RAB10 Interacts with ABCB4 and Regulates Its Intracellular Traffic
Abstract
:1. Introduction
2. Results
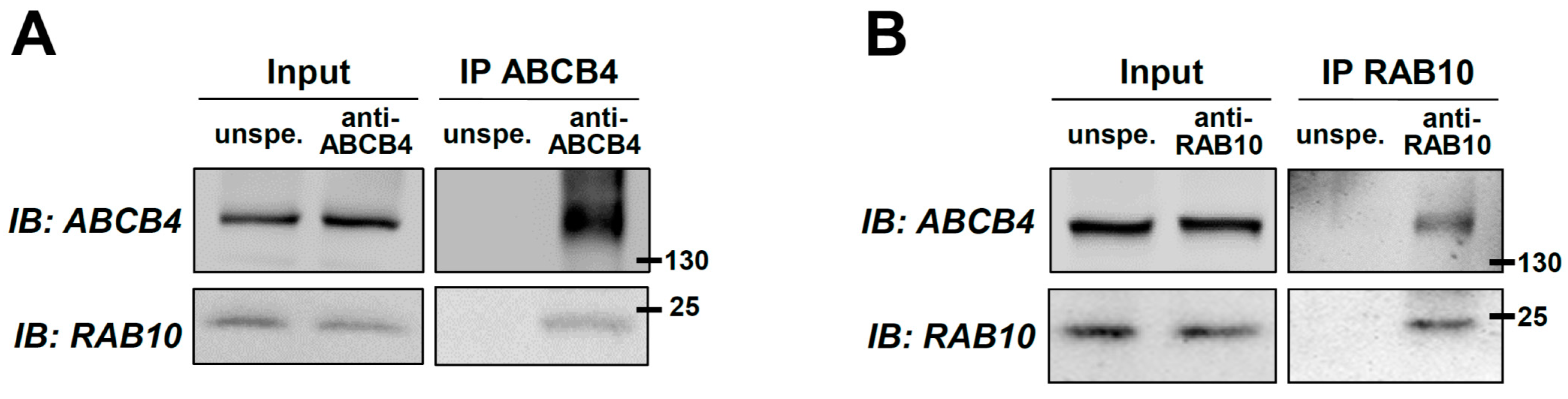
2.1. Identification of RAB10 as a New Molecular Partner of ABCB4
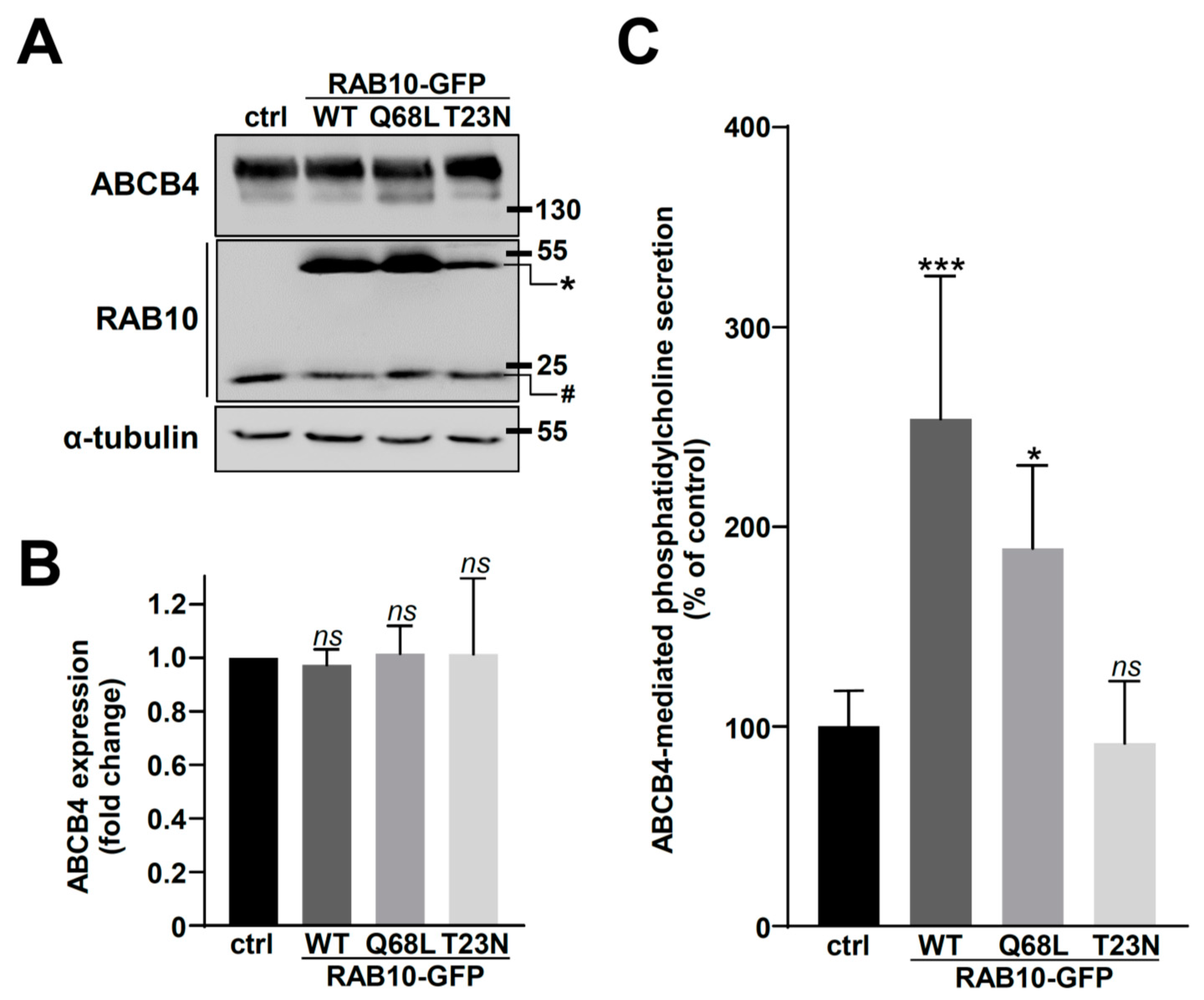
2.2. Overexpression of RAB10-WT or Its Dominant-Active Mutant Increases ABCB4 Plasma Membrane Expression and Function
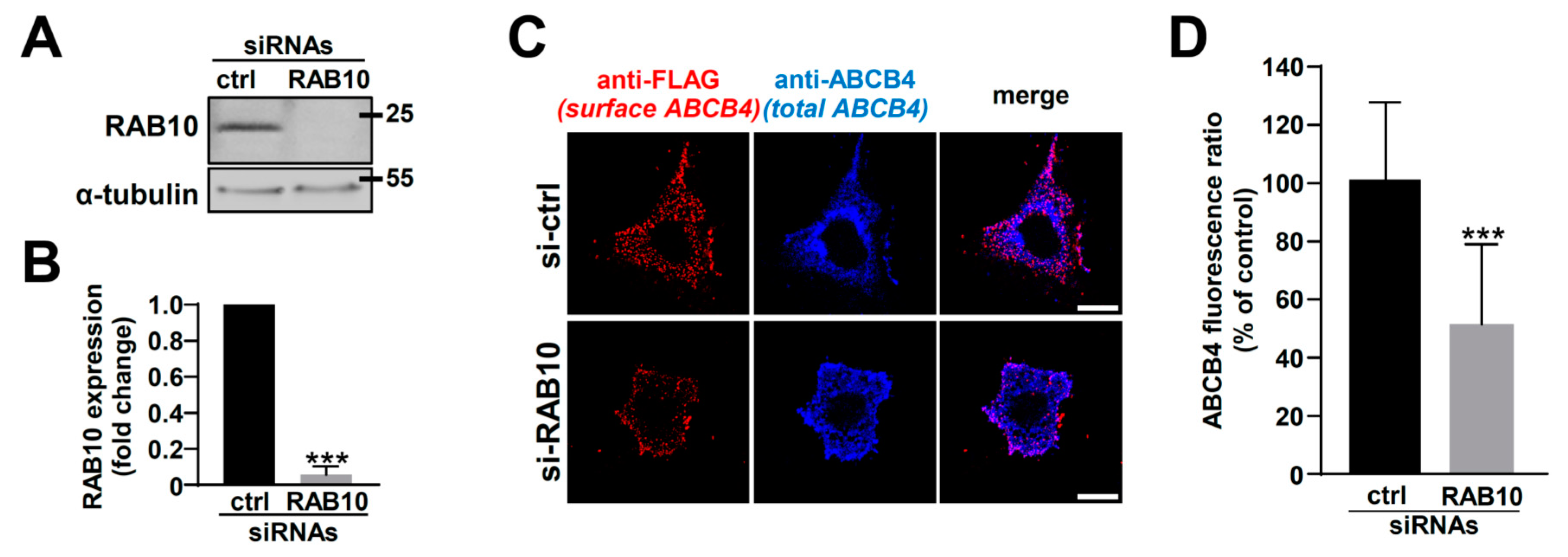
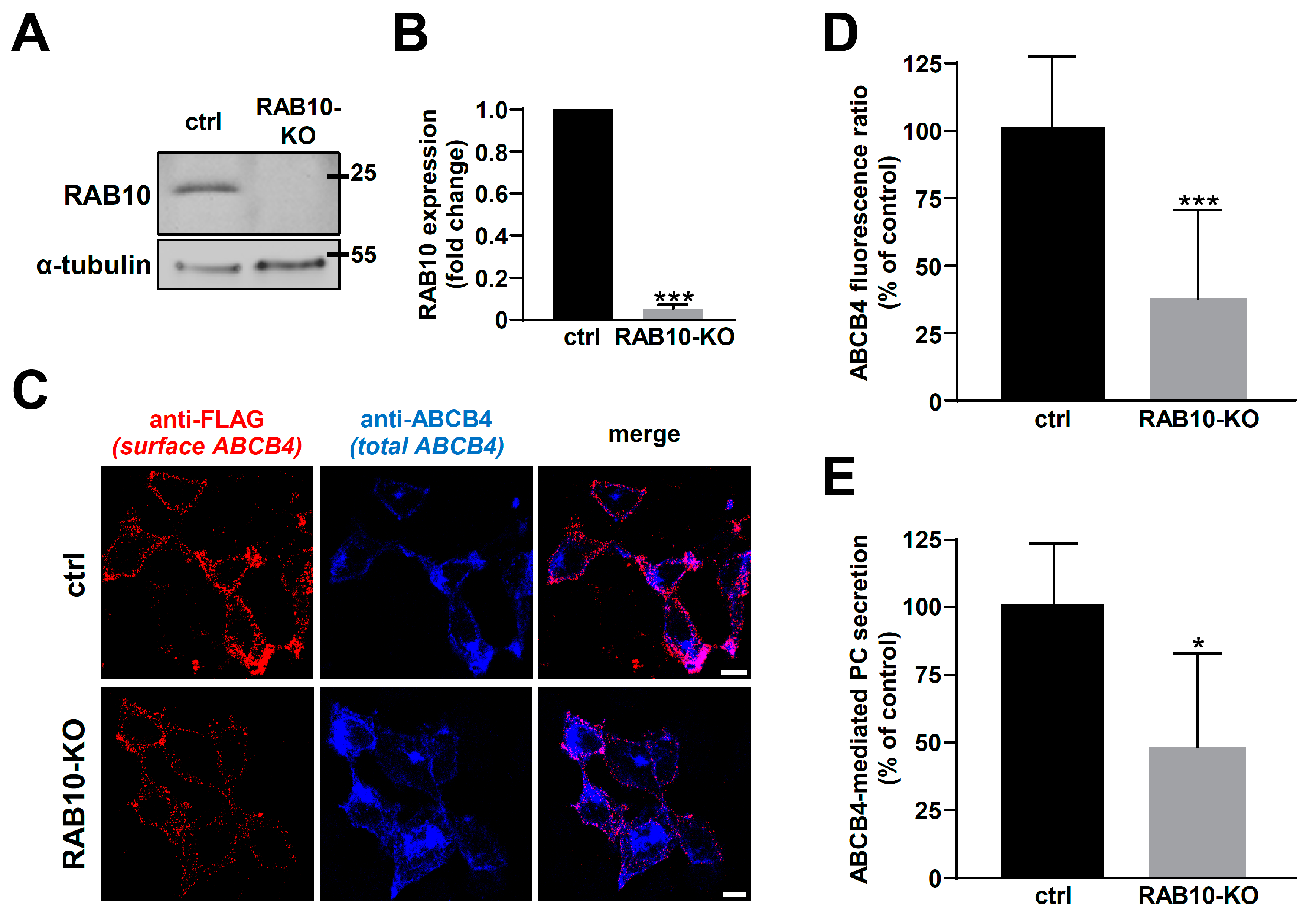
2.3. RAB10 Silencing Reduces ABCB4 Plasma Membrane Expression and Decreases Its Function
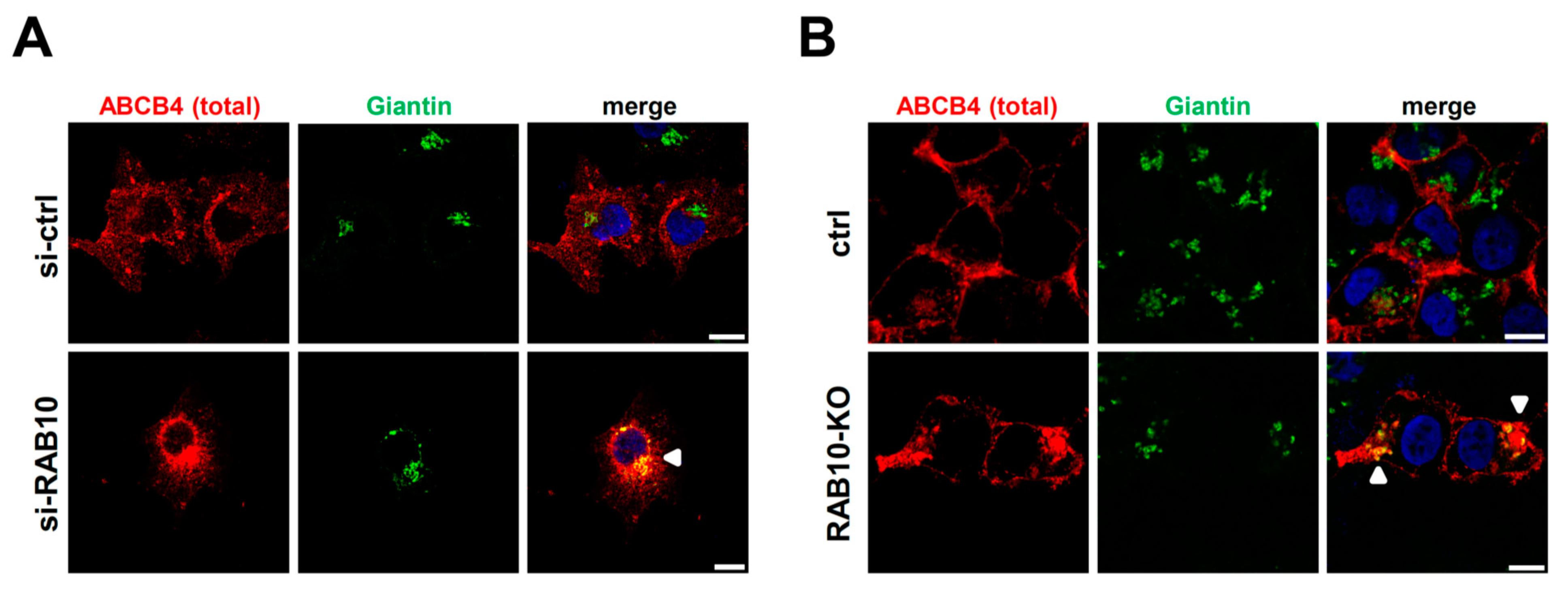
2.4. RAB10 Promotes ABCB4 Trafficking from the Golgi Apparatus to the Plasma Membrane
3. Discussion
4. Materials and Methods
4.1. Plasmids, Cell Culture and Transfection
4.2. RNA Interference and CRISPR/Cas9 System
4.3. Immunoprecipitation and Mass Spectrometry Analyses (AP-MS)
4.4. Immunoblots, Immunofluorescence and Measurement of ABCB4-Mediated Phosphatidylcholine Secretion
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kroll, T.; Prescher, M.; Smits, S.H.J.; Schmitt, L. Structure and function of hepatobiliary atp binding cassette transporters. Chem. Rev. 2020, 121, 5240–5288. [Google Scholar] [CrossRef]
- Falguières, T.; Aït-Slimane, T.; Housset, C.; Maurice, M. Abcb4: Insights from pathobiology into therapy. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 557–563. [Google Scholar] [CrossRef]
- Boyer, J.L. Bile formation and secretion. Compr. Physiol. 2013, 3, 1035–1078. [Google Scholar] [PubMed] [Green Version]
- Davit-Spraul, A.; Gonzales, E.; Baussan, C.; Jacquemin, E. The spectrum of liver diseases related to abcb4 gene mutations: Pathophysiology and clinical aspects. Semin. Liver Dis. 2010, 30, 134–146. [Google Scholar] [CrossRef]
- Stättermayer, A.F.; Halilbasic, E.; Wrba, F.; Ferenci, P.; Trauner, M. Variants in abcb4 (mdr3) across the spectrum of cholestatic liver diseases in adults. J. Hepatol. 2020, 73, 651–663. [Google Scholar] [CrossRef]
- Jacquemin, E.; Hermans, D.; Myara, A.; Habes, D.; Debray, D.; Hadchouel, M.; Sokal, E.M.; Bernard, O. Ursodeoxycholic acid therapy in pediatric patients with progressive familial intrahepatic cholestasis. Hepatology 1997, 25, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Poupon, R. Ursodeoxycholic acid and bile-acid mimetics as therapeutic agents for cholestatic liver diseases: An overview of their mechanisms of action. Clin. Res. Hepatol. Gastroenterol. 2012, 36 (Suppl. 1), S3–S12. [Google Scholar] [CrossRef]
- Delaunay, J.L.; Bruneau, A.; Hoffmann, B.; Durand-Schneider, A.M.; Barbu, V.; Jacquemin, E.; Maurice, M.; Housset, C.; Callebaut, I.; Ait-Slimane, T. Functional defect of variants in the adenosine triphosphate-binding sites of abcb4 and their rescue by the cystic fibrosis transmembrane conductance regulator potentiator, ivacaftor (vx-770). Hepatology 2017, 65, 560–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vauthier, V.; Ben Saad, A.; Elie, J.; Oumata, N.; Durand-Schneider, A.M.; Bruneau, A.; Delaunay, J.L.; Housset, C.; Aït-Slimane, T.; Meijer, L.; et al. Structural analogues of roscovitine rescue the intracellular traffic and the function of er-retained abcb4 variants in cell models. Sci. Rep. 2019, 9, 6653. [Google Scholar] [CrossRef]
- Aronson, S.J.; Bakker, R.S.; Shi, X.; Duijst, S.; Ten Bloemendaal, L.; de Waart, D.R.; Verheij, J.; Elferink, R.P.O.; Beuers, U.; Paulusma, C.C.; et al. Liver-directed gene therapy results in long term correction of progressive familial intrahepatic cholestasis type 3 in mice. J. Hepatol. 2019, 71, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Siew, S.M.; Cunningham, S.C.; Zhu, E.; Tay, S.S.; Venuti, E.; Bolitho, C.; Alexander, I.E. Prevention of cholestatic liver disease and reduced tumorigenicity in a murine model of pfic type 3 using hybrid aav-piggybac gene therapy. Hepatology 2019, 70, 2047–2061. [Google Scholar] [CrossRef]
- Weber, N.D.; Odriozola, L.; Martínez-García, J.; Ferrer, V.; Douar, A.; Bénichou, B.; González-Aseguinolaza, G.; Smerdou, C. Gene therapy for progressive familial intrahepatic cholestasis type 3 in a clinically relevant mouse model. Nat. Commun. 2019, 10, 5694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Cao, J.; Huang, P.; An, P.; Badlani, D.; Vaid, K.A.; Zhao, S.; Wang, D.Q.; Zhuo, J.; Yin, L.; et al. Synthetic human abcb4 mrna therapy rescues severe liver disease phenotype in a balb/c.Abcb4(-/-) mouse model of pfic3. J. Hepatol. 2020, 74, 1416–1428. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, D.F.; Moseley, J.; Calderon, G.; Swift, A.L.; Li, S.; Arias, I.M. Identification of hax-1 as a protein that binds bile salt export protein and regulates its abundance in the apical membrane of madin-darby canine kidney cells. J. Biol. Chem. 2004, 279, 32761–32770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, W.; Calderon, G.; Swift, A.L.; Moseley, J.; Li, S.; Hosoya, H.; Arias, I.M.; Ortiz, D.F. Myosin ii regulatory light chain is required for trafficking of bile salt export protein to the apical membrane in madin-darby canine kidney cells. J. Biol. Chem. 2005, 280, 23741–23747. [Google Scholar] [CrossRef] [Green Version]
- Ikebuchi, Y.; Takada, T.; Ito, K.; Yoshikado, T.; Anzai, N.; Kanai, Y.; Suzuki, H. Receptor for activated c-kinase 1 regulates the cellular localization and function of abcb4. Hepatol. Res. 2009, 39, 1091–1107. [Google Scholar] [CrossRef]
- Venot, Q.; Delaunay, J.L.; Fouassier, L.; Delautier, D.; Falguieres, T.; Housset, C.; Maurice, M.; Ait-Slimane, T. A pdz-like motif in the biliary transporter abcb4 interacts with the scaffold protein ebp50 and regulates abcb4 cell surface expression. PLoS ONE 2016, 11, e0146962. [Google Scholar] [CrossRef] [Green Version]
- Ben Saad, A.; Bruneau, A.; Mareux, E.; Lapalus, M.; Delaunay, J.L.; Gonzales, E.; Jacquemin, E.; Aït-Slimane, T.; Falguières, T. Molecular regulation of canalicular abc transporters. Int. J. Mol. Sci. 2021, 22, 2113. [Google Scholar] [CrossRef]
- Stenmark, H. Rab gtpases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab family of small gtpases: An updated view on their regulation and functions. FEBS J. 2021, 288, 36–55. [Google Scholar] [CrossRef]
- Chavrier, P.; Vingron, M.; Sander, C.; Simons, K.; Zerial, M. Molecular cloning of ypt1/sec4-related cdnas from an epithelial cell line. Mol. Cell Biol. 1990, 10, 6578–6585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, C.E.L.; Tang, B.L. Rab 10-a traffic controller in multiple cellular pathways and locations. J. Cell Physiol. 2018, 233, 6483–6494. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lou, J.; Ouyang, C.; Chen, W.; Liu, Y.; Liu, X.; Cao, X.; Wang, J.; Lu, L. Ras-related protein rab10 facilitates tlr4 signaling by promoting replenishment of tlr4 onto the plasma membrane. Proc. Natl. Acad. Sci. USA 2010, 107, 13806–13811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Lippincott-Schwartz, J. Rab10 delivers glut4 storage vesicles to the plasma membrane. Commun. Integr. Biol. 2013, 6, e23779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, P.; Pearson, C.L.; Soroka, C.J.; Xu, S.; Mennone, A.; Boyer, J.L. Levels of plasma membrane expression in progressive and benign mutations of the bile salt export pump (bsep/abcb11) correlate with severity of cholestatic diseases. Am. J. Physiol. Cell Physiol. 2007, 293, C1709–C1716. [Google Scholar] [CrossRef] [Green Version]
- Szeri, F.; Niaziorimi, F.; Donnelly, S.; Orndorff, J.; van de Wetering, K. Generation of fully functional fluorescent fusion proteins to gain insights into abcc6 biology. FEBS Lett. 2020, 595, 799–810. [Google Scholar] [CrossRef]
- Wakabayashi, Y.; Lippincott-Schwartz, J.; Arias, I.M. Intracellular trafficking of bile salt export pump (abcb11) in polarized hepatic cells: Constitutive cycling between the canalicular membrane and rab11-positive endosomes. Mol. Biol. Cell 2004, 15, 3485–3496. [Google Scholar] [CrossRef] [Green Version]
- Delaunay, J.L.; Durand-Schneider, A.M.; Delautier, D.; Rada, A.; Gautherot, J.; Jacquemin, E.; Ait-Slimane, T.; Maurice, M. A missense mutation in abcb4 gene involved in progressive familial intrahepatic cholestasis type 3 leads to a folding defect that can be rescued by low temperature. Hepatology 2009, 49, 1218–1227. [Google Scholar] [CrossRef]
- Gautherot, J.; Durand-Schneider, A.M.; Delautier, D.; Delaunay, J.L.; Rada, A.; Gabillet, J.; Housset, C.; Maurice, M.; Aït-Slimane, T. Effects of cellular, chemical and pharmacological chaperones on the rescue of a trafficking-defective mutant of the atp-binding cassette transporters abcb1/abcb4. J. Biol. Chem. 2012, 287, 5070–5078. [Google Scholar] [CrossRef] [Green Version]
- Przybylla, S.; Stindt, J.; Kleinschrodt, D.; Schulte Am Esch, J.; Haussinger, D.; Keitel, V.; Smits, S.H.; Schmitt, L. Analysis of the bile salt export pump (abcb11) interactome employing complementary approaches. PLoS ONE 2016, 11, e0159778. [Google Scholar] [CrossRef]
- Park, S.W.; Schonhoff, C.M.; Webster, C.R.; Anwer, M.S. Rab11, but not rab4, facilitates cyclic amp- and tauroursodeoxycholate-induced mrp2 translocation to the plasma membrane. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G863–G870. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; van Dam, E.M.; Brymora, A.; Duggin, I.G.; Robinson, P.J.; Roufogalis, B.D. The small gtpases rab5 and rala regulate intracellular traffic of p-glycoprotein. Biochim. Biophys. Acta 2007, 1773, 1062–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrándiz-Huertas, C.; Fernández-Carvajal, A.; Ferrer-Montiel, A. Rab4 interacts with the human p-glycoprotein and modulates its surface expression in multidrug resistant k562 cells. Int. J. Cancer 2011, 128, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Farinha, C.M.; Matos, P. Rab gtpases regulate the trafficking of channels and transporters—A focus on cystic fibrosis. Small GTPases 2018, 9, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Lucken-Ardjomande Häsler, S.; Vallis, Y.; Pasche, M.; McMahon, H.T. Graf2, wdr44, and mical1 mediate rab8/10/11-dependent export of e-cadherin, mmp14, and cftr δf508. J. Cell Biol. 2020, 219, e201811014. [Google Scholar] [CrossRef]
- Schuck, S.; Gerl, M.J.; Ang, A.; Manninen, A.; Keller, P.; Mellman, I.; Simons, K. Rab10 is involved in basolateral transport in polarized madin-darby canine kidney cells. Traffic 2007, 8, 47–60. [Google Scholar] [CrossRef]
- Morita, S.Y.; Tsuda, T.; Horikami, M.; Teraoka, R.; Kitagawa, S.; Terada, T. Bile salt-stimulated phospholipid efflux mediated by abcb4 localized in nonraft membranes. J. Lipid Res. 2013, 54, 1221–1230. [Google Scholar] [CrossRef] [Green Version]
- Gissen, P.; Johnson, C.A.; Morgan, N.V.; Stapelbroek, J.M.; Forshew, T.; Cooper, W.N.; McKiernan, P.J.; Klomp, L.W.; Morris, A.A.; Wraith, J.E.; et al. Mutations in vps33b, encoding a regulator of snare-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (arc) syndrome. Nat. Genet. 2004, 36, 400–404. [Google Scholar] [CrossRef] [Green Version]
- Cullinane, A.R.; Straatman-Iwanowska, A.; Zaucker, A.; Wakabayashi, Y.; Bruce, C.K.; Luo, G.; Rahman, F.; Gürakan, F.; Utine, E.; Ozkan, T.B.; et al. Mutations in vipar cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat. Genet. 2010, 42, 303–312. [Google Scholar] [CrossRef]
- Gonzales, E.; Taylor, S.A.; Davit-Spraul, A.; Thébaut, A.; Thomassin, N.; Guettier, C.; Whitington, P.F.; Jacquemin, E. Myo5b mutations cause cholestasis with normal serum gamma-glutamyl transferase activity in children without microvillous inclusion disease. Hepatology 2017, 65, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, Y.; van Ijzendoorn, S.C. A link between intrahepatic cholestasis and genetic variations in intracellular trafficking regulators. Biology 2021, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Dai, F.; Yu, L.; She, X.; Zhao, Y.; Jiang, J.; Chen, X.; Zhao, S. Identification and characterization of nine novel human small gtpases showing variable expressions in liver cancer tissues. Gene Expr. 2002, 10, 231–242. [Google Scholar] [CrossRef]
- Li, Z.; Schulze, R.J.; Weller, S.G.; Krueger, E.W.; Schott, M.B.; Zhang, X.; Casey, C.A.; Liu, J.; Stöckli, J.; James, D.E.; et al. A novel rab10-ehbp1-ehd2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2016, 2, e1601470. [Google Scholar] [CrossRef] [Green Version]
- Aoudjehane, L.; Gautheron, J.; Le Goff, W.; Goumard, C.; Gilaizeau, J.; Nget, C.S.; Savier, E.; Atif, M.; Lesnik, P.; Morichon, R.; et al. Novel defatting strategies reduce lipid accumulation in primary human culture models of liver steatosis. Dis. Models Mech. 2020, 13, dmm042663. [Google Scholar] [CrossRef] [Green Version]
- Ben Saad, A.; Vauthier, V.; Tóth, Á.; Janaszkiewicz, A.; Durand-Schneider, A.M.; Bruneau, A.; Delaunay, J.L.; Lapalus, M.; Mareux, E.; Garcin, I.; et al. Effect of cftr correctors on the traffic and the function of intracellularly retained abcb4 variants. Liver Int. 2021, 41, 1344–1357. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the crispr-cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Poullet, P.; Carpentier, S.; Barillot, E. Myproms, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 2007, 7, 2553–2556. [Google Scholar] [CrossRef] [PubMed]
- Gautherot, J.; Delautier, D.; Maubert, M.A.; Aït-Slimane, T.; Bolbach, G.; Delaunay, J.L.; Durand-Schneider, A.M.; Firrincieli, D.; Barbu, V.; Chignard, N.; et al. Phosphorylation of abcb4 impacts its function: Insights from disease-causing mutations. Hepatology 2014, 60, 610–621. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Saad, A.; Vauthier, V.; Lapalus, M.; Mareux, E.; Bennana, E.; Durand-Schneider, A.-M.; Bruneau, A.; Delaunay, J.-L.; Gonzales, E.; Housset, C.; et al. RAB10 Interacts with ABCB4 and Regulates Its Intracellular Traffic. Int. J. Mol. Sci. 2021, 22, 7087. https://doi.org/10.3390/ijms22137087
Ben Saad A, Vauthier V, Lapalus M, Mareux E, Bennana E, Durand-Schneider A-M, Bruneau A, Delaunay J-L, Gonzales E, Housset C, et al. RAB10 Interacts with ABCB4 and Regulates Its Intracellular Traffic. International Journal of Molecular Sciences. 2021; 22(13):7087. https://doi.org/10.3390/ijms22137087
Chicago/Turabian StyleBen Saad, Amel, Virginie Vauthier, Martine Lapalus, Elodie Mareux, Evangéline Bennana, Anne-Marie Durand-Schneider, Alix Bruneau, Jean-Louis Delaunay, Emmanuel Gonzales, Chantal Housset, and et al. 2021. "RAB10 Interacts with ABCB4 and Regulates Its Intracellular Traffic" International Journal of Molecular Sciences 22, no. 13: 7087. https://doi.org/10.3390/ijms22137087
APA StyleBen Saad, A., Vauthier, V., Lapalus, M., Mareux, E., Bennana, E., Durand-Schneider, A.-M., Bruneau, A., Delaunay, J.-L., Gonzales, E., Housset, C., Aït-Slimane, T., Guillonneau, F., Jacquemin, E., & Falguières, T. (2021). RAB10 Interacts with ABCB4 and Regulates Its Intracellular Traffic. International Journal of Molecular Sciences, 22(13), 7087. https://doi.org/10.3390/ijms22137087









