; Imai, T.; Okada, A.; Sekiguchi, K. Identification of a novel cell-adhesive protein spatiotemporally expressed in the basement membrane of mouse developing hair follicle. Exp. Cell Res. 2005, 306, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Staub, E.; Hinzmann, B.; Rosenthal, A. A novel repeat in the melanoma-associated chondroitin sulfate proteoglycan defines a new protein family. FEBS Lett. 2002, 527, 114–118. [Google Scholar] [CrossRef]
- Yuan, X.Y.; Liu, L.R.; Krawchenko, A.; Sainsbury, J.; Zhao, L.; Plummer, F.; Yang, X.; Luo, M. Development of monoclonal antibodies to interrogate functional domains and isoforms of FREM1 protein. Monoclon. Antib. Immunodiagn. Immunother. 2014, 33, 129–140. [Google Scholar] [CrossRef]
- Nishiyama, A.; Dahlin, K.J.; Prince, J.T.; Johnstone, S.R.; Stallcup, W.B. The primary structure of NG2, a novel membrane-spanning proteoglycan. J. Cell Biol. 1991, 114, 359–371. [Google Scholar] [CrossRef]
- Burg, M.A.; Tillet, E.; Timpl, R.; Stallcup, W.B. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J. Biol. Chem. 1996, 271, 26110–26116. [Google Scholar] [CrossRef] [PubMed]
- Tillet, E.; Gential, B.; Garrone, R.; Stallcup, W.B. NG2 proteoglycan mediates beta1 integrin-independent cell adhesion and spreading on collagen VI. J. Cell Biochem. 2002, 86, 726–736. [Google Scholar] [CrossRef]
- Nishiyama, A.; Stallcup, W.B. Expression of NG2 proteoglycan causes retention of type VI collagen on the cell surface. Mol. Biol. Cell 1993, 4, 1097–1108. [Google Scholar] [CrossRef]
- Goretzki, L.; Burg, M.A.; Grako, K.A.; Stallcup, W.B. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J. Biol. Chem. 1999, 274, 16831–16837. [Google Scholar] [CrossRef]
- Nathanson, J.; Swarr, D.T.; Singer, A.; Liu, M.; Chinn, A.; Jones, W.; Hurst, J.; Khalek, N.; Zackai, E.; Slavotinek, A. Novel FREM1 mutations expand the phenotypic spectrum associated with Manitoba-oculo-tricho-anal (MOTA) syndrome and bifid nose renal agenesis anorectal malformations (BNAR) syndrome. Am. J. Med. Genet. A 2013, 161A, 473–478. [Google Scholar] [CrossRef]
- Schwarz, E.M.; Benzer, S. Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1997, 94, 10249–10254. [Google Scholar] [CrossRef]
- Thierry-Mieg, D.; Thierry-Mieg, J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome. Biol. 2006, 7, 11–14. [Google Scholar] [CrossRef]
- NCBI. The AceView Genes. Available online: https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ (accessed on 20 April 2020).
- Beck, T.F.; Veenma, D.; Shchelochkov, O.A.; Yu, Z.; Kim, B.J.; Zaveri, H.P.; van Bever, Y.; Choi, S.; Douben, H.; Bertin, T.K.; et al. Deficiency of FRAS1-related extracellular matrix 1 (FREM1) causes congenital diaphragmatic hernia in humans and mice. Hum. Mol. Genet. 2013, 22, 1026–1038. [Google Scholar] [CrossRef]
- Petrou, P.; Chiotaki, R.; Dalezios, Y.; Chalepakis, G. Overlapping and divergent localization of Frem1 and Fras1 and its functional implications during mouse embryonic development. Exp. Cell Res. 2007, 313, 910–920. [Google Scholar] [CrossRef]
- Vissers, L.E.; Cox, T.C.; Maga, A.M.; Short, K.M.; Wiradjaja, F.; Janssen, I.M.; Jehee, F.; Bertola, D.; Liu, J.; Yagnik, G.; et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet. 2011, 7, e1002278. [Google Scholar] [CrossRef] [PubMed]
- Alazami, A.M.; Shaheen, R.; Alzahrani, F.; Snape, K.; Saggar, A.; Brinkmann, B.; Bavi, P.; Al-Gazali, L.I.; Alkuraya, F.S. FREM1 mutations cause bifid nose, renal agenesis, and anorectal malformations syndrome. Am. J. Hum. Genet. 2009, 85, 414–418. [Google Scholar] [CrossRef]
- Arroyo, A.G.; Iruela-Arispe, M.L. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc. Res. 2010, 86, 226–235. [Google Scholar] [CrossRef]
- Adair-Kirk, T.L.; Senior, R.M. Fragments of extracellular matrix as mediators of inflammation. Int. J. Biochem. Cell Biol. 2008, 40, 1101–1110. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Popovich, P.G. Extracellular matrix regulation of inflammation in the healthy and injured spinal cord. Exp. Neurol. 2014, 258, 24–34. [Google Scholar] [CrossRef]
- Gopal, S. Syndecans in Inflammation at a Glance. Front. Immunol. 2020, 11, 227. [Google Scholar] [CrossRef]
- Li, H.N.; Li, X.R.; Lv, Z.T.; Cai, M.M.; Wang, G.; Yang, Z.F. Elevated expression of FREM1 in breast cancer indicates favorable prognosis and high-level immune infiltration status. Cancer Med. 2020. [Google Scholar] [CrossRef]
- Kim, C.H. Crawling of effector T cells on extracellular matrix: Role of integrins in interstitial migration in inflamed tissues. Cell Mol. Immunol. 2014, 11, 1–4. [Google Scholar] [CrossRef]
- Iqbal, S.M.; Ball, T.B.; Kimani, J.; Kiama, P.; Thottingal, P.; Embree, J.E.; Fowke, K.R.; Plummer, F.A. Elevated T cell counts and RANTES expression in the genital mucosa of HIV-1-resistant Kenyan commercial sex workers. J. Infect. Dis. 2005, 192, 728–738. [Google Scholar] [CrossRef]
- Burgener, A.; Boutilier, J.; Wachihi, C.; Kimani, J.; Carpenter, M.; Westmacott, G.; Cheng, K.; Ball, T.B.; Plummer, F. Identification of differentially expressed proteins in the cervical mucosa of HIV-1-resistant sex workers. J. Proteome. Res. 2008, 7, 4446–4454. [Google Scholar] [CrossRef]
- Iqbal, S.M.; Ball, T.B.; Levinson, P.; Maranan, L.; Jaoko, W.; Wachihi, C.; Pak, B.J.; Podust, V.N.; Broliden, K.; Hirbod, T.; et al. Elevated elafin/trappin-2 in the female genital tract is associated with protection against HIV acquisition. AIDS 2009, 23, 1669–1677. [Google Scholar] [CrossRef]
- Alimonti, J.B.; Kimani, J.; Matu, L.; Wachihi, C.; Kaul, R.; Plummer, F.A.; Fowke, K.R. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 2006, 84, 482–485. [Google Scholar] [CrossRef]
- Alimonti, J.B.; Koesters, S.A.; Kimani, J.; Matu, L.; Wachihi, C.; Plummer, F.A.; Fowke, K.R. CD4+ T cell responses in HIV-exposed seronegative women are qualitatively distinct from those in HIV-infected women. J. Infect. Dis. 2005, 191, 20–24. [Google Scholar] [CrossRef]
- Luo, M.; Daniuk, C.A.; Diallo, T.O.; Capina, R.E.; Kimani, J.; Wachihi, C.; Kimani, M.; Bielawny, T.; Peterson, T.; Mendoza, M.G.; et al. For protection from HIV-1 infection, more might not be better: A systematic analysis of HIV Gag epitopes of two alleles associated with different outcomes of HIV-1 infection. J. Virol. 2012, 86, 1166–1180. [Google Scholar] [CrossRef]
- Rowland-Jones, S.L.; Dong, T.; Fowke, K.R.; Kimani, J.; Krausa, P.; Newell, H.; Blanchard, T.; Ariyoshi, K.; Oyugi, J.; Ngugi, E.; et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 1998, 102, 1758–1765. [Google Scholar] [CrossRef] [PubMed]
- Ball, T.B.; Ji, H.; Kimani, J.; McLaren, P.; Marlin, C.; Hill, A.V.; Plummer, F.A. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS 2007, 21, 1091–1101. [Google Scholar] [CrossRef]
- Salazar, G.; Zhang, N.; Fu, T.M.; An, Z. Antibody therapies for the prevention and treatment of viral infections. NPJ Vaccines 2017, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.Y.; Guo, W.J.; Pan, S.H.; Zhang, Y.; Gao, F.L.; Wang, J.T.; Zhang, S.; Li, H.Y.; Wang, R.; Zhang, X. TILRR (FREM1 isoform 2) is a prognostic biomarker correlated with immune infiltration in breast cancer. Aging 2020, 12, 19335–19351. [Google Scholar] [CrossRef] [PubMed]
- Hudson, R.C.; Gray, C.; Kiss-Toth, E.; Chico, T.J.; Qwarnstrom, E.E. Bioinformatics Analysis of the FREM1 Gene-Evolutionary Development of the IL-1R1 Co-Receptor, TILRR. Biology 2012, 1, 484–494. [Google Scholar] [CrossRef]
- Smith, S.A.; Samokhin, A.O.; Alfadi, M.; Murphy, E.C.; Rhodes, D.; Holcombe, W.M.L.; Kiss-Toth, E.; Storey, R.F.; Yee, S.P.; Francis, S.E.; et al. The IL-1RI Co-Receptor TILRR (FREM1 Isoform 2) Controls Aberrant Inflammatory Responses and Development of Vascular Disease. JACC Basic Transl. Sci. 2017, 2, 398–414. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Bowie, A.G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007, 7, 353–364. [Google Scholar] [CrossRef]
- O’Neill, L.A. How Toll-like receptors signal: What we know and what we don’t know. Curr. Opin. Immunol. 2006, 18, 3–9. [Google Scholar] [CrossRef]
- Rhodes, D.M.; Smith, S.A.; Holcombe, M.; Qwarnstrom, E.E. Computational Modelling of NF-kappaB Activation by IL-1RI and Its Co-Receptor TILRR, Predicts a Role for Cytoskeletal Sequestration of IkappaBalpha in Inflammatory Signalling. PLoS ONE 2015, 10, e0129888. [Google Scholar] [CrossRef]
- Mac Gabhann, F. TILRR Steers Interleukin-1 Signaling: Co-Receptor Provides Context and a Therapeutic Target. JACC Basic Transl. Sci. 2017, 2, 415–417. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2. [Google Scholar] [CrossRef]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (MCP-1): An overview. J. Interferon Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Kashem, M.A.; Ren, X.; Li, H.; Liang, B.; Li, L.; Lin, F.; Plummer, F.; Luo, M. TILRR Promotes Migration of Immune Cells Through Induction of Soluble Inflammatory Mediators. Front. Cell Dev. Biol. 2020, 8, 1–13. [Google Scholar] [CrossRef]
- McKinnon, L.R.; Liebenberg, L.J.; Yende-Zuma, N.; Archary, D.; Ngcapu, S.; Sivro, A.; Nagelkerke, N.; Garcia Lerma, J.G.; Kashuba, A.D.; Masson, L.; et al. Genital inflammation undermines the effectiveness of tenofovir gel in preventing HIV acquisition in women. Nat. Med. 2018, 24, 491–496. [Google Scholar] [CrossRef]
- Barnes, P.J.; Karin, M. Nuclear factor-kappaB: A pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 1997, 336, 1066–1071. [Google Scholar] [CrossRef]
- Arnold, K.B.; Burgener, A.; Birse, K.; Romas, L.; Dunphy, L.J.; Shahabi, K.; Abou, M.; Westmacott, G.R.; McCorrister, S.; Kwatampora, J.; et al. Increased levels of inflammatory cytokines in the female reproductive tract are associated with altered expression of proteases, mucosal barrier proteins, and an influx of HIV-susceptible target cells. Mucosal. Immunol. 2016, 9, 194–205. [Google Scholar] [CrossRef]
- Pavlakis, E.; Chiotaki, R.; Chalepakis, G. The role of Fras1/Frem proteins in the structure and function of basement membrane. Int. J. Biochem. Cell Biol. 2011, 43, 487–495. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashem, M.A.; Li, H.; Liu, L.R.; Liang, B.; Omange, R.W.; Plummer, F.A.; Luo, M. The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation. Int. J. Mol. Sci. 2021, 22, 7825. https://doi.org/10.3390/ijms22157825
Kashem MA, Li H, Liu LR, Liang B, Omange RW, Plummer FA, Luo M. The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation. International Journal of Molecular Sciences. 2021; 22(15):7825. https://doi.org/10.3390/ijms22157825
Chicago/Turabian StyleKashem, Mohammad Abul, Hongzhao Li, Lewis Ruxi Liu, Binhua Liang, Robert Were Omange, Francis A. Plummer, and Ma Luo. 2021. "The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation" International Journal of Molecular Sciences 22, no. 15: 7825. https://doi.org/10.3390/ijms22157825
APA StyleKashem, M. A., Li, H., Liu, L. R., Liang, B., Omange, R. W., Plummer, F. A., & Luo, M. (2021). The Potential Role of FREM1 and Its Isoform TILRR in HIV-1 Acquisition through Mediating Inflammation. International Journal of Molecular Sciences, 22(15), 7825. https://doi.org/10.3390/ijms22157825





