Assessment of Biotechnologically Important Filamentous Fungal Biomass by Fourier Transform Raman Spectroscopy
Abstract
:1. Introduction
2. Results and Discussion
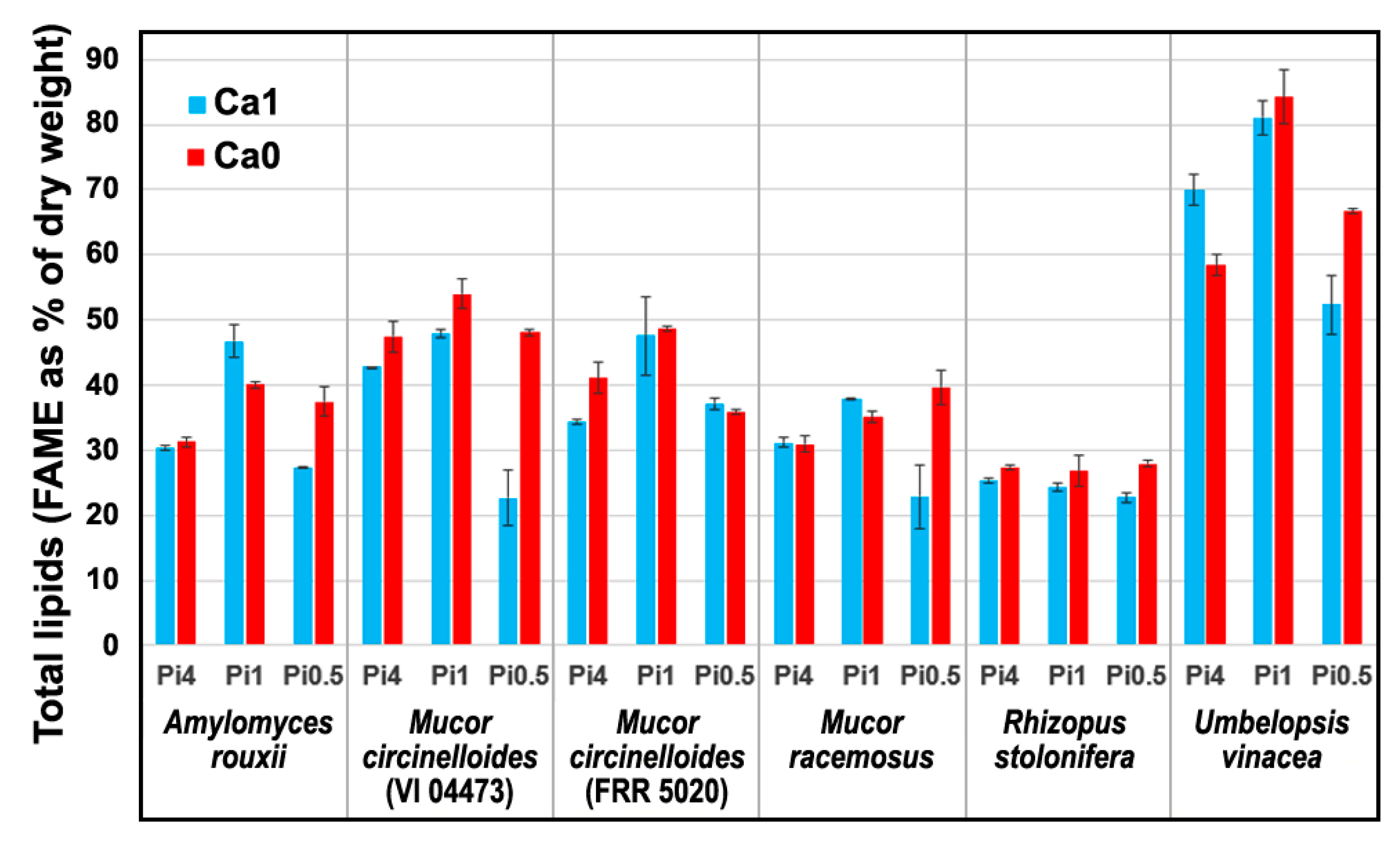
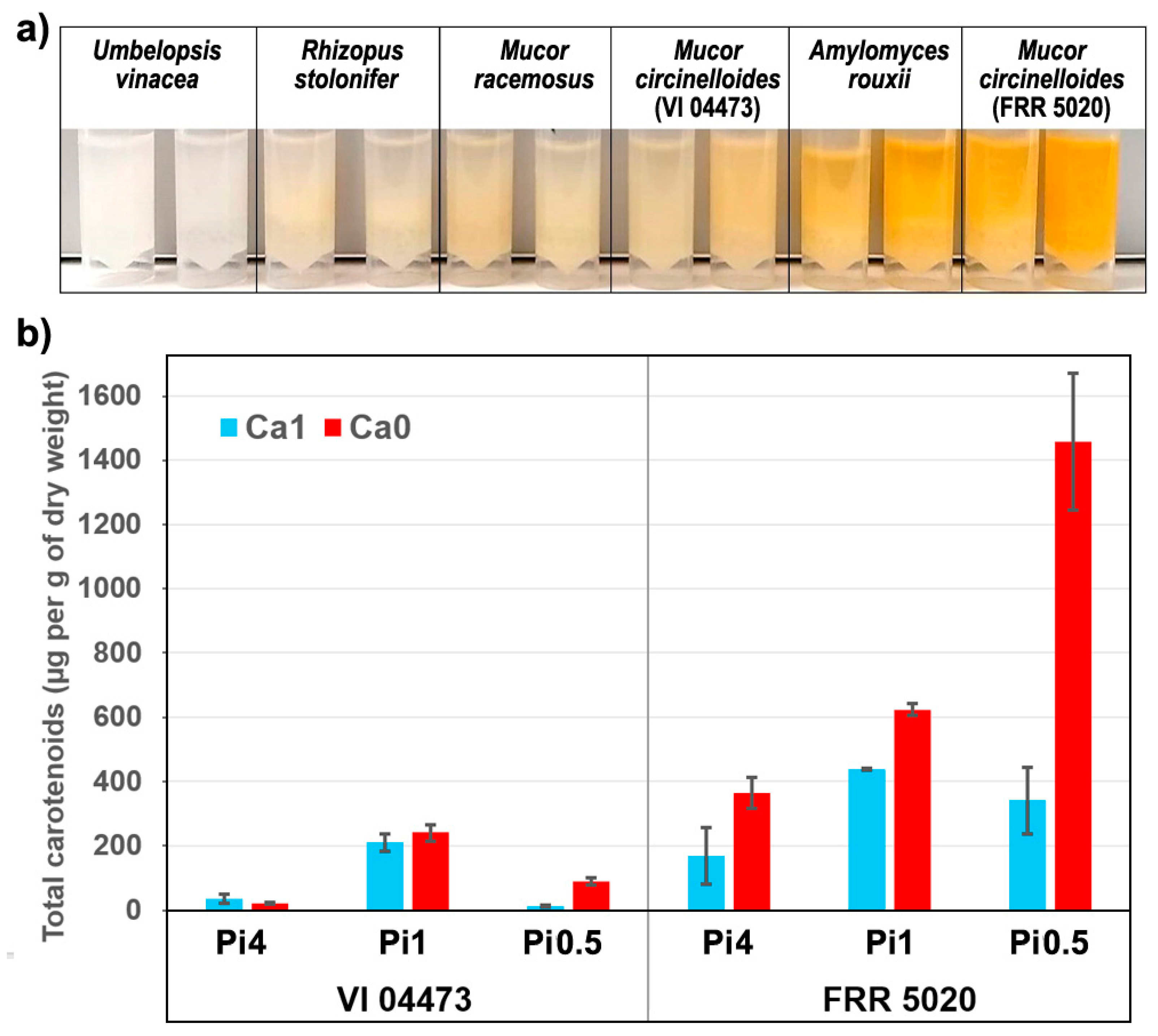
2.1. Chemical Composition of Fungal Biomass
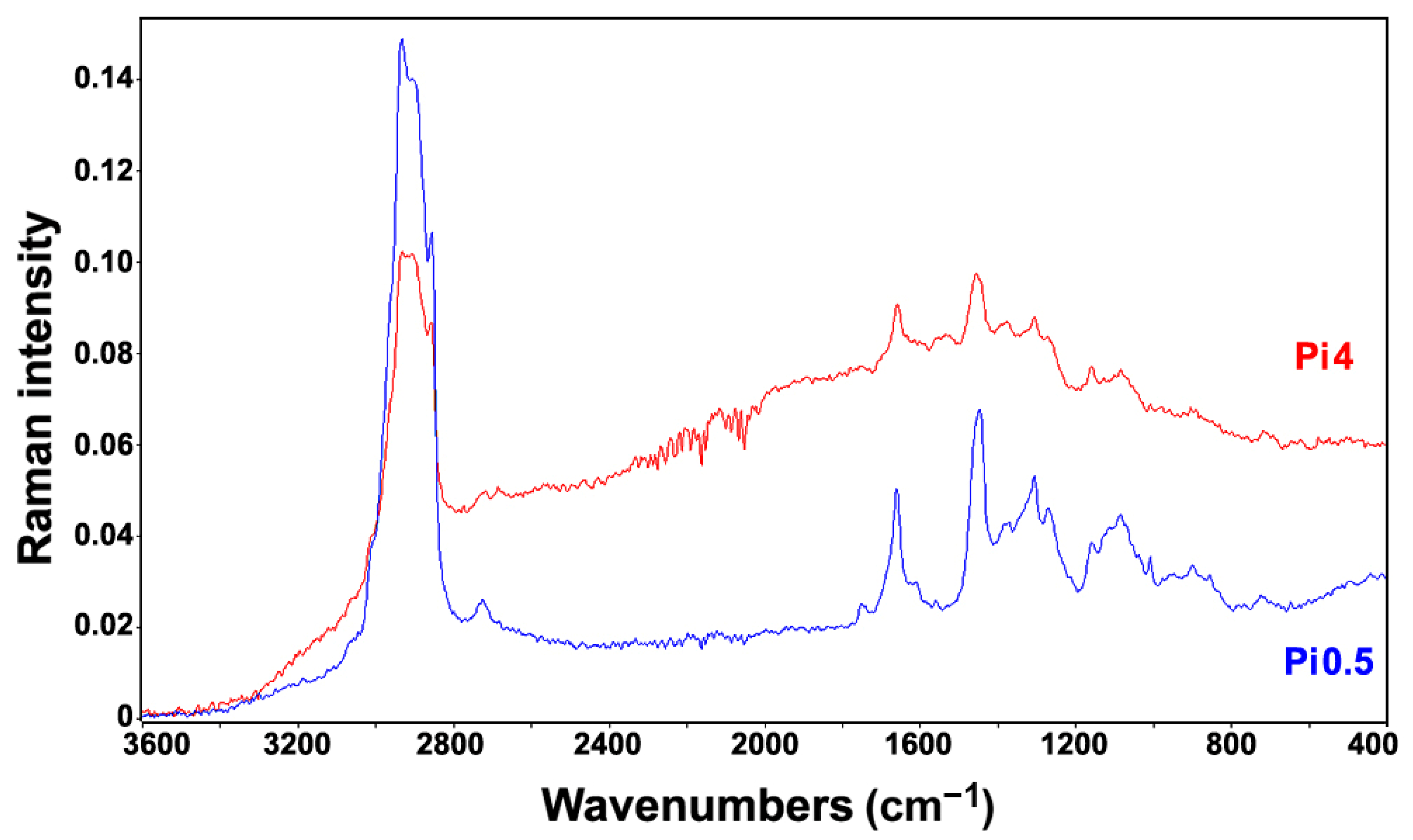
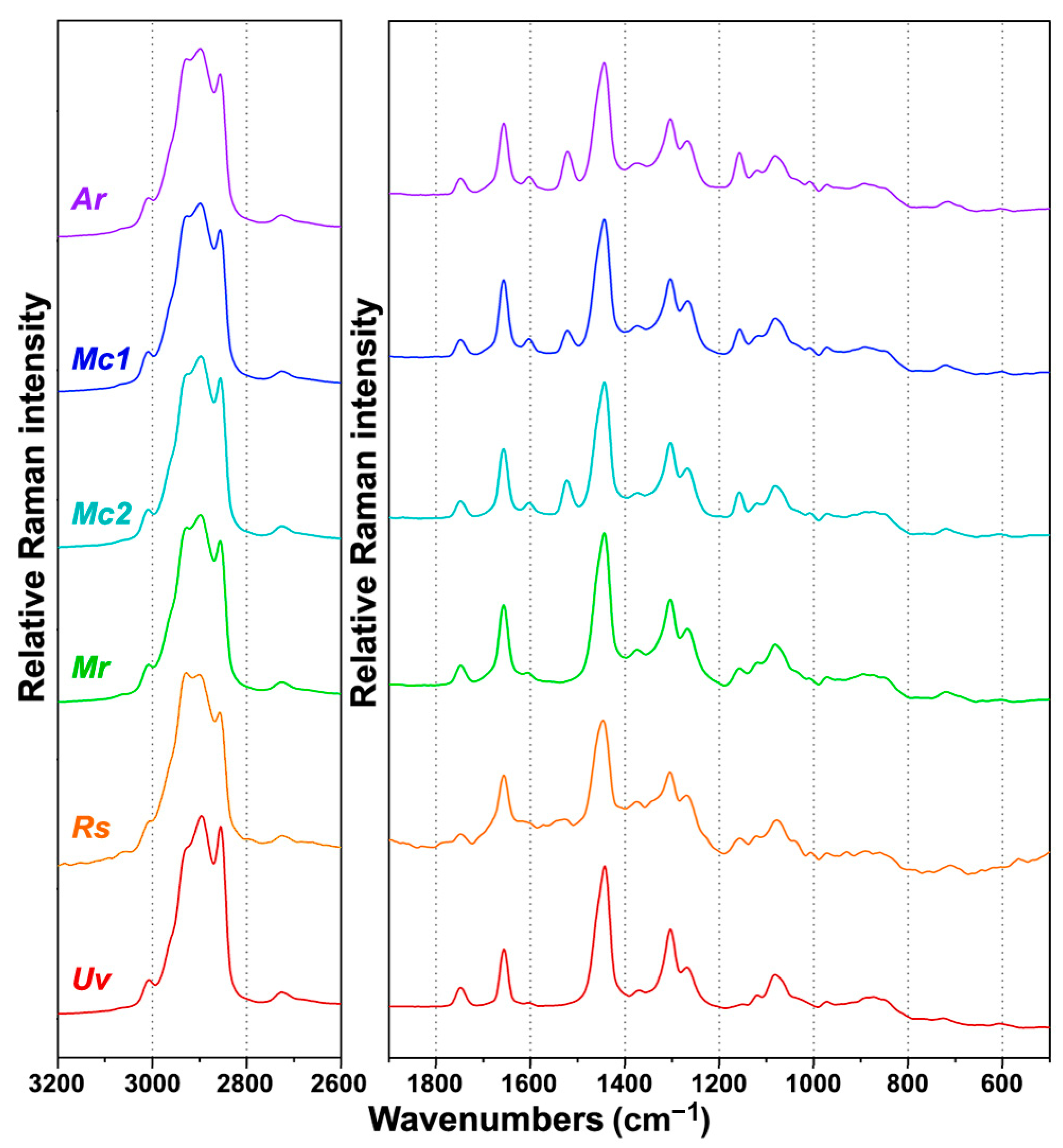
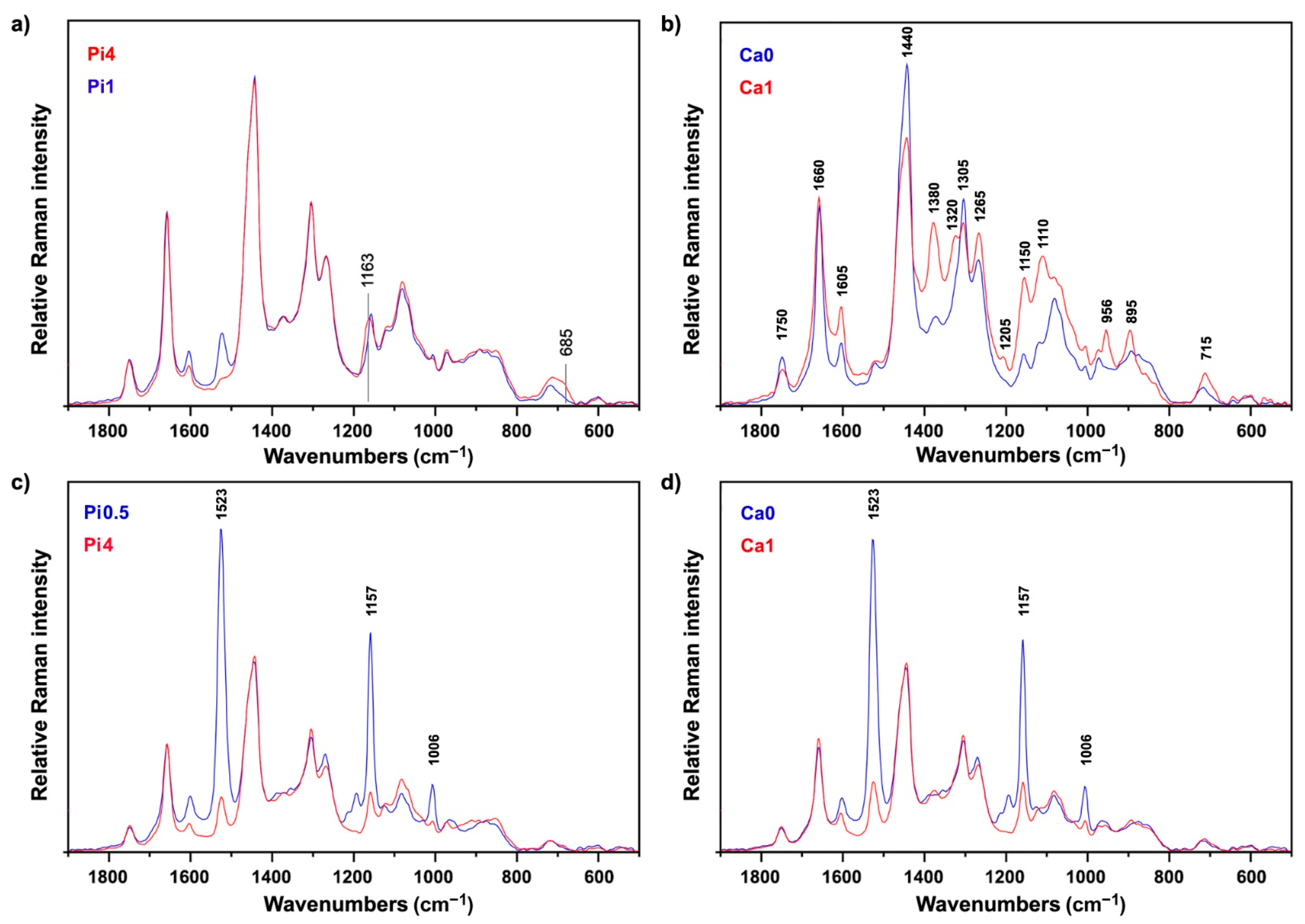
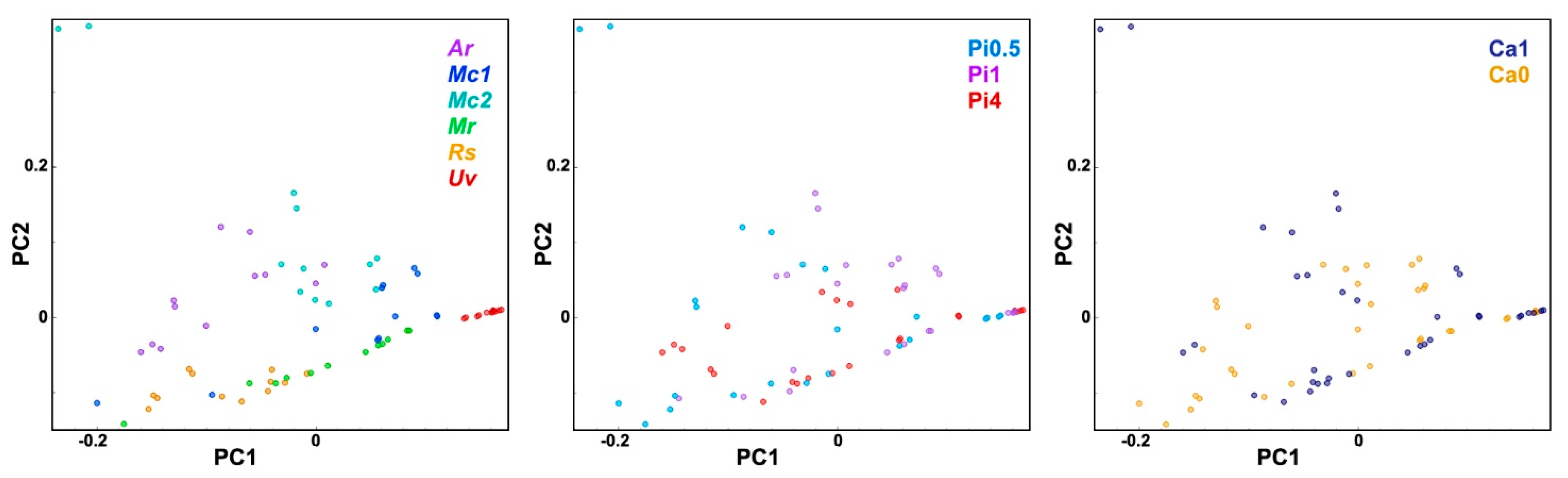
2.2. FT-Raman Chemical Profiling of Fungal Biomass
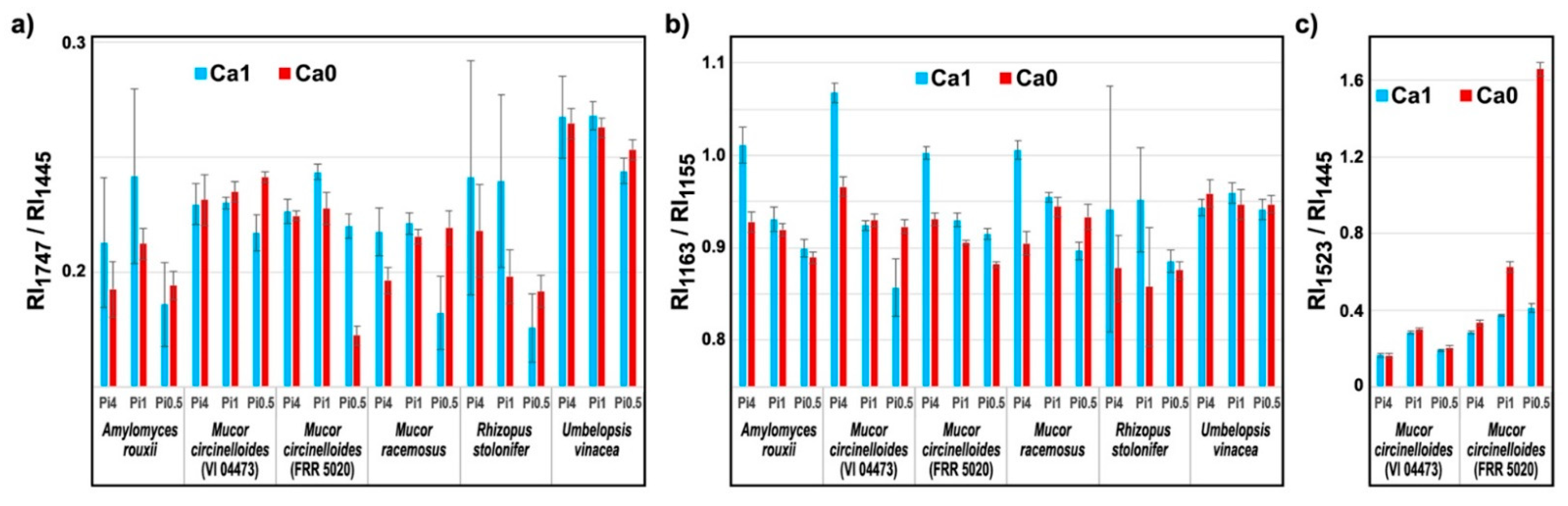
2.3. Quantitative Determination of Chemical Constituents of Fungal Biomass Based on Vibrational Spectra
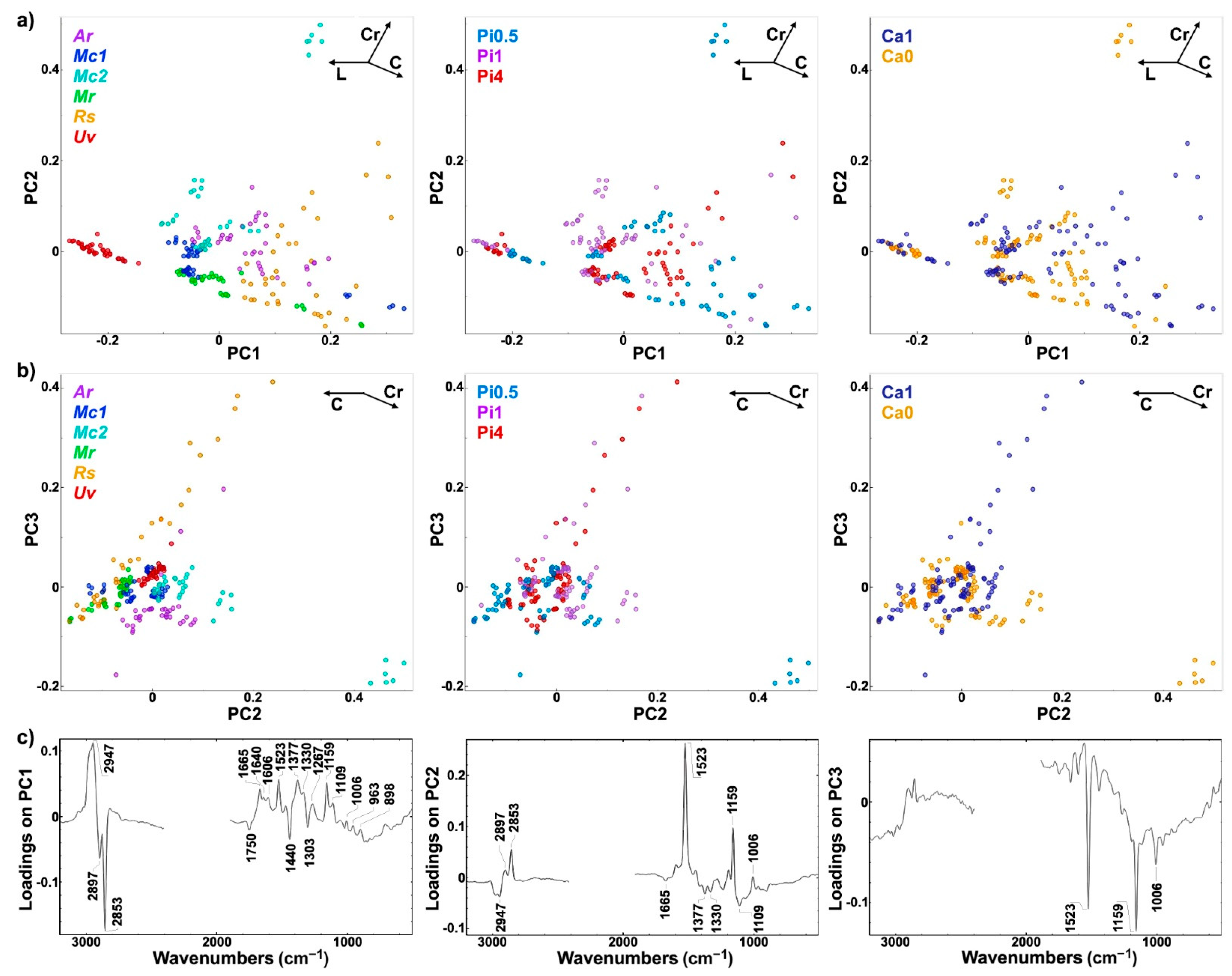
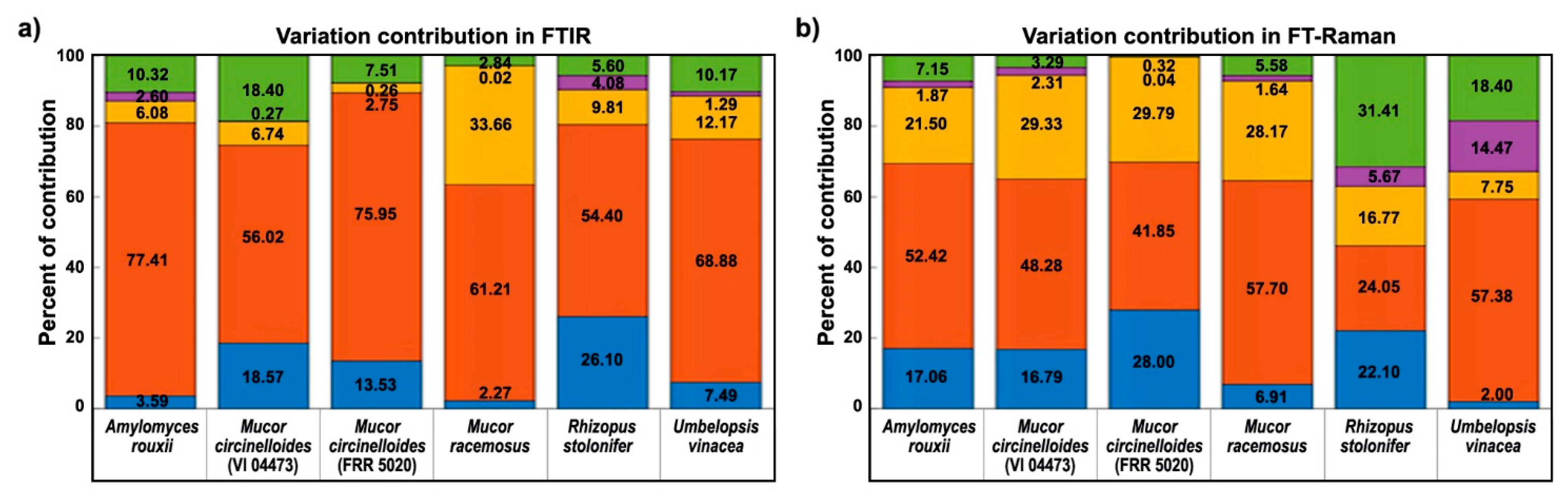
2.4. Multiblock and Analysis of Spectral Variation by the ANOVA Model of FTIR and FT-Raman Data
3. Materials and Methods
3.1. Fungal Strains
3.2. Cultivation of Fungi
3.3. Preparation of Fungal Biomass for Vibrational Spectroscopy Analyses
3.4. FT-Raman Spectroscopy Analysis
3.5. FTIR Spectroscopy Analysis
3.6. Spectral Preprocessing and Data Analysis
3.6.1. Spectral Preprocessing
3.6.2. Principal Component Analysis
3.6.3. Quantitative Determination of Chemical Constituents of Fungal Biomass Based on Vibrational Spectra
3.6.4. Multiblock and Analysis of Spectral Variation by the ANOVA Model of FTIR and FT-Raman Data
3.7. Reference Compounds and Reference Chemical Analyses
3.7.1. Reference Spectra
3.7.2. Carotenoid Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, T.C.; de Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: A white paper. Fungal Biol. Biotechnol. 2016, 3, 6. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.K.; Treichel, H.; Shapaval, V.; Oliveira, L.A.d.; Tuohy, M.G. Microbial Functional Foods and Nutraceuticals; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 1–309. [Google Scholar]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Fakas, S.; Komaitis, M.; Aggelis, G. Lipid production by oleaginous Mucorales cultivated on renewable carbon sources. Eur. J. Lipid Sci. Technol. 2007, 109, 1060–1070. [Google Scholar] [CrossRef]
- Qiao, W.C.; Tao, J.Q.; Luo, Y.; Tang, T.H.; Miao, J.H.; Yang, Q.W. Microbial oil production from solid-state fermentation by a newly isolated oleaginous fungus, Mucor circinelloides Q531 from mulberry branches. R. Soc. Open Sci. 2018, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Yang, J.M.; Xu, X.; Zhang, L.; Nie, Q.J.; Xian, M. Biodiesel production from oleaginous microorganisms. Renew. Energy 2009, 34, 1–5. [Google Scholar] [CrossRef]
- Formenti, L.R.; Norregaard, A.; Bolic, A.; Hernandez, D.Q.; Hagemann, T.; Heins, A.L.; Larsson, H.; Mears, L.; Mauricio-Iglesias, M.; Kruhne, U.; et al. Challenges in industrial fermentation technology research. Biotechnol. J. 2014, 9, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Kuhar, N.; Sil, S.; Verma, T.; Umapathy, S. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 2018, 8, 25888–25908. [Google Scholar] [CrossRef] [Green Version]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Potocki, L.; Depciuch, J.; Kuna, E.; Worek, M.; Lewinska, A.; Wnuk, M. FTIR and Raman Spectroscopy-Based Biochemical Profiling Reflects Genomic Diversity of Clinical Candida Isolates That May Be Useful for Diagnosis and Targeted Therapy of Candidiasis. Int. J. Mol. Sci. 2019, 20, 988. [Google Scholar] [CrossRef] [Green Version]
- Schalk, R.; Braun, F.; Frank, R.; Rädle, M.; Gretz, N.; Methner, F.-J.; Beuermann, T. Non-contact Raman spectroscopy for in-line monitoring of glucose and ethanol during yeast fermentations. Bioprocess. Biosyst. Eng. 2017, 40, 1519–1527. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, E.H.; Liakopoulou-Kyriakides, M.; Christofilos, D.; Arvanitidis, I.; Kourouklis, G. Raman Spectroscopy for Intracellular Monitoring of Carotenoid in Blakeslea trispora. Appl. Biochem. Biotechnol. 2009, 159, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Rebrošová, K.; Šiler, M.; Samek, O.; Růžička, F.; Bernatová, S.; Holá, V.; Ježek, J.; Zemánek, P.; Sokolová, J.; Petráš, P. Rapid identification of staphylococci by Raman spectroscopy. Sci. Rep. UK 2017, 7, 14846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizovský, M.; Pilát, Z.; Mylenko, M.; Hrouzek, P.; Kuta, J.; Skoupý, R.; Krzyžánek, V.; Hrubanová, K.; Adamczyk, O.; Ježek, J.; et al. Raman Microspectroscopic Analysis of Selenium Bioaccumulation by Green Alga Chlorella vulgaris. Biosensors 2021, 11, 115. [Google Scholar] [CrossRef]
- Moudříková, Š.; Sadowsky, A.; Metzger, S.; Nedbal, L.; Mettler-Altmann, T.; Mojzeš, P. Quantification of Polyphosphate in Microalgae by Raman Microscopy and by a Reference Enzymatic Assay. Anal. Chem. 2017, 89, 12006–12013. [Google Scholar] [CrossRef] [PubMed]
- Grace, C.E.E.; Lakshmi, P.K.; Meenakshi, S.; Vaidyanathan, S.; Srisudha, S.; Mary, M.B. Biomolecular transitions and lipid accumulation in green microalgae monitored by FTIR and Raman analysis. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 224, 117382. [Google Scholar] [CrossRef] [PubMed]
- Pilát, Z.; Bernatová, S.; Ježek, J.; Kirchhoff, J.; Tannert, A.; Neugebauer, U.; Samek, O.; Zemánek, P. Microfluidic Cultivation and Laser Tweezers Raman Spectroscopy of E. coli under Antibiotic Stress. Sensors 2018, 18, 1623. [Google Scholar] [CrossRef] [Green Version]
- Tafintseva, V.; Shapaval, V.; Smirnova, M.; Kohler, A. Extended multiplicative signal correction for FTIR spectral quality test and pre-processing of infrared imaging data. J. Biophotonics 2020, 13, e201960112. [Google Scholar] [CrossRef]
- Tahir, H.E.; Zou, X.B.; Xiao, J.B.; Mahunu, G.K.; Shi, J.Y.; Xu, J.L.; Sun, D.W. Recent Progress in Rapid Analyses of Vitamins, Phenolic, and Volatile Compounds in Foods Using Vibrational Spectroscopy Combined with Chemometrics: A Review. Food Anal. Method 2019, 12, 2361–2382. [Google Scholar] [CrossRef]
- Biancolillo, A.; Marini, F. Chemometric Methods for Spectroscopy-Based Pharmaceutical Analysis. Front. Chem. 2018, 6, 576. [Google Scholar] [CrossRef]
- Salzer, R.; Siesler, H.W. Infrared and Raman Spectroscopic Imaging; Wiley-VCH: Weinheim, Germany, 2009; pp. 65–112. [Google Scholar]
- Tafintseva, V.; Vigneau, E.; Shapaval, V.; Cariou, V.; Qannari, E.; Kohler, A. Hierarchical classification of microorganisms based on high-dimensional phenotypic data. J. Biophotonics 2018, 11, e201700047. [Google Scholar] [CrossRef]
- Zhang, X.L.; Lin, T.; Xu, J.F.; Luo, X.; Ying, Y.B. DeepSpectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta 2019, 1058, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Liland, K.H.; Kohler, A.; Shapaval, V. Hot PLS-a framework for hierarchically ordered taxonomic classification by partial least squares. Chemometr. Intell. Lab. 2014, 138, 41–47. [Google Scholar] [CrossRef]
- Cannizzaro, C.; Rhiel, M.; Marison, I.; von Stockar, U. On-line monitoring of Phaffia rhodozyma fed-batch process with in situ dispersive Raman spectroscopy. Biotechnol. Bioeng. 2003, 83, 668–680. [Google Scholar] [CrossRef]
- Horiue, H.; Sasaki, M.; Yoshikawa, Y.; Toyofuku, M.; Shigeto, S. Raman spectroscopic signatures of carotenoids and polyenes enable label-free visualization of microbial distributions within pink biofilms. Sci. Rep. UK 2020, 10. [Google Scholar] [CrossRef]
- Tauber, J.P.; Matthaus, C.; Lenz, C.; Hoffmeister, D.; Popp, J. Analysis of basidiomycete pigments in situ by Raman spectroscopy. J. Biophotonics 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Xue, F.; Yu, X.H. GC-MS, FTIR and Raman Analysis of Antioxidant Components of Red Pigments from Stemphylium lycopersici. Curr. Microbiol. 2017, 74, 532–539. [Google Scholar] [CrossRef]
- Li, K.; Cheng, J.; Ye, Q.; He, Y.; Zhou, J.H.; Cen, K.F. In vivo kinetics of lipids and astaxanthin evolution in Haematococcus pluvialis mutant under 15% CO2 using Raman microspectroscopy. Bioresour. Technol. 2017, 244, 1439–1444. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.F.C.; Le Hyaric, M.; Berg, M.M.; de Almeida, M.V.; Edwards, H.G.M. Raman spectroscopic characterization of cinnabarin produced by the fungus Pycnoporus sanguineus (Fr.) Murr. J. Raman Spectrosc. 2007, 38, 1628–1632. [Google Scholar] [CrossRef]
- Culka, A.; Jehlicka, J.; Ascaso, C.; Artieda, O.; Casero, C.M.; Wierzchos, J. Raman microspectrometric study of pigments in melanized fungi from the hyperarid Atacama desert gypsum crust. J. Raman Spectrosc. 2017, 48, 1487–1493. [Google Scholar] [CrossRef]
- Arcangeli, C.; Cannistraro, S. In situ Raman microspectroscopic identification and localization of carotenoids: Approach to monitoring of UV-B irradiation stress on antarctic fungus. Biopolymers 2000, 57, 179–186. [Google Scholar] [CrossRef]
- Munchberg, U.; Wagner, L.; Spielberg, E.T.; Voigt, K.; Rosch, P.; Popp, J. Spatially resolved investigation of the oil composition in single intact hyphae of Mortierella spp. with micro-Raman spectroscopy. BBA-Mol. Cell Biol. L 2013, 1831, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.F.; Huang, C.K.; Shigeto, S. In Vivo Probing of the Temperature Responses of Intracellular Biomolecules in Yeast Cells by Label-Free Raman Microspectroscopy. ChemBioChem 2013, 14, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Munchberg, U.; Wagner, L.; Rohrer, C.; Voigt, K.; Rosch, P.; Jahreis, G.; Popp, J. Quantitative assessment of the degree of lipid unsaturation in intact Mortierella by Raman microspectroscopy. Anal. Bioanal. Chem. 2015, 407, 3303–3311. [Google Scholar] [CrossRef]
- Kochan, K.; Peng, H.D.; Gwee, E.S.H.; Izgorodina, E.; Haritos, V.; Wood, B.R. Raman spectroscopy as a tool for tracking cyclopropane fatty acids in genetically engineered Saccharomyces cerevisiae. Analyst 2019, 144, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Gherman, A.M.R.; Dina, N.E.; Chis, V.; Wieser, A.; Haisch, C. Yeast cell wall–Silver nanoparticles interaction: A synergistic approach between surface-enhanced Raman scattering and computational spectroscopy tools. Spectrochim. Acta A 2019, 222. [Google Scholar] [CrossRef] [PubMed]
- Noothalapati, H.; Sasaki, T.; Kaino, T.; Kawamukai, M.; Ando, M.; Hamaguchi, H.; Yamamoto, T. Label-free Chemical Imaging of Fungal Spore Walls by Raman Microscopy and Multivariate Curve Resolution Analysis. Sci. Rep. UK 2016, 6, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Edwards, H.G.M.; Russell, N.C.; Weinstein, R.; Wynnwilliams, D.D. Fourier-Transform Raman-Spectroscopic Study of Fungi. J. Raman Spectrosc. 1995, 26, 911–916. [Google Scholar] [CrossRef]
- Esmonde-White, K.A.; Cuellar, M.; Uerpmann, C.; Lenain, B.; Lewis, I.R. Raman spectroscopy as a process analytical technology for pharmaceutical manufacturing and bioprocessing. Anal. Bioanal. Chem. 2017, 409, 637–649. [Google Scholar] [CrossRef] [Green Version]
- De Gussem, K.; Vandenabeele, P.; Verbeken, A.; Moens, L. Raman spectroscopic study of Lactarius spores (Russulales, Fungi). Spectrochim. Acta A 2005, 61, 2896–2908. [Google Scholar] [CrossRef]
- McGovern, A.C.; Broadhurst, D.; Taylor, J.; Kaderbhai, N.; Winson, M.K.; Small, D.A.; Rowland, J.J.; Kell, D.B.; Goodacre, R. Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: Application to gibberellic acid production. Biotechnol. Bioeng. 2002, 78, 527–538. [Google Scholar] [CrossRef]
- De Gussem, K.; Vandenabeele, P.; Verbeken, A.; Moens, L. Chemotaxonomical identification of spores of macrofungi: Possibilities of Raman spectroscopy. Anal. Bioanal. Chem. 2007, 387, 2823–2832. [Google Scholar] [CrossRef]
- Meenu, M.; Xu, B.J. Application of vibrational spectroscopy for classification, authentication and quality analysis of mushroom: A concise review. Food Chem. 2019, 289, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Witkowska, E.; Jagielski, T.; Kaminska, A. Genus- and species-level identification of dermatophyte fungi by surface-enhanced Raman spectroscopy. Spectrochim. Acta A 2018, 192, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Dina, N.E.; Gherman, A.M.R.; Chis, V.; Sarbu, C.; Wieser, A.; Bauer, D.; Haisch, C. Characterization of Clinically Relevant Fungi via SERS Fingerprinting Assisted by Novel Chemometric Models. Anal. Chem. 2018, 90, 2484–2492. [Google Scholar] [CrossRef]
- Lee, C.M.; Cho, E.M.; Ochir, E.G.; Dembereldorj, U.; Yang, S.I. Chemotaxonomic Raman Spectroscopy Investigation of Ascomycetes and Zygomycetes. Bull. Korean Chem. Soc. 2013, 34, 1240–1242. [Google Scholar] [CrossRef] [Green Version]
- Baranska, M.; Roman, M.; Dobrowolski, J.C.; Schulz, H.; Baranski, R. Recent Advances in Raman Analysis of Plants: Alkaloids, Carotenoids, and Polyacetylenes. Curr. Anal. Chem. 2013, 9, 108–127. [Google Scholar] [CrossRef]
- Bowie, B.T.; Chase, D.B.; Griffiths, P.R. Factors affecting the performance of bench-top Raman spectrometers. Part II: Effect of sample. Appl. Spectrosc. 2000, 54, 200a–207a. [Google Scholar] [CrossRef]
- Bowie, B.T.; Chase, D.B.; Griffiths, P.R. Factors affecting the performance of bench-top Raman spectrometers. Part I: Instrumental effects. Appl. Spectrosc. 2000, 54, 164a–173a. [Google Scholar] [CrossRef]
- Moester, M.J.B.; Zada, L.; Fokker, B.; Ariese, F.; de Boer, J.F. Stimulated Raman scattering microscopy with long wavelengths for improved imaging depth. J. Raman Spectrosc. 2019, 50, 1321–1328. [Google Scholar] [CrossRef] [Green Version]
- Boyaci, I.H.; Temiz, H.T.; Genis, H.E.; Soykut, E.A.; Yazgan, N.N.; Guven, B.; Uysal, R.S.; Bozkurt, A.G.; Ilaslan, K.; Torun, O.; et al. Dispersive and FT-Raman spectroscopic methods in food analysis. RSC Adv. 2015, 5, 56606–56624. [Google Scholar] [CrossRef]
- He, H.R.; Sun, D.W.; Pu, H.B.; Chen, L.J.; Lin, L. Applications of Raman spectroscopic techniques for quality and safety evaluation of milk: A review of recent developments. Crit. Rev. Food Sci. 2019, 59, 770–793. [Google Scholar] [CrossRef]
- Agarwal, U.P. 1064 nm FT-Raman spectroscopy for investigations of plant cell walls and other biomass materials. Front. Plant. Sci. 2014, 5, 490. [Google Scholar] [CrossRef]
- Kendel, A.; Zimmermann, B. Chemical Analysis of Pollen by FT-Raman and FTIR Spectroscopies. Front. Plant. Sci. 2020, 11, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzurendova, S.; Zimmermann, B.; Kohler, A.; Tafintseva, V.; Slany, O.; Certik, M.; Shapaval, V. Microcultivation and FTIR spectroscopy-based screening revealed a nutrient-induced co-production of high-value metabolites in oleaginous Mucoromycota fungi. PLoS ONE 2020, 15, e0234870. [Google Scholar] [CrossRef] [PubMed]
- Kosa, G.; Kohler, A.; Tafintseva, V.; Zimmermann, B.; Forfang, K.; Afseth, N.K.; Tzimorotas, D.; Vuoristo, K.S.; Horn, S.J.; Mounier, J.; et al. Microtiter plate cultivation of oleaginous fungi and monitoring of lipogenesis by high-throughput FTIR spectroscopy. Microb. Cell Fact. 2017, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosa, G.; Shapaval, V.; Kohler, A.; Zimmermann, B. FTIR spectroscopy as a unified method for simultaneous analysis of intra- and extracellular metabolites in high-throughput screening of microbial bioprocesses. Microb. Cell Fact. 2017, 16. [Google Scholar] [CrossRef] [Green Version]
- Kosa, G.; Zimmermann, B.; Kohler, A.; Ekeberg, D.; Afseth, N.K.; Mounier, J.; Shapaval, V. High-throughput screening of Mucoromycota fungi for production of low- and high-value lipids. Biotechnol. Biofuels 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Dzurendova, S.; Zimmermann, B.; Kohler, A.; Reitzel, K.; Nielsen, U.G.; Dupuy-Galet, B.X.; Leivers, S.; Horn, S.J.; Shapaval, V. Calcium Affects Polyphosphate and Lipid Accumulation in Mucoromycota Fungi. J. Fungi 2021, 7, 300. [Google Scholar] [CrossRef]
- Forfang, K.; Zimmermann, B.; Kosa, G.; Kohler, A.; Shapaval, V. FTIR Spectroscopy for Evaluation and Monitoring of Lipid Extraction Efficiency for Oleaginous Fungi. PLoS ONE 2017, 12, e0170611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beever, R.E.; Burns, D.J.W. Phosphorus Uptake, Storage and Utilization by Fungi. In Advances in Botanical Research; Woolhouse, H.W., Ed.; Academic Press: San Diego, CA, USA, 1981; Volume 8, pp. 127–219. [Google Scholar]
- Ye, Y.L.; Gan, J.; Hu, B. Screening of Phosphorus-Accumulating Fungi and Their Potential for Phosphorus Removal from Waste Streams. Appl. Biochem. Biotechnol. 2015, 177, 1127–1136. [Google Scholar] [CrossRef]
- Dzurendova, S.; Zimmermann, B.; Tafintseva, V.; Kohler, A.; Horn, S.J.; Shapaval, V. Metal and Phosphate Ions Show Remarkable Influence on the Biomass Production and Lipid Accumulation in Oleaginous Mucor circinelloides. J. Fungi 2020, 6, 260. [Google Scholar] [CrossRef] [PubMed]
- Ramos, I.B.; Miranda, K.; Ulrich, P.; Ingram, P.; LeFurgey, A.; Machado, E.A.; de Souza, W.; Docampo, R. Calcium- and polyphosphate-containing acidocalcisomes in chicken egg yolk. Biol. Cell 2010, 102, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Mouyna, I.; Hartland, R.P.; Paris, S.; Latge, J.P. From the surface to the inner layer of the fungal cell wall. Biochem. Soc. T 1997, 25, 194–199. [Google Scholar] [CrossRef]
- Cabib, E.; Bowers, B.; Sburlati, A.; Silverman, S.J. Fungal Cell-Wall Synthesis—The Construction of a Biological Structure. Microbiol. Sci. 1988, 5, 370–375. [Google Scholar]
- Bartnick, S. Cell Wall Chemistry Morphogenesis and Taxonomy of Fungi. Annu. Rev. Microbiol. 1968, 22, 87–108. [Google Scholar] [CrossRef]
- Dzurendova, S.; Zimmermann, B.; Tafintseva, V.; Kohler, A.; Ekeberg, D.; Shapaval, V. The influence of phosphorus source and the nature of nitrogen substrate on the biomass production and lipid accumulation in oleaginous Mucoromycota fungi. Appl. Microbiol. Biotechnol. 2020, 104, 8065–8076. [Google Scholar] [CrossRef] [PubMed]
- Martinezcadena, G.; Ruizherrera, J. Activation of Chitin Synthetase from Phycomyces-Blakesleeanus by Calcium and Calmodulin. Arch. Microbiol. 1987, 148, 280–285. [Google Scholar] [CrossRef]
- Papp, T.; Velayos, A.; Bartok, T.; Eslava, A.; Vagvolgyi, C.; Iturriaga, E. Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl. Microbiol. Biotechnol. 2006, 69, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Papp, T.; Nagy, G.; Csernetics, Á.; Szekeres, A.; Vágvölgyi, C. Beta-carotene production by Mucoralean fungi. J. Eng. Anim. 2009, 7, 173–176. [Google Scholar]
- Naz, T.; Nosheen, S.; Li, S.; Nazir, Y.; Mustafa, K.; Liu, Q.; Garre, V.; Song, Y. Comparative Analysis of β-Carotene Production by Mucor circinelloides Strains CBS 277.49 and WJ11 under Light and Dark Conditions. Metabolites 2020, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- Zajac, A.; Hanuza, J.; Wandas, M.; Dyminska, L. Determination of N-acetylation degree in chitosan using Raman spectroscopy. Spectrochim. Acta A 2015, 134, 114–120. [Google Scholar] [CrossRef]
- Jehlicka, J.; Edwards, H.G.M.; Orenc, A. Raman Spectroscopy of Microbial Pigments. Appl. Environ. Microb. 2014, 80, 3286–3295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avalos, J.; Limon, M.C. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Hassani, S.; Martens, H.; Qannari, E.M.; Hanafi, M.; Borge, G.I.; Kohler, A. Analysis of -omics data: Graphical interpretation- and validation tools in multi-block methods. Chemometr. Intell. Lab. 2010, 104, 140–153. [Google Scholar] [CrossRef]
- Diehn, S.; Zimmermann, B.; Tafintseva, V.; Seifert, S.; Bagcioglu, M.; Ohlson, M.; Weidner, S.; Fjellheim, S.; Kohler, A.; Kneipp, J. Combining Chemical Information From Grass Pollen in Multimodal Characterization. Front. Plant. Sci. 2020, 10, 1788. [Google Scholar] [CrossRef] [Green Version]
- Hassani, S.; Hanafi, M.; Qannari, E.; Kohler, A. Deflation strategies for multi-block principal component analysis revisited. Chemometr. Intell. Lab. 2013, 120, 154–168. [Google Scholar] [CrossRef]
- Kavadia, A.; Komaitis, M.; Chevalot, I.; Blanchard, F.; Marc, I.; Aggelis, G. Lipid and γ-linolenic acid accumulation in strains of Zygomycetes growing on glucose. J. Am. Oil Chem. Soc. 2001, 78, 341–346. [Google Scholar] [CrossRef]
- Kosa, G.; Vuoristo, K.S.; Horn, S.J.; Zimmermann, B.; Afseth, N.K.; Kohler, A.; Shapaval, V. Assessment of the scalability of a microtiter plate system for screening of oleaginous microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 4915–4925. [Google Scholar] [CrossRef] [Green Version]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Staric, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Toplak, M.; Birarda, G.; Read, S.; Sandt, C.; Rosendahl, S.M.; Vaccari, L.; Demšar, J.; Borondics, F. Infrared Orange: Connecting Hyperspectral Data with Machine Learning. Synchrotron Radiat. News 2017, 30, 40–45. [Google Scholar] [CrossRef]
- Guo, S.X.; Kohler, A.; Zimmermann, B.; Heinke, R.; Stockel, S.; Rosch, P.; Popp, J.; Bocklitz, T. Extended Multiplicative Signal Correction Based Model Transfer for Raman Spectroscopy in Biological Applications. Anal. Chem. 2018, 90, 9787–9795. [Google Scholar] [CrossRef]
- Zimmermann, B.; Kohler, A. Optimizing Savitzky-Golay Parameters for Improving Spectral Resolution and Quantification in Infrared Spectroscopy. Appl. Spectrosc. 2013, 67, 892–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtasu, M.V.; Tafintseva, V.; Bendiks, Z.A.; Marco, M.L.; Kohler, A.; Xu, Y.T.; Norskov, N.P.; Laerke, H.N.; Knudsen, K.E.B.; Hedemann, M.S. Obesity-Related Metabolome and Gut Microbiota Profiles of Juvenile Gottingen Minipigs-Long-Term Intake of Fructose and Resistant Starch. Metabolites 2020, 10, 456. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Kourti, T.; MacGregor, J.F. Analysis of multiblock and hierarchical PCA and PLS models. J. Chemometr. 1998, 12, 301–321. [Google Scholar] [CrossRef]
- Harrington, P.D.; Vieira, N.E.; Espinoza, J.; Nien, J.K.; Romero, R.; Yergey, A.L. Analysis of variance-principal component analysis: A soft tool for proteomic discovery. Anal. Chim. Acta 2005, 544, 118–127. [Google Scholar] [CrossRef]
- Smilde, A.K.; Jansen, J.J.; Hoefsloot, H.C.J.; Lamers, R.J.A.N.; van der Greef, J.; Timmerman, M.E. ANOVA-simultaneous component analysis (ASCA): A new tool for analyzing designed metabolomics data. Bioinformatics 2005, 21, 3043–3048. [Google Scholar] [CrossRef]
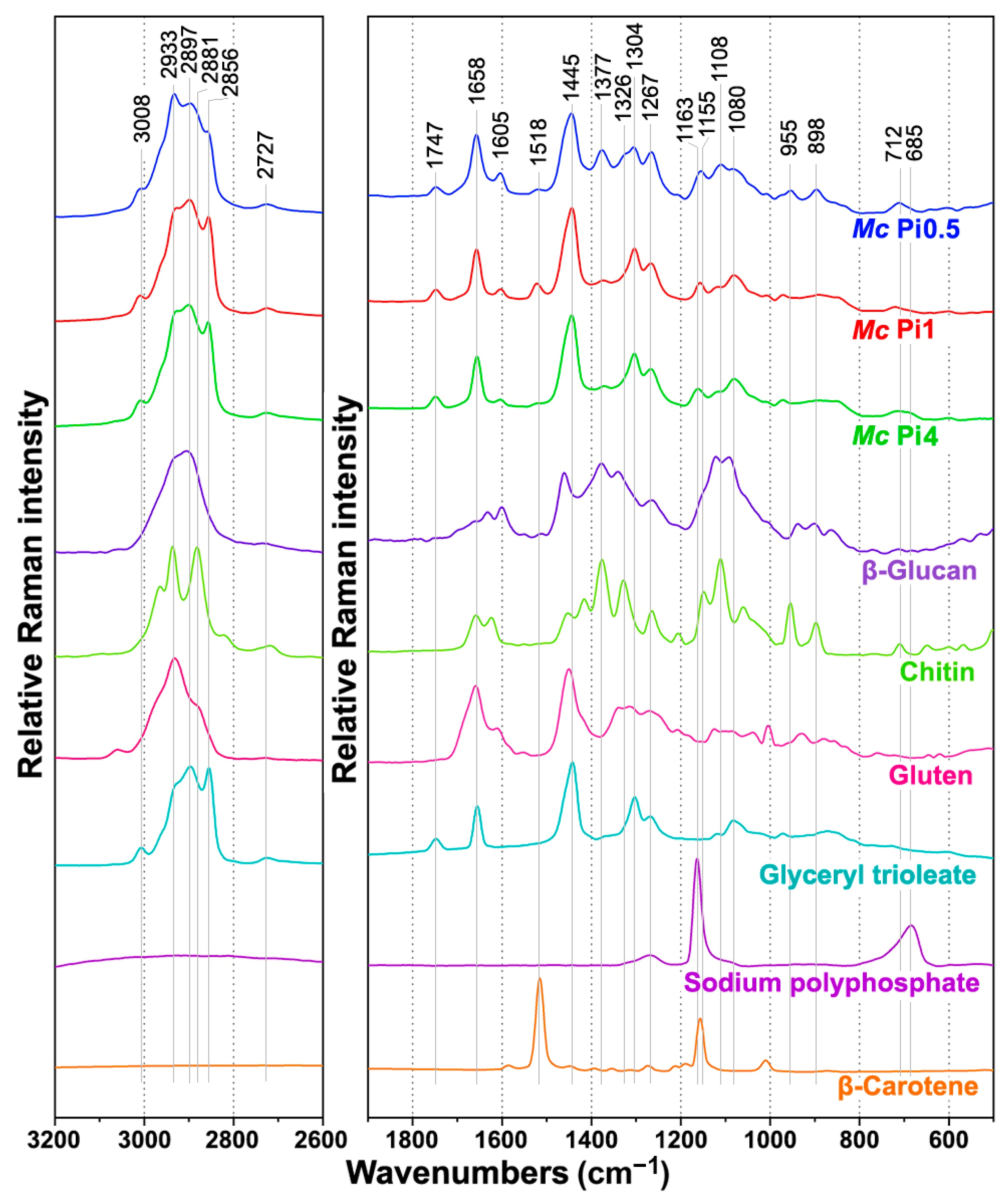
| Cell Component | Infrared | Raman | ||
|---|---|---|---|---|
| Wavenumbers (cm−1) | Molecular Vibration | Wavenumbers (cm−1) | Molecular Vibration | |
| Carbohydrates (glucosamines, glucans, glucuronans) | 3300 | O-H str. | 2933 and 2895 | -C-H str. (CH3) |
| 3400–3100 | N-H str., N-H2 str. | 2855 | -C-H str. (CH2, glucan) | |
| 2879 | -C-H str. (CH3) | 1680–1620 | -C=O str. (Amide I, chitin) | |
| 1730 | -C=O str. (glucuronans) | 1755 | -C=O str. (glucuronan) | |
| 1680–1620 | -C=O str. (Amide I, chitin) | 1620–1570 | NH2 def. (chitosan) | |
| 1600–1550 | NH2 def. (chitosan) | 1460–1440 | CH2 and CH3 def. | |
| 1554 | C-N str. & NH def. (Amide II, chitin) | 1377 | CH2, CH, COH def. | |
| 1375 | -CH3 def. | 1327 | CH2, CH, COH def. | |
| 1305 | C-N-H def. (Amide III, chitin) | 1256 | C-C, C-O, CH, CH2 | |
| 1200–1000 | C-O-C str., COH def. COC def. | 1200–1150 | C-O-C str. | |
| 950 | -CH3 def. | 1050–1150 | C-N str. & C-C str. | |
| 950–850 | C-C str, C-O-C str. & def., COH def. | |||
| 715 | O-C-O str. & CH def. | |||
| Acylglycerol lipids (triglycerides) | 3010 | =C-H str. | 3008 | =C-H str. |
| 2921 | -C-H str. (CH3) | 2933 and 2895 | -C-H str. (CH3) | |
| 2852 | -C-H str. (CH2) | 2855 | -C-H str. (CH2) | |
| 1743 | -C=O str. | 1750 | C=O str. | |
| 1463 | -CH2 def. | 1660 | C=C str. | |
| 1160 | C-O-C str. | 1460–1440 | CH2 and CH3 def. | |
| 723 | -CH2 def. | 1305 | CH2 def. | |
| 1080–1060 | C-C str. C-O str. | |||
| Polyphosphates | 1263 | P=O str (PO2-) | 1165 | P=O str. (PO2-) |
| 885 | P-O-P str. | 685 | P-O-P str. | |
| Proteins | 1680–1630 | -C=O str. (Amide I) | 1660 | -C=O str. (Amide I) |
| 1560–1530 | C-N-H def. (Amide II) | 1620–1580 | NH2 def. | |
| 1310–1250 | C-N-H def. (Amide III) | 1605 | C=C str. (phenyl ring) | |
| 1460–1440 | CH2 and CH3 def. | |||
| 1310–1250 | C-N-H def. (Amide III) | |||
| 1005 | phenyl ring def. | |||
| Carotenoids | Not detectable at concentrations present in fungal biomass | 1525 | C=C str. (polyene chain) | |
| 1155 | C-C str. & CH def. | |||
| 1005 | C-CH3 def. | |||
| Analysis | Range | Nonderivative | Derivative | ||
|---|---|---|---|---|---|
| R2 (Aopt) | RMSE | R2 (Aopt) | RMSE | ||
| Total lipids (6 strains) | 19.42–87.13%dry weight | 0.83 (5) | 6.60%dry weight | 0.75 (3) | 8.06%dry weight |
| Total lipids (Mucor) | 19.42–55.57%dry weight | 0.88 (5) | 2.94%dry weight | 0.88 (5) | 2.90%dry weight |
| Total phosphorus (6 strains) | 0.64–6.24%dry weight | 0.86 (7) | 0.50%dry weight | 0.79 (5) | 0.60%dry weight |
| Total phosphorus (Mucor) | 1.40–5.20%dry weight | 0.89 (6) | 0.38%dry weight | 0.89 (5) | 0.37%dry weight |
| Total carotenoids | 10.21–1669.88 µg/gdry weight | 0.84 (1) | 134.69 µg/gdry weight | 0.84 (2) | 137.34 µg/gdry weight |
| Analysis | Range | Nonderivative | Derivative | ||
|---|---|---|---|---|---|
| R2 (Aopt) | RMSE | R2 (Aopt) | RMSE | ||
| Total lipids (6 strains) | 19.42–87.13%dry weight | 0.86 (2) | 6.02%dry weight | 0.85 (8) | 6.12%dry weight |
| Total lipids (Mucor) | 19.42–55.57%dry weight | 0.79 (7) | 3.93%dry weight | 0.82 (5) | 3.59%dry weight |
| Total phosphorus (6 strains) | 0.64–6.24%dry weight | 0.87 (9) | 0.47%dry weight | 0.84 (5) | 0.53%dry weight |
| Total phosphorus (Mucor) | 1.40–5.20%dry weight | 0.94 (6) | 0.29%dry weight | 0.84 (4) | 0.46%dry weight |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzurendová, S.; Shapaval, V.; Tafintseva, V.; Kohler, A.; Byrtusová, D.; Szotkowski, M.; Márová, I.; Zimmermann, B. Assessment of Biotechnologically Important Filamentous Fungal Biomass by Fourier Transform Raman Spectroscopy. Int. J. Mol. Sci. 2021, 22, 6710. https://doi.org/10.3390/ijms22136710
Dzurendová S, Shapaval V, Tafintseva V, Kohler A, Byrtusová D, Szotkowski M, Márová I, Zimmermann B. Assessment of Biotechnologically Important Filamentous Fungal Biomass by Fourier Transform Raman Spectroscopy. International Journal of Molecular Sciences. 2021; 22(13):6710. https://doi.org/10.3390/ijms22136710
Chicago/Turabian StyleDzurendová, Simona, Volha Shapaval, Valeria Tafintseva, Achim Kohler, Dana Byrtusová, Martin Szotkowski, Ivana Márová, and Boris Zimmermann. 2021. "Assessment of Biotechnologically Important Filamentous Fungal Biomass by Fourier Transform Raman Spectroscopy" International Journal of Molecular Sciences 22, no. 13: 6710. https://doi.org/10.3390/ijms22136710
APA StyleDzurendová, S., Shapaval, V., Tafintseva, V., Kohler, A., Byrtusová, D., Szotkowski, M., Márová, I., & Zimmermann, B. (2021). Assessment of Biotechnologically Important Filamentous Fungal Biomass by Fourier Transform Raman Spectroscopy. International Journal of Molecular Sciences, 22(13), 6710. https://doi.org/10.3390/ijms22136710






