1. Introduction
Since the beginning of the 21st century, researchers have focused their efforts on using dendritic cells (DC) as a powerful tool to control altered immune responses and restore the homeostatic balance. DC are a heterogeneous group of potent antigen-presenting cells (APC) with the capacity of processing and presenting antigens to naïve T cells, thus priming adaptive immune response [
1]. Importantly, DC are able to control T cell activation, making them a promising treatment not only for cancer therapy, but also for restoring immune tolerance in autoimmune diseases and avoiding allograft rejection [
2,
3].
In this context, tolerogenic dendritic cells (tolDC) represent a recent emerging strategy to induce a specific and long-term re-establishment of the immune tolerance towards self-antigens causing the immunogenic response of autoimmune diseases such as multiple sclerosis (MS), rheumatoid arthritis (RA) or type-1 diabetes (T1D). In this way, a tolDC strategy avoids the chronic and general immunosuppression of current treatments producing different serious adverse effects in patients [
4].
TolDC are defined as semi-mature DC, an intermediate state of maturation between immature DC (iDC) and mature DC (mDC), combining characteristics of both states. Phenotypically, tolDC express low levels of co-stimulatory molecules (CD80, CD83, CD86) and HLA-DR compared to mDC. Functionally, tolDC secrete anti-inflammatory cytokines (IL-10, TGF-β) and have reduced capacity to trigger T cell proliferation due to the induction of regulatory T cells (Treg), T cell anergy/depletion or T cell hyporesponsiveness [
5,
6,
7,
8]. Importantly, in contrast to iDC, tolDC are phenotypically stable and upon pro-inflammatory stimuli they do not become mDC. Therefore, tolDC are not potentially harmful when being injected in patients with an inflammatory disease.
DC can be derived ex vivo from autologous peripheral blood monocytes in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL4 [
9], and different protocols have been described to generate tolDC by using anti-inflammatory cytokines (IL-10, TGF-β), pharmacological agents (vitamin D3, dexamethasone, rapamycin), commercialized drugs (aspirin, simvastatin) or by genetic engineering [
5,
10,
11,
12]. From all these protocols, the use of the active form of vitamin D3 (VitD3, 1,25-dyhydroxyvitamin D3) is one of the approaches most widely used to generate tolDC due to their reported characteristics: VitD3-tolDC show a semi-mature phenotype and can induce CD4
+ Foxp3
+ Treg, T cell hyporesponsiveness and a switch of the immune response towards a Th2 profile [
10,
13,
14].
To date, several phase I clinical trials have been conducted or are currently ongoing using tolDC in patients with RA, MS, optic neuromyelitis, Crohn’s disease and T1D [
4,
15]. All these studies indicate that tolDC therapies are safe, do not present relevant side effects and are well tolerated by patients. However, in-depth characterization of tolDC is still required to improve their effectiveness and their translation into the clinic.
To date, no universal tolDC biomarker has been identified [
16]. Currently, quality control studies to evaluate the proper tolDC production prior to their administration into the patient are based on phenotyping and/or allogeneic T cell proliferation assays, which are unreliable or time-consuming methods, respectively. An appropriate selection of biomarkers of tolerogenicity would allow us to guarantee the therapeutical success and biosafety of these treatments by providing a faster and more reliable way of validating the effectiveness of the generated cellular product.
To improve the efficacy of VitD3-tolDC therapy for future phase II clinical trials, we have been investigating transcriptional differences between mDC and VitD3-tolDC to identify the molecules and pathways involved in tolerogenicity. Through a microarray analysis we found upregulated in VitD3-tolDC, compared to mDC, the genes
CD115 (or
CSF1R, that encodes for macrophage colony-stimulating factor receptor) as well as
CSF1 (colony-stimulating factor 1, also known as macrophage colony-stimulating factor, M-CSF, one of the two possible CSF1R ligands), among others [
17]. It has been reported that CSF1R regulates macrophage survival and differentiation [
18] and, interestingly, in the presence of IL-4 CSF1-CSF1R signaling promotes the differentiation of CD14⁺ monocytes into suppressive macrophages that inhibit CD8
+ T cell proliferation and expand CD4
+ Foxp3
+ Treg in vitro [
19]. Consistently, CSF1 blockade prevents the in vivo expansion of CD4
+ Foxp3
+ Treg cells and abrogates the induction of transplantation tolerance [
19]. Indeed, pharmacological CSF1R inhibition is under investigation as a therapeutic strategy for cancer therapy application [
20,
21,
22].
On the other hand, our microarray study also revealed
CD209 (or
DC-SIGN, which encodes for DC-specific intracellular adhesion molecule 3-grabbing non-integrin) as another upregulated gene in VitD3-tolDC. It has been reported that DC-SIGN has a role in the regulation of T cell proliferation through its interaction with ICAM-3 (intercellular adhesion molecule-3), expressed by T cells [
23]. Moreover, DC-SIGN expression is induced by CSF1 + IL-4 and is required for the development of the above-mentioned suppressive capacity of macrophages through IL-10 production [
19,
24]. All these findings suggest a possible role of CD115 and CD209 molecules in the tolerogenic potential of VitD3-tolDC.
Gene silencing using small interfering ribonucleic acids (siRNA) is a powerful and widely used method to investigate the functional relevance of specific genes. However, strategies available to deliver specific siRNA into cells are not effective for all cell types and require optimization. In this regard, primary human monocyte-derived DC are cells which are difficult to transfect without inducing cell maturation. Some studies have reported optimized protocols for the transfection of monocyte-derived DC [
25,
26,
27]. However, none of them have been optimized so far to deliver siRNA into tolDC during their process of differentiation.
In this study, two different approaches, a lipid-based (HiPerFect®) method and a polymer mimicking influenza hemagglutinin virus (Viromer® blue, ViroB) transfection were optimized and tested to downregulate the expression of CD115 and CD209 molecules during the differentiation process of VitD3-tolDC to determine their role in tolerogenicity. However, only ViroB was shown to be a fast and easy strategy to deliver siRNA into monocytes on day 1 of culture without altering viability, phenotype and functionality of VitD3-tolDC.
The application of the ViroB protocol to analyze the role of
CD115 and
CD209 genes revealed that the molecule CSF1R (CD115), but not DC-SIGN (CD209), plays a partial role in the induction of tolerance through VitD3 in tolDC by enhancing glycolysis and glucose consumption to produce lactate, a novel suppressor mechanism of T cell proliferation recently found in autologous tolerogenic dendritic cells (ATDCs) [
28].
3. Discussion
TolDC cell-based therapy is a promising strategy to restore immune tolerance in autoimmune diseases such as MS. Many in vitro and in vivo studies have demonstrated their ability to control autoreactive T cell proliferation by different mechanisms (induction of Treg and Breg, IL-10 and TGF-β secretion, induction of T cell hyporesponsiveness, among others), as well as their beneficial effect in reducing inflammation and clinical signs when administrated in vivo in animal models. In addition, some phase I clinical trials have reported their safety and good tolerability in RA, T1D, MS and Crohn’s patients [
4].
Unfortunately, to date, there is not a universal biomarker for tolDC. Therefore, currently tolDC can be characterized phenotypically, but levels of expression of co-stimulatory molecules and HLA-DR are difficult to standardize and are sometimes unreliable. On the other hand, functional assays are trustworthy but time-consuming because they require around 5 days of co-culture. Consequently, there is an unmet need to find fast and robust functional biomarkers of tolDC to improve their efficacy and translation into the clinic.
In this context, our group has developed an autologous antigen-specific cell therapy based on autologous antigen-specific VitD3-tolDC, which is currently being administrated to active MS patients from a phase I clinical trial (TOLERVIT-MS: Tolerogenic Dendritic Cells as a Therapeutic Strategy for the Treatment of Multiple Sclerosis Patients, EudraCT: 2015-003541-26, ClinicalTrials.gov ID: NCT02903537). To go a step further and try to improve the efficacy of VitD3-tolDC therapy prior to initiating a phase II clinical trial in MS patients, our group has searched for genes differentially expressed in VitD3-tolDC compared to mDC with the objective to identify molecules and pathways involved in the tolerogenicity of VitD3-tolDC.
To investigate the tolerogenic role of the selected differentially expressed genes in VitD3-tolDC, a fast and easy technique is needed. Since most of genes were upregulated in VitD3-tolDC, gene silencing using siRNA was chosen as the method to analyze the effects of loss-of-function of those genes in VitD3-tolDC. Although there are different transfection strategies to introduce siRNA into primary cells (viral vectors, electroporation and lipid-based vectors), monocytes and DC are highly sensitive and especially difficult to transfect. Over recent years, several studies have reported optimized protocols to transfect DC [
25,
26,
27], and tolDC have been generated even by genetic inhibition of co-stimulatory molecules expression [
30]. However, these protocols are aimed at transfecting already differentiated DC, but not monocytes during their differentiation culture to tolDC.
In this study, we optimized two different transfection methods, a lipid-based (HiPerFect) and a polymer-based (ViroB) transfection strategy, to deliver siRNA into VitD3-tolDC with high efficiency (>80% transfected cells) and without altering their viability, phenotype and functional characteristics.
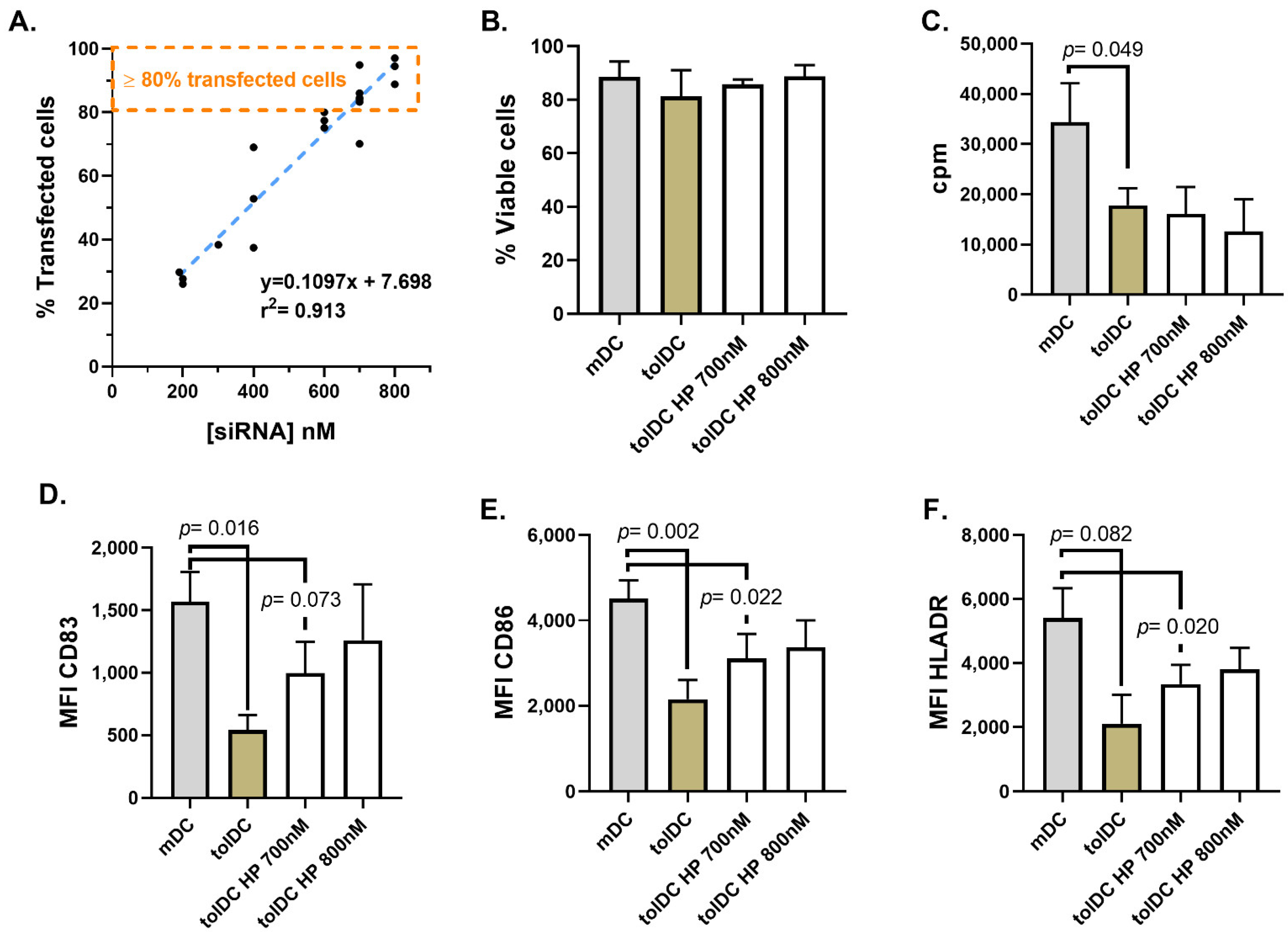
HiPerFect is a lipid transfection reagent based on the formation of siRNA packaging vesicles or liposomes structurally similar to cell or organelles membranes, therefore facilitating the cellular uptake and protecting siRNA from enzymatic degradation during cellular endocytosis [
31]. In 2014, Troegeler and colleagues developed a protocol using HiPerfect to achieve gene silencing in primary blood monocytes, DC and macrophages without altering their cell functions [
29]. We optimized the reported protocol to achieve a high transfection efficiency in monocytes during their culture differentiation into VitD3-tolDC. Although high concentrations of siRNA were needed to achieve >80% transfected cells (700 nM and 800 nM siRNA), the protocol did not cause relevant viability, phenotypical or functional alterations in monocytes transfected on day 5 of differentiation, after receiving the maturation stimulus (at the end of the differentiation culture). However, we cannot discard other alterations caused by the high concentration of siRNA required for the transfection (700 nM), such as the induction of a virus infection-like response triggering apoptosis and inflammasome response in the VitD3-tolDC. Future studies will be required to address this issue. On the other hand, when the HiPerfect protocol was used to transfect monocytes on day 4 of culture (prior to maturation stimulus), cells were sensitive to the manipulation required during the HiPerfect transfection procedure, preventing the effect of VitD3 and inducing phenotypical and functional characteristics typical of mDC in transfected VitD3-tolDC. Therefore, this protocol does not allow the analysis of the tolerogenic function of potential genes during the differentiation of monocytes into VitD3-tolDC.
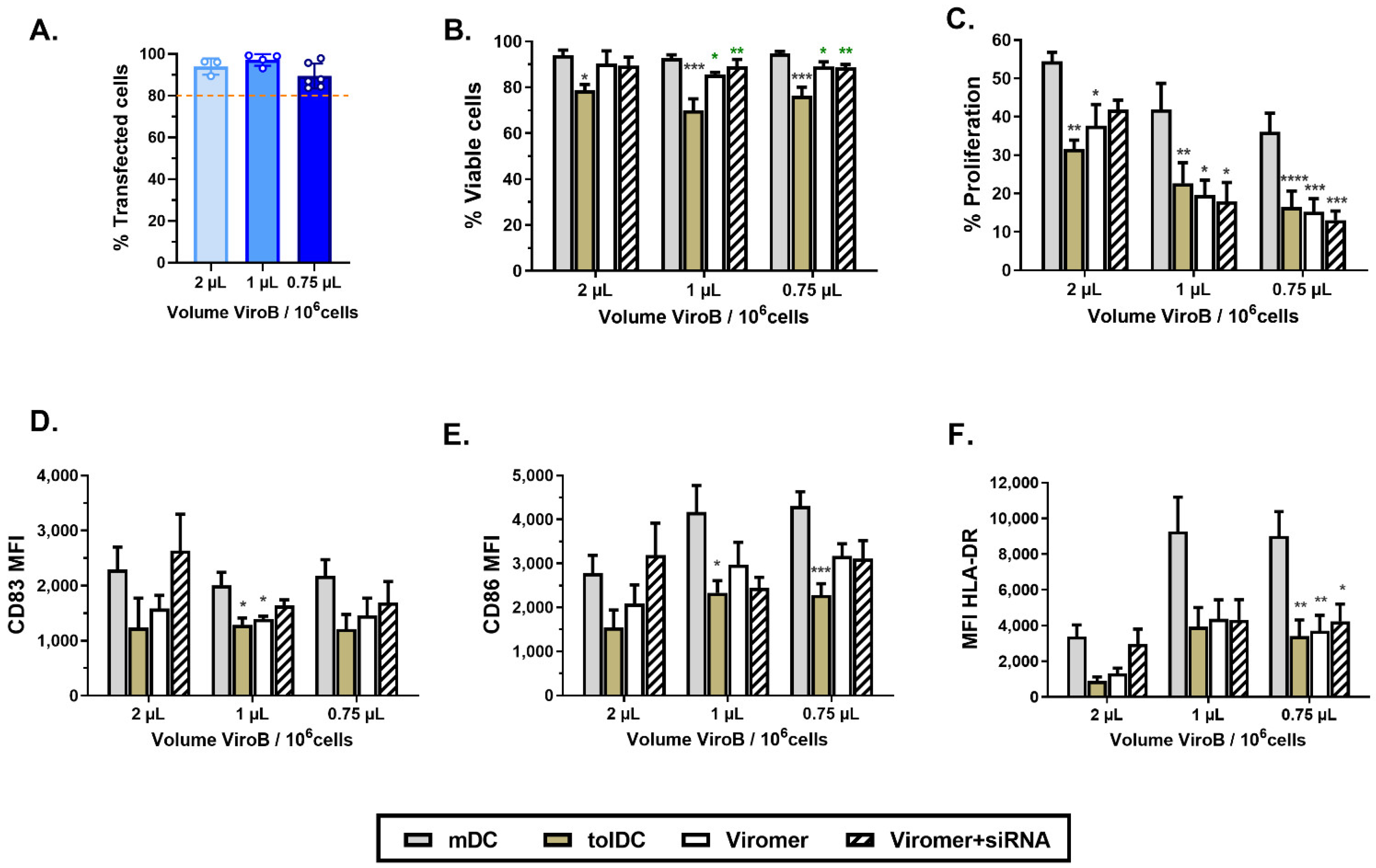
In recent years, Viromer has been commercialized and is an alternative transfection method taking advantage of the use of a mix of cationic and anionic polymers combined with the endosomal escape mechanism of influenza virus to deliver siRNA inside cells. One of the reasons why the influenza virus is highly infective is the presence of a surface glycoprotein called hemagglutinin, which is responsible for the fusion of the viral envelope with the endocytic vesicle once exposed to low pH, allowing the exit of the genetic content. Viromer mimics this mechanism by using both cationic and anionic polymers, resulting in a neutrally surface-charged vesicle. When these vesicles are exposed to the low pH of the endosome, they become more hydrophobic and are then able to escape from the endosome and to spread their content in the cytoplasm of the cell [
32]. Among the different Viromer reagents available, ViroB is specially designed to deliver siRNA. Although both HiPerFect and ViroB are easy-to-use protocols, ViroB requires less manipulation of cells. Thus, it was possible to optimize the ViroB transfection procedure to deliver siRNA in monocytes at the beginning of their differentiation (day 1 of culture) with high efficiency, using a low concentration of siRNA (275 nM siRNA) and without altering viability and tolerogenic characteristics of VitD3-tolDC. Consequently, we established a gene silencing protocol using ViroB + siRNA to evaluate the tolerogenic function of potential genes during the process of monocyte differentiation into VitD3-tolDC.
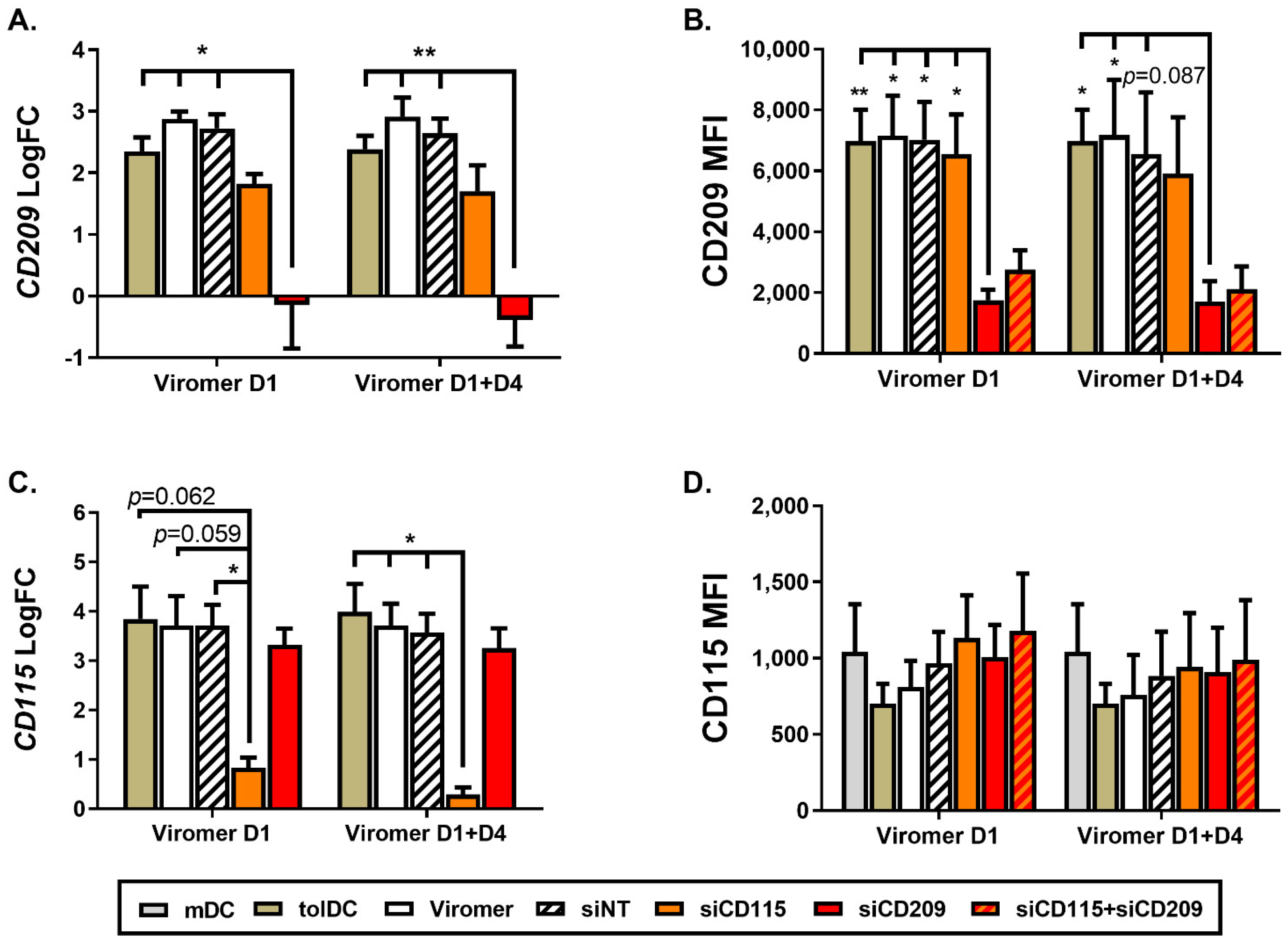
A study comparing the transcriptomic profile of mDC and VitD3-tolDC revealed an over-expression of
CD115 and
CD209 genes on VitD3-tolDC that was validated by qPCR, although at the protein level only CD209 was found upregulated. It is likely that CD115 protein expression was not found overexpressed in VitD3-tolDC because after CSF1 engagement to CD115 (CSF1R), the cytokine–receptor complex is internalized and degraded [
33], with surface determination not a reliable protein expression value for CD115.
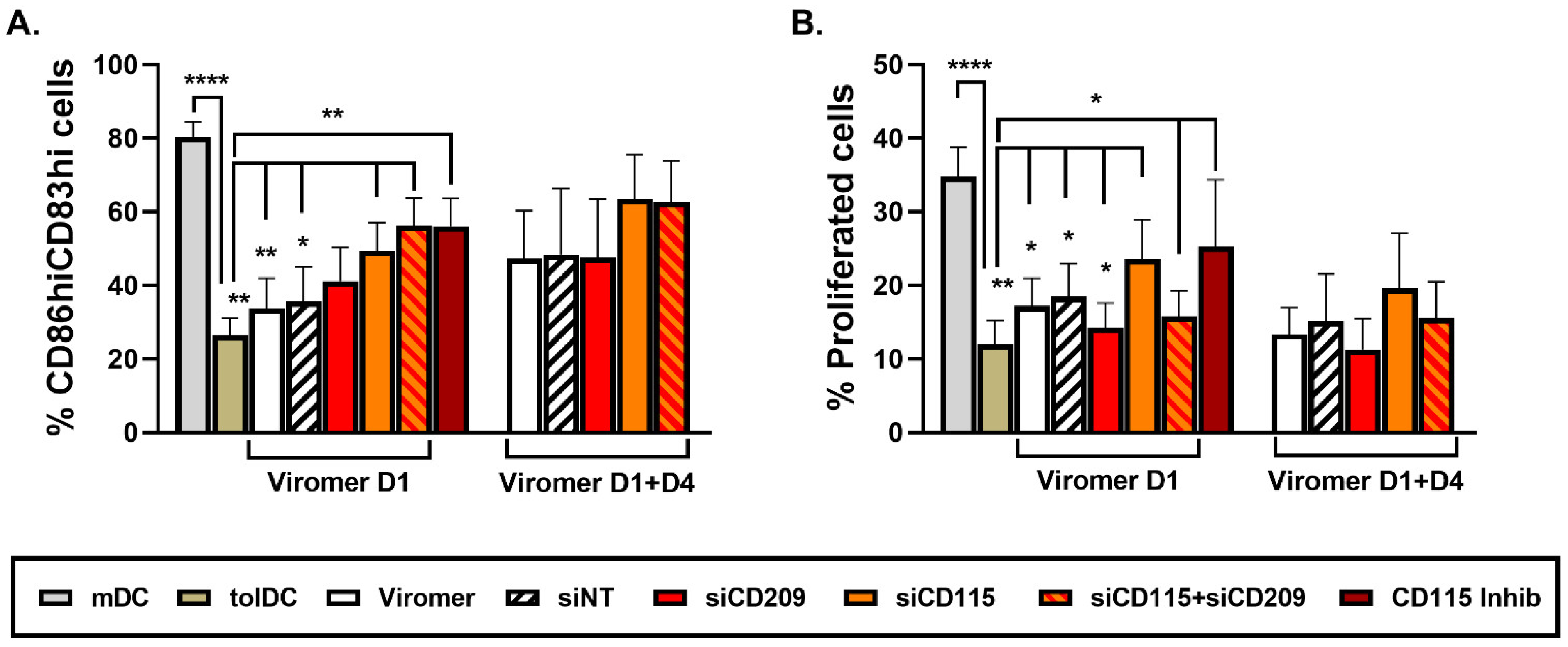
When gene and protein expression of CD209 was down-modulated by using Viro + siRNA on day 1 of culture differentiation, no effect on phenotype and functional characteristics of VitD3-tolDC were observed, indicating that CD209 is not directly involved in the tolerogenicity of VitD3-tolDC. Since the effect of siRNA is transient and functional assays require 5 additional days of co-culture after VitD3-tolDC obtention, to confirm results observed following CD209 gene silencing on day 1 of culture, monocytes were treated with ViroB + siRNA on day 1 + day 4 to achieve a longer knock-down effect. Interestingly, results were similar when using ViroB + siRNA only on day 1 or on day 1 + day 4, meaning that gene silencing is stable after at least 10 days of treatment, and CD209 or DC-SIGN is not involved in the induction of tolerogenic characteristics of VitD3-tolDC. In contrast, gene silencing of CD115 on day 1 caused a partial reversion of the tolerogenic properties of VitD3-tolDC. These results were similar when CD115 down-modulation was performed on day 1 + day 4. In addition, double transfection of VitD3-tolDC with siRNA of CD115 and CD209 did not increase the effect of CD115 down-modulation. At the phenotype level, the effect of double CD115 + CD209 siRNA transfection was comparable to the one observed using only CD115 siRNA, but double gene transfection did not alter functionality of VitD3-tolDC, probably because the effect of using half of the quantity of siRNA for CD115 is shorter and was not enough to find effects after 5 days of co-culture assay.
Since gene expression results of CD115 were not correlated with the protein surface expression of CD115 (CSF1R), VitD3-tolDC treated with a specific CD115 cell signaling inhibitor, PLX5622, were included in the study and compared with VitD3-tolDC treated with CD115 siRNA. Interestingly, results found using the CD115 inhibitor and CD115 siRNA in VitD3-tolDC were almost identical, validating the partial role of CD115 in the tolerogenicity of VitD3-tolDC.
In addition to
CD115 (CSF1R) overexpression in VitD3-tolDC, an increase of its ligand
CSF1 was also found upregulated in the transcriptional study of VitD3-tolDC compared to mDC. Recently, IL-34 has been identified as a novel CSF1R ligand [
34]. It has been reported that CSF1 and IL-34 promote monocyte survival and differentiation into M2-like macrophages with protumoral and immunoregulatory properties [
35]. These M2-like macrophages are characterized by producing high levels of IL-10 and low levels of IL-12 [
35]. In addition, the expression of CSF1 in the tumor microenvironment is associated with the recruitment, accumulation and polarization of tumor-associated macrophages [
18,
20,
22]. In this context, elevated levels of CSF1 and expression of CSF1R have been associated with poor prognosis in different cancers such as breast and ovarian cancer and, consequently, different strategies have emerged to neutralize or inhibit the CSF1/CSF1R signaling in several cancers [
20,
21,
22].
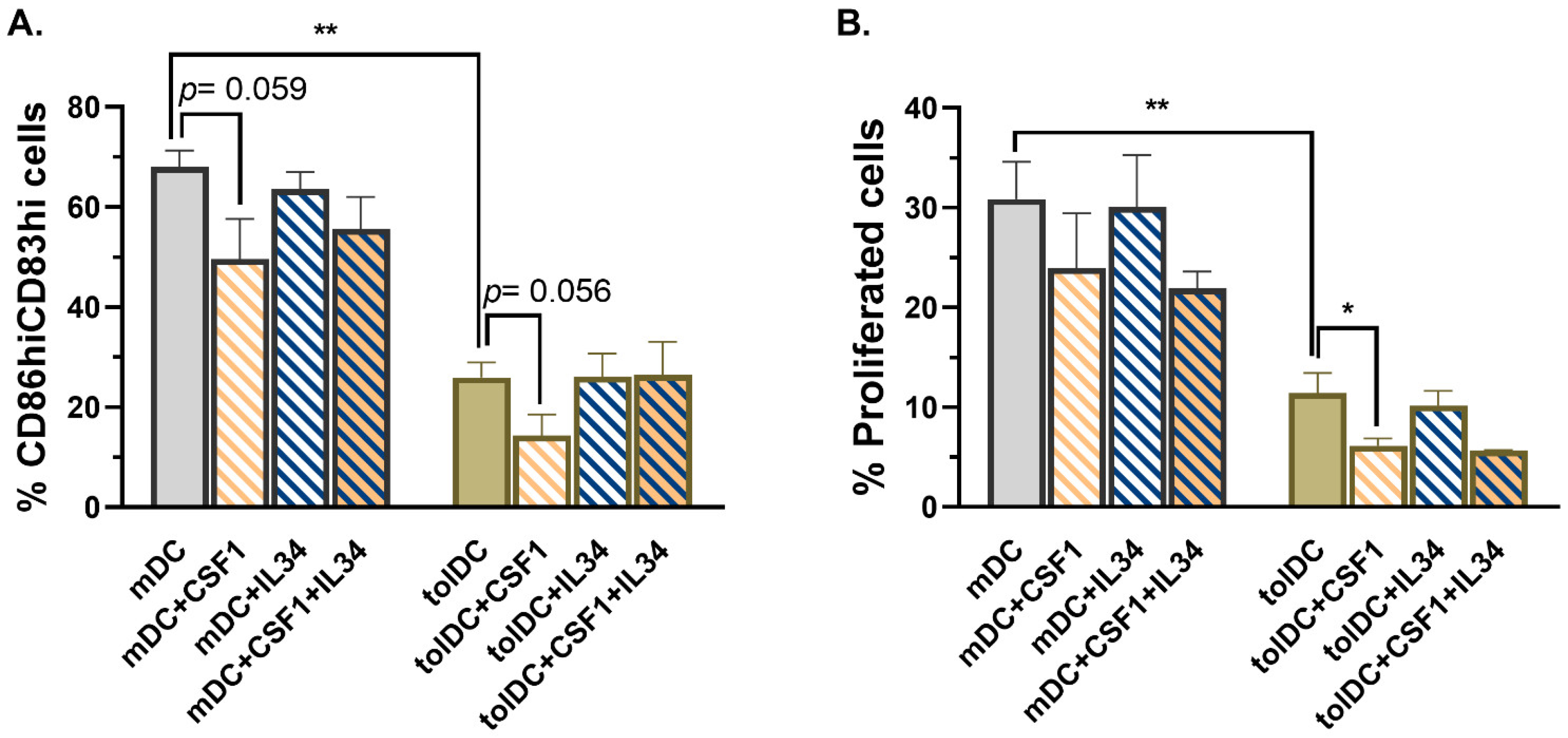
To further investigate the mechanism induced by CD115 (CSF1R) in VitD3-tolDC, the role of the two CSF1R ligands, CSF1 and IL-34, was examined. Remarkably, only the incubation of mDC and VitD3-tolDC with CSF1 during their process of differentiation reduced the percentage of CD83
hiCD86
hi cells and alloproliferation in both mDC and VitD3-tolDC, further supporting the idea that CSF1-CSF1R cell signaling is involved in the induction of tolerogenic properties of monocytes. Interestingly, Zhu and colleagues reported in 2002 that the incubation of monocytes with the active form of VitD3 (1,25(OH)
2D
3) induced a fast (after 1 h) and strong upregulation and secretion of CSF1, which can act in autocrine fashion, blocking the normal DC differentiation mediated by GM-CSF + IL-4 towards a macrophage-like phenotype [
36]. Although IL-34 is also involved in M2-like macrophage differentiation, the expression of IL-34 is reduced compared to CSF1, it has two additional receptors (PTPζ and syndecan-1, CD138) and it is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. In contrast, CSF1 can only bind to CSF1R and exerts an ubiquitous immunoregulatory function, thus explaining why CSF1 but not IL-34 could be involved in tolerogenicity of VitD3-tolDC.
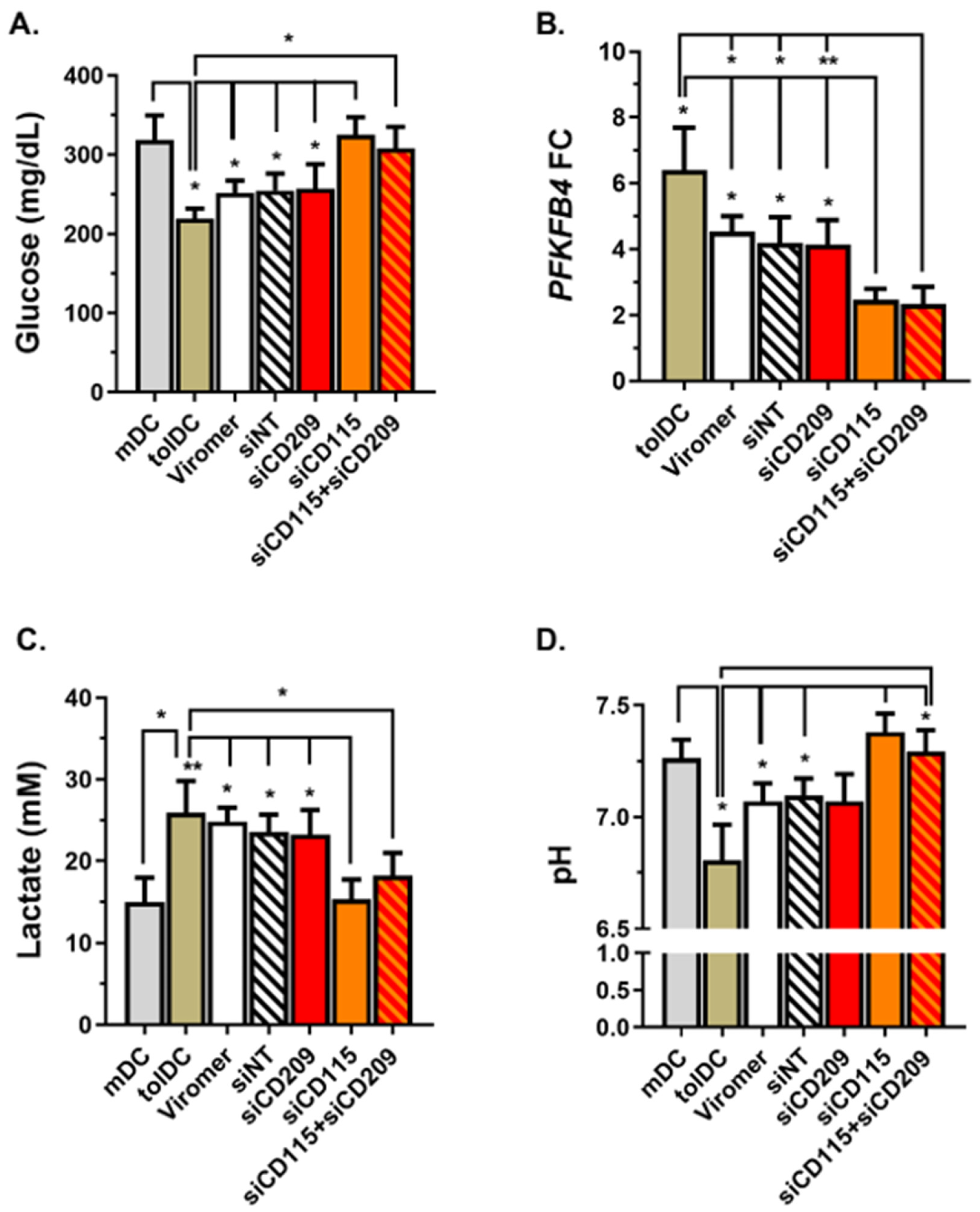
Previous studies investigating mechanisms involved in the tolerogenic functions gained by monocytes exposed to VitD3 revealed the relevance of metabolic cell reprogramming, with glucose oxidation and glycolysis essential for the VitD3-tolDC function [
37,
38]. Indeed, the glycolytic enzyme PFKFB4 has been recently identified as a critical checkpoint and direct transcriptional target of VitD3, which is early and strongly upregulated to promote the establishment of DC tolerogenicity [
38]. As a result of glucose consumption and glycolysis induction, high amounts of lactate are produced by VitD3-tolDC. Interestingly, lactate has been reported as a novel mechanism producing suppression of T cell proliferation by human autologous tolerogenic dendritic cells (ATDCs) [
28]. Along these lines, the metabolic analysis of cells containing
CD115 siRNA exhibited a reversion of the metabolic reprogramming of VitD3-tolDC, showing low glucose consumption, a partial blockage of the PFKFB4 gene expression induction, reduced lactate secretion and low acidification of the medium. Collectively, results highlight CD115 (CSF1R) signaling through CSF1 as one of the key regulators of VitD3-tolDC functions by inducing metabolic reprogramming of VitD3-tolDC towards glycolysis and lactic acid production. Of note, the role of CSF1-CSF1R in the tolerogenicity of VitD3-tolDC is partial and further analysis of potential molecules is required to identify tolerogenic pathways induced in VitD3-tolDC and discover functional biomarkers of these cells.
In this study, we established an optimized method based on ViroB to transfect monocytes with siRNA on day 1 of culture to evaluate their tolerogenic function on VitD3-tolDC. By studying other genes found differentially expressed in VitD3-tolDC compared to mDC, we expect to unravel the tolerogenic signaling of VitD3-tolDC to improve their efficacy for their translation into the clinic.
4. Material and Methods
4.1. Monocyte Isolation
To obtain monocytes, buffy coat samples from the Banc de Sang i Teixits (Barcelona, Spain) were obtained following the institutional standard operating procedures for blood donation.
First, samples were depleted of CD3+ cells using RosetteSep® Human Monocyte Enrichment Cocktail kit (StemCell Technologies, Vancouver, Burnaby, BC, Canada) followed by a ficoll-hypaque (Rafer, Zaragoza, Spain) density gradient separation. After that, isolation of CD14+ cells was performed by using EasySep® Human CD14 Positive Selection kit (StemCell) following the manufacturer’s instructions. Finally, viability and cell counting of viable cells were determined by staining (20 min at 4 °C in dark) with 7-amino-actinomycin D (7-AAD) (BD Biosciences, Franklin Lakes, NJ, USA) and phycoerythrin (PE)-conjugated annexin V (Immunotools, Friesoythe, Germany) and the use of PerfectCount microspheres (Cytognos, Salamanca, Spain) for cell quantification. Samples were acquired on a FACSCanto II flow cytometer and analyzed using the FACSDiva software (BD Biosciences).
4.2. Generation of VitD3-tolDC
Viable monocytes were cultured at a density of 1 × 106 cells/mL in 24-well plates in IMDM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 2% heat-inactivated human AB serum (Sigma-Aldrich, St. Louis, MO, USA), 250 U/mL IL-4 and 200 U/mL granulocyte macrophage colony-stimulating factor (GM-CSF) (both from Peprotech, London, UK) for 6 days at 37 °C and 5% CO2 atmosphere. On day 4, culture cell medium and cytokines were refreshed and a maturation cocktail containing 1000 U/mL IL-1β, 1000 U/mL TNF-α (both from Peprotech) and 1 μM prostaglandin-E2 (PGE2) (Pfizer, New York, NY, USA) was added to obtain mDC. For the generation of VitD3-tolDC, monocytes were additionally treated with 1 nM 1α,25-dihydroxyvitamin D3 (Calcitriol, Kern Pharma, Barcelona, Spain) on days 0 and 4. On day 6, cells were harvested by 25 min of accutase incubation at 37 °C to detach the cells from the plate. After washing, cell counts and viability were assessed by flow cytometry as previously mentioned, and phenotype and functional characterization were determined (see below). Dry pellets of each condition with the remaining cells were stored at −80 °C.
4.3. Transfection of Monocytes Using HiPerFect Reagent
For the transfection of monocytes on day 5 of culture (after the maturation stimulus), the reverse protocol described by Troegeles et al. [
29] was followed and optimized. Briefly, a supernatant of culture was kept and cells were harvested using accutase incubation. After determining the number of viable cells, 1 × 10
6 cells were resuspended in non-supplemented RPMI (Thermo Fisher Scientific) at a cell density of 1 × 10
6 cells in 250 µL of culture medium. For the formation of the lipid–siRNA complexes, 3.75 µL of HiPerFect transfectant reagent (Qiagen, Hilden, Germany) were added to 117.5 µL of warm and non-supplemented RPMI medium containing the DY-547-labeled non-targeting siRNA (siGLO, Thermo Scientific, Dharmacon, Illkirch, France). Different volumes of siGLO were tested to determine the optimal siRNA concentration (200 nM, 300 nM, 400 nM, 600 nM, 700 nM or 800 nM) required to achieve a high transfection efficacy of cells. Each lipid and siRNA mix was incubated for 15 min at room temperature in the dark and the tube was gently inverted several times during the incubation to increase the formation of lipid–siGLO complexes. Following the incubation, lipid–siGLO complexes were spinned down and 117.5 µL was transferred to a 24-well plate well. Then, 250 µL of cells (1 million) were pipetted directly into the siRNA complexes and mixed by a gentle swirl of the plate. After a 4 h incubation at 37 °C in the incubator, transfection was stopped by adding 500 µL of previously collected supernatants plus 125 µL IMDM with 10% human AB serum. Finally, cells were incubated for additional 36 h and the phenotype and functionality of the cells were assessed (see below).
4.4. Transfection of Monocytes Using Viromer Blue Reagent
To transfect monocytes on day 1, at the beginning of the process of differentiation to VitD3-tolDC, Viromer Blue transfection reagent (ViroB, Lipocalyx, Halle, Germany) was used following manufacturer’s instructions for forward transfection. Briefly, ON-TARGETplus SMARTpool siRNA targeting CD115 or CD209 (Dharmacon) was diluted in buffer blue to a final concentration of 2.75 μM. In a separate tube, 2 µL, 1 µL and 0.75 µL of ViroB were diluted with 180 µL, 90 µL and 67.5 µL buffer blue, respectively, and mixed by 3–5 s of vortex. For complexation, 180 µL, 90 µL and 67.5 µL of diluted ViroB were added into 20 µL, 10 µL and 7.5 µL of siRNA, respectively, mixed gently and incubated for 15 min at room temperature. Finally, 200 µL, 100 µL or 75 µL of siRNA-containing polymer vesicles were added to each well containing monocytes on day 1 of culture differentiation. For inducing the downregulation of CD115 and CD209 (double knockdown), the working siRNA concentration combined for the 2 genes remained as 2.75 µM (half from each specific gene). In some experiments, two steps of transfection were carried out on day 1 and again on day 4 to ensure the maximum effect of gene silencing during VitD3-tolDC generation and characterization. Cell transfection without siRNA (containing only blue buffer) or with ON-TARGETplus non-targeting control siRNA (Dharmacon) were used as controls to determine unspecific effects of ViroB and siRNA molecules, respectively. In addition, cell transfection with the red transfection indicator, siGLO, was used to analyze transfection efficacy.
4.5. Chemical Inhibition of CSF1R
As a complementary control, monocytes were treated on day 0 and day 4 with 4 µM of PLX5622 (kindly provided by MedChemExpress, Mo nMouth Junction, NJ, USA), which inhibits the receptor tyrosine kinase activity of CSF1R (CD115). On day 6, the effect of blocking CD115 cell signaling during monocyte differentiation into VitD3-tolDC was analyzed by phenotype and functional characterization.
4.6. CSF1 and IL34 Treatment
To further analyze the role of CD115 signaling during monocyte differentiation, 100 ng/mL of their ligands, CSF1 and IL34 (both from Peprotech), was added independently or in combination to the culture of mDC and tolDC on day 0 and day 4. Phenotype and functional characterization of CSF1- and/or IL34-treated cells were analyzed on day 6.
4.7. Phenotype Analysis
Once the cultured cells were harvested on day 6, the expression of the surface molecules CD11c, CD14, CD83, CD86, CD115, CD209 and HLA-DR was determined by flow cytometry. Cells were incubated for 20 min at room temperature in dark with the proper amount of the corresponding monoclonal antibodies (anti-): CD11c phycoerythrin (PE)-Cyanine dye 7 (PE-Cy7), CD14 Violet 450 (V450), CD83 allophycocyanin (APC), CD86 fluorescein isothiocyanate (FITC), CD115 PE, CD209 APC and HLA-DR Violet 500 (V500) (all of them from BD Biosciences, except CD209 APC from BioLegend, San Diego, CA, USA). Samples were acquired on a FACSCanto II flow cytometer and analyzed using the FACSDiva software (BD Biosciences).
4.8. Allogeneic Proliferation Assay
Fresh allogeneic PBMCs were obtained from whole blood samples following ficoll-hypaque density gradient separation. Subsequently, cells were resuspended in PBS at concentration of 8 × 106 PBMC/mL and were incubated with 1 mM BD Horizon Violet Proliferation Dye 450 (VPD450) (BD Biosciences) for 12 min at 37 °C in dark. In order to evaluate the ability of the generated DC to stimulate allogeneic PBMC, a proliferation assay was performed in 96-well round bottom plates with co-cultures of 105 allogeneic PBMCs and 5000 DCs (1:20 ratio) or 10,000 DCs (1:10 ratio) in a total volume of 200 μL of IMDM supplemented with 2% human AB serum. Four replicates of each condition were performed. Finally, cells were incubated for 5 days at 37 °C in a 5% CO2. Negative and positive proliferative controls were also included by culturing VPD450-PBMC only with supplemented IMDM medium or adding 5 µg/mL phytohemagglutinin (PHA) (Sigma-Aldrich), respectively. On day 5, samples were placed in a FACSCanto II or FACSLyric flow cytometer (BD Biosciences) and the percentage of proliferated cells was analyzed by VPD450 fluorescence dilution.
In some experiments, unlabeled allogeneic PBMC were co-cultured with DC for 4 days. Then, 1 µCi [3H]-thymidine (PerkinElmer, Waltham, MA, USA) was added to each well and cells were incubated for additional 18h. Cells were finally collected using an automated cell harvester (Micro Cell Harvester Skatron) (Tomtec Inc, Hamdem, CT, USA) and [3H]-thymidine incorporation was measured using a 1450 MicroBeta TriLux liquid scintillation counter (Wallac, Turku, Finland). Six replicas for each condition were performed. Results were expressed as mean of counts per minute (cpm) of replicas.
4.9. Gene Expression Analysis
Total RNA was isolated from frozen dry pellets samples using the RNeasy Mini Kit (Qiagen) complemented with DNase digestion with the RNase-free DNase set (Qiagen), following instructions provided by the manufacturer. RNA obtained from each condition was immediately quantified in a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific) and retrotranscribed into complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Finally, quantitative PCR (qPCR) was carried out to determine the relative expression of CD209, CSF1R and PFKFB4 (6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 4) genes after 50 cycles of cDNA amplification using the TaqMan Gene Expression Master Mix and specific TaqMan Gene Expression Assays (all of them from Applied Biosystems) in a LightCycler 480 System thermocycler (Roche, Basel, Switzerland). The quantitative expression of each gene was calculated based on the 2−∆Cp method and relative expression was assessed using the mean crossing point (Cp) values of the housekeeping genes Cyclophilin A (CYPA), TATA-Box Binding Protein (TBP) and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH).
4.10. Metabolic Analysis of Culture Supernatants
To analyze metabolism of cells, consumption of glucose, secretion of lactate and pH of supernatants on day 6 of culture of mDC, VitD3-tolDC and VitD3-tolDC treated with siRNA were examined. Glucose and lactate concentrations were determined in an AU5800 platform (Bekman Coulter; Clare, Ireland) using a standard hexokinase method and a lactate oxidase reaction, respectively. For the detection of pH, a direct potentiometry method was used in a Gem Premier 4000 analyzer (Werfen, MA, USA).
4.11. Statistical Analysis
Differences between the studied conditions were analyzed using Prism v6.0 software (GraphPad, La Jolla, CA, USA). For comparisons between two groups, a paired Student t-test or a Wilcoxon test were used if the samples were normally distributed or not, respectively. For multiple comparisons, a one-way ANOVA test was used. For the analysis of the relationship between % of HiPerFect transfected cells and RNA concentration, linear regression was performed. Results of all these analyses were expressed as mean ± standard deviation (SD), unless noted otherwise, and the results were considered statistically significant when p-value < 0.05.