Strategies and Perspectives for UV Resonance Raman Applicability in Clinical Analyses of Human Sperm RNA
Abstract
:1. Introduction
2. Results
2.1. Spermiograms
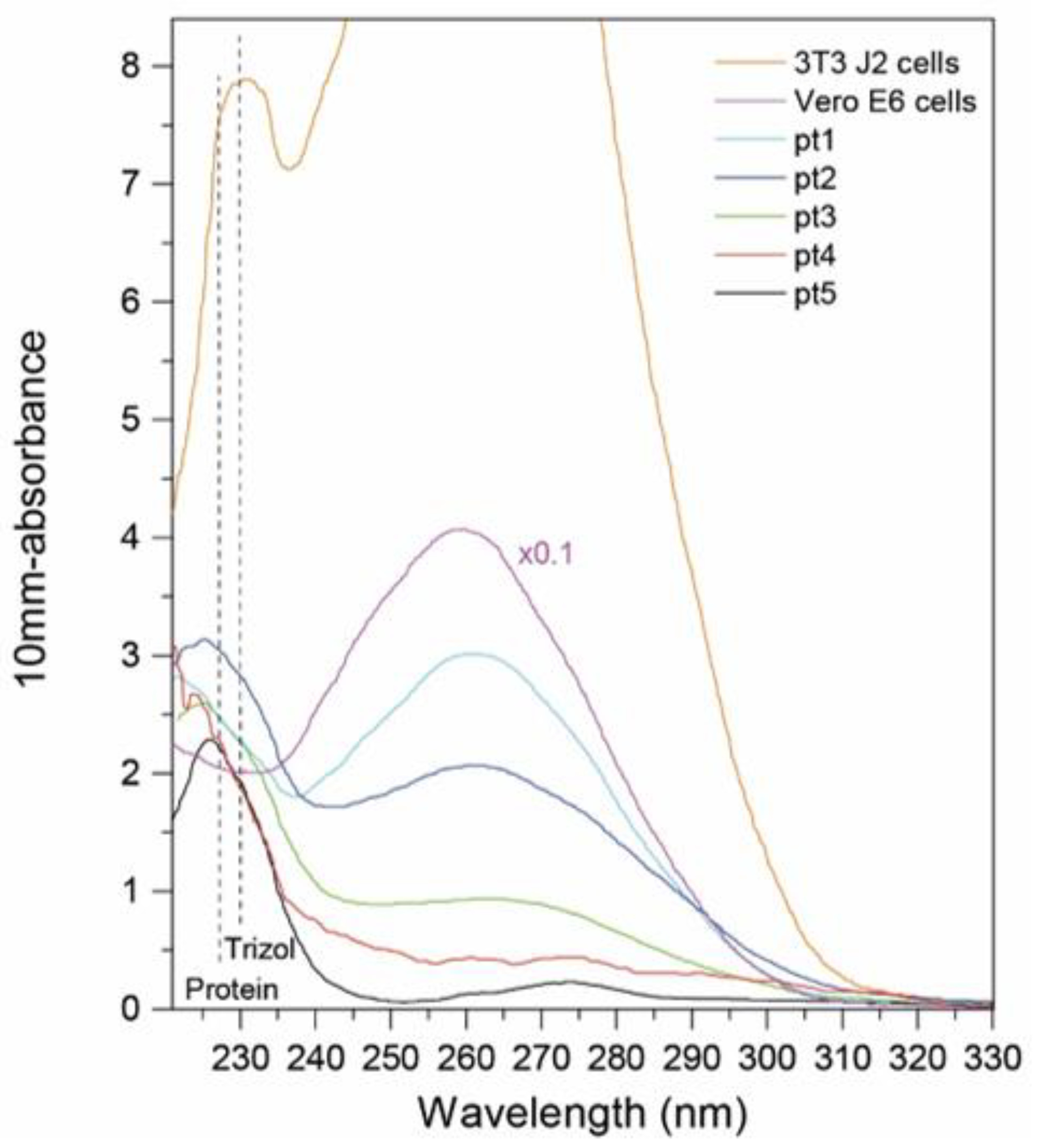
2.2. UV/Vis Absorbance Measurement of RNA
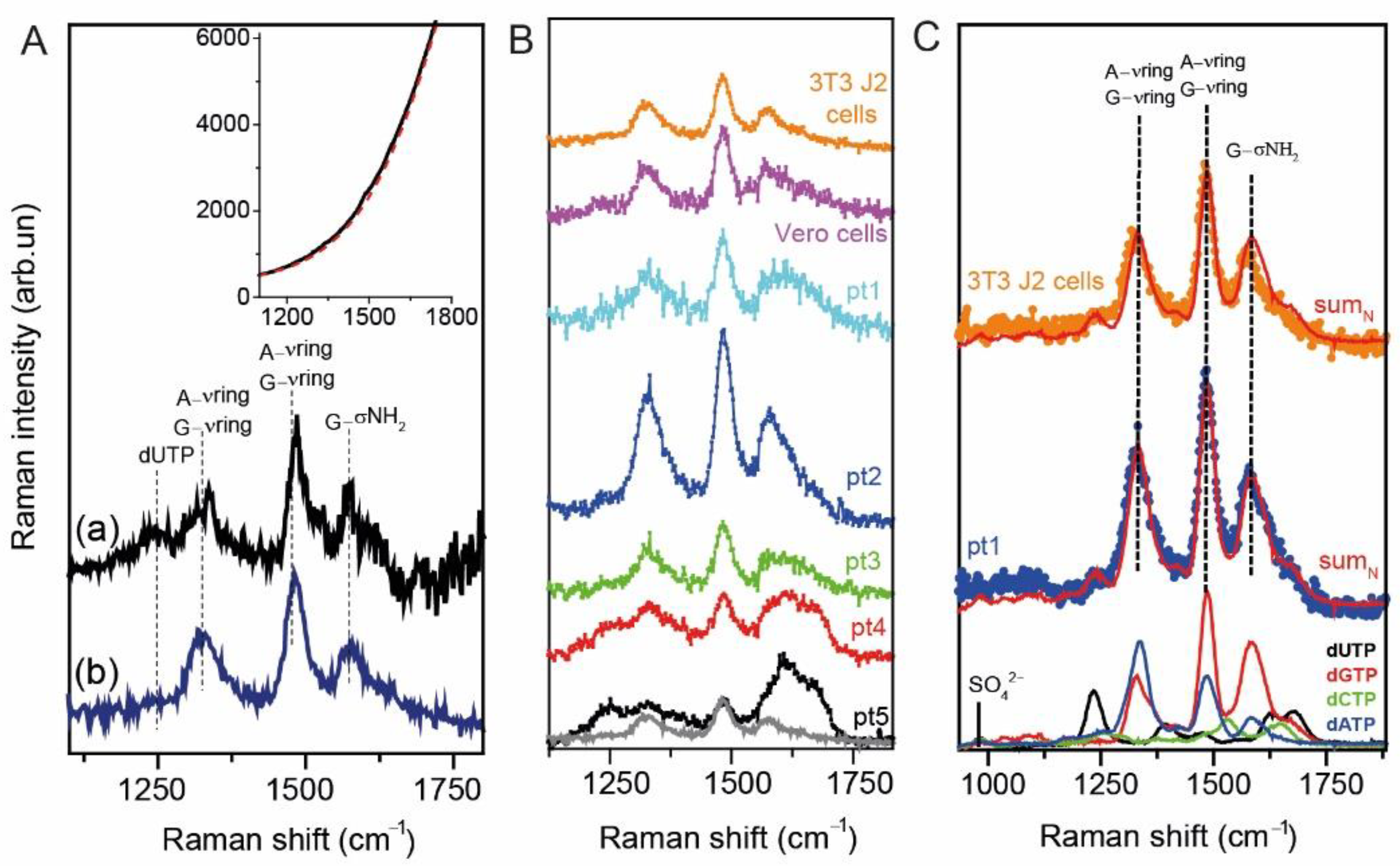
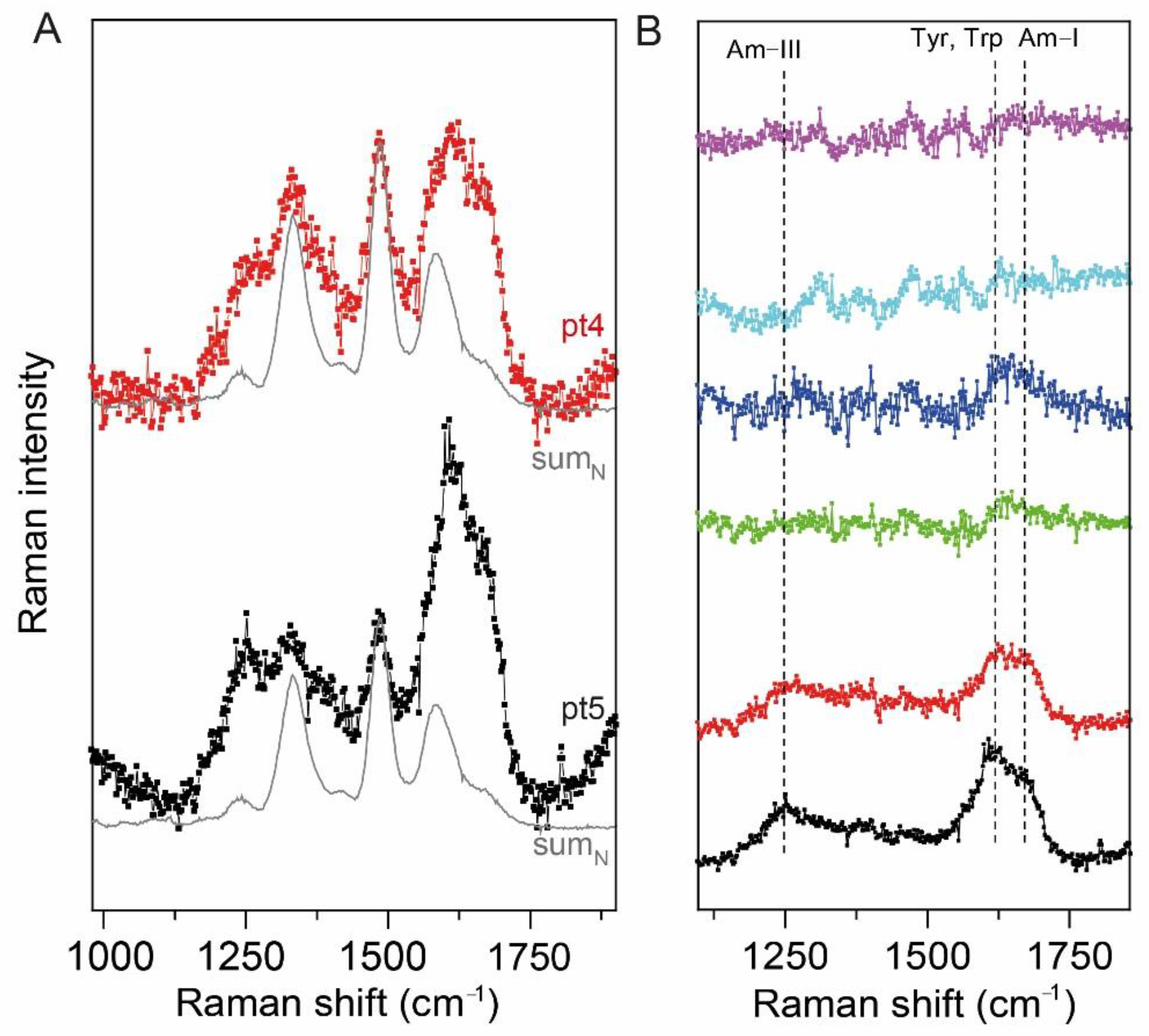
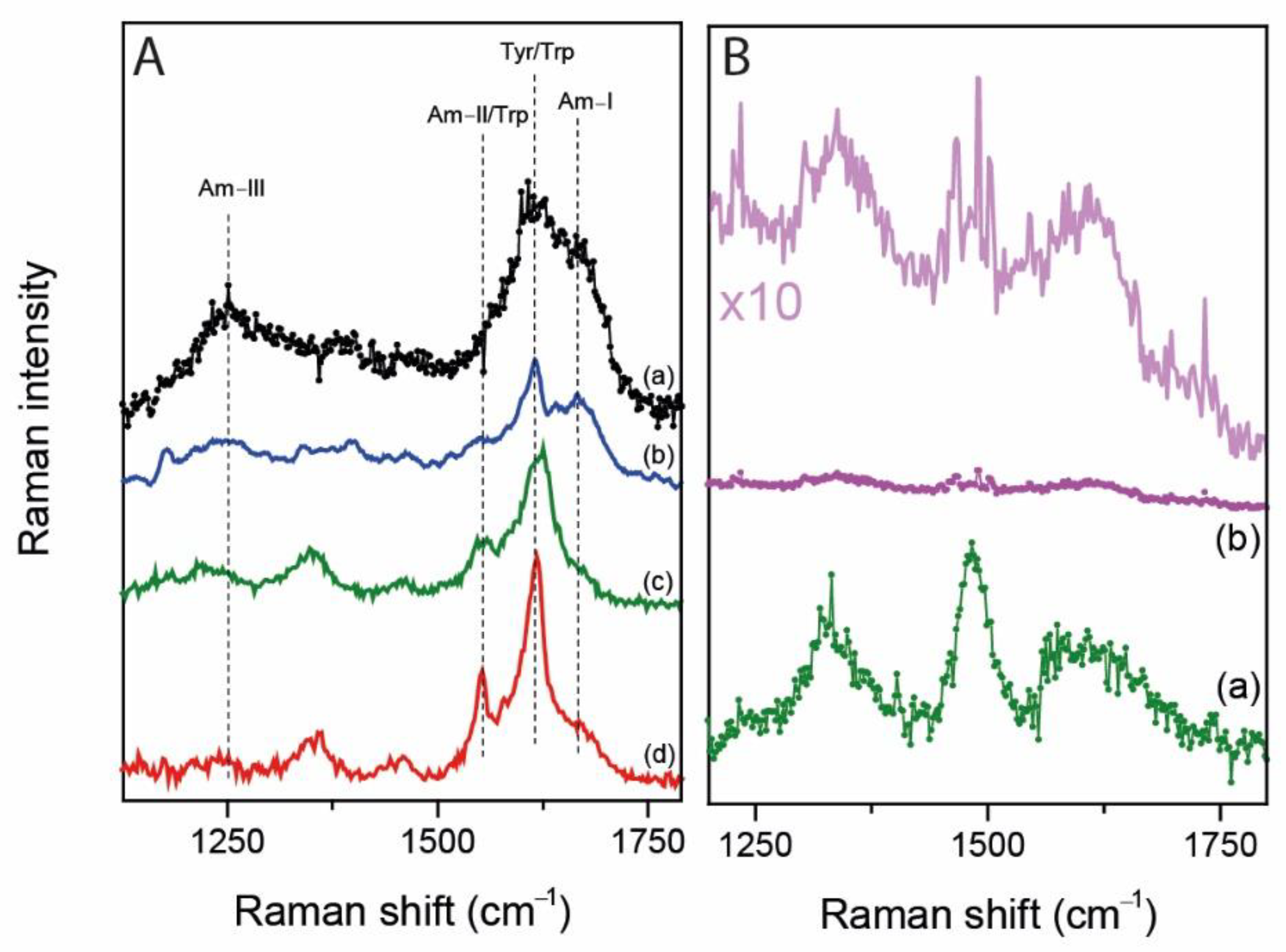
2.3. UV Resonance Raman Spectroscopy
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Subjects and Sample Preparation
5.2. UV Resonance Raman Spectroscopy Measurements
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pascolo, L.; Bedolla, D.E.; Vaccari, L.; Venturin, I.; Cammisuli, F.; Gianoncelli, A.; Mitri, E.; Giolo, E.; Luppi, S.; Martinelli, M.; et al. Pitfalls and Promises in FTIR Spectromicroscopy Analyses to Monitor Iron-Mediated DNA Damage in Sperm. Reprod. Toxicol. 2016, 61, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, M.; Zupin, L.; Venturin, I.; Mitri, E.; Boscolo, R.; D’Amico, F.; Vaccari, L.; Crovella, S.; Ricci, G.; Pascolo, L. FTIR Spectroscopy to Reveal Lipid and Protein Changes Induced on Sperm by Capacitation: Bases for an Improvement of Sample Selection in ART. Int. J. Mol. Sci. 2020, 21, 8659. [Google Scholar] [CrossRef]
- Lotti, F.; Maggi, M. Sexual Dysfunction and Male Infertility. Nat. Rev. Urol. 2018, 15, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Naz, M.; Kamal, M. Classification, Causes, Diagnosis and Treatment of Male Infertility: A Review. Orient. Pharm. Exp. Med. 2017, 17, 89–109. [Google Scholar] [CrossRef]
- Zupin, L.; Pascolo, L.; Luppi, S.; Ottaviani, G.; Crovella, S.; Ricci, G. Photobiomodulation Therapy for Male Infertility. Lasers Med. Sci. 2020, 35, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Zupin, L.; Pascolo, L.; Gianoncelli, A.; Gariani, G.; Luppi, S.; Giolo, E.; Ottaviani, G.; Crovella, S.; Ricci, G. Synchrotron radiation soft X-ray microscopy and low energy X-ray fluorescence to reveal elemental changes in spermatozoa treated with photobiomodulation therapy. Anal. Methods 2020, 12, 3691–3696. [Google Scholar] [CrossRef]
- Le, M.T.; Nguyen, T.; Nguyen, H.; Nguyen, T.; Nguyen, V.T.; Le, D.D.; Nguyen, V.; Cao, N.T. Does sperm DNA fragmentation correlate with semen parameters? Reprod. Med. Biol. 2019, 18, 390–396. [Google Scholar] [CrossRef] [Green Version]
- Dada, R.; Kumar, M.; Jesudasan, R.; Fernández, J.L.; Gosálvez, J.; Agarwal, A. Epigenetics and Its Role in Male Infertility. J. Assist. Reprod. Genet. 2012, 29, 213–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Flynn O’Brien, K.L.; Varghese, A.C.; Agarwal, A. The Genetic Causes of Male Factor Infertility: A Review. Fertil. Steril. 2010, 93, 1–12. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Lanzillotti, C.; Mazziotta, C.; Tognon, M.; Martini, F. Epigenetics of Male Infertility: The Role of DNA Methylation. Front. Cell Dev. Biol. 2021, 9, 689624. [Google Scholar] [CrossRef]
- Gapp, K.; van Steenwyk, G.; Germain, P.L.; Matsushima, W.; Rudolph, K.L.M.; Manuella, F.; Roszkowski, M.; Vernaz, G.; Ghosh, T.; Pelczar, P.; et al. Alterations in Sperm Long RNA Contribute to the Epigenetic Inheritance of the Effects of Postnatal Trauma. Mol. Psychiatry 2020, 25, 2162–2174. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Shi, J.; Rassoulzadegan, M.; Tuorto, F.; Chen, Q. Sperm RNA code programmes the metabolic health of offspring. Nat. Rev. Endocrinol. 2019, 15, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.H.; Wang, A.; Song, C.; Shankar, G.; Srivastava, R.K.; Au, K.F.; Li, X.Z. Single-Molecule Long-Read Sequencing Reveals a Conserved Intact Long RNA Profile in Sperm. Nat. Commun. 2021, 12, 1361. [Google Scholar] [CrossRef]
- Mendel, M.; Chen, K.-M.; Homolka, D.; Gos, P.; Pandey, R.R.; McCarthy, A.A.; Pillai, R.S. Methylation of Structured RNA by the M6A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell 2018, 71, 986–1000.e11. [Google Scholar] [CrossRef] [Green Version]
- Sharma, U. Paternal Contributions to Offspring Health: Role of Sperm Small RNAs in Intergenerational Transmission of Epigenetic Information. Front. Cell Dev. Biol. 2019, 7, 215. [Google Scholar] [CrossRef]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef]
- Wagner, K.D.; Wagner, N.; Ghanbarian, H.; Grandjean, V.; Gounon, P.; Cuzin, F.; Rassoulzadegan, M. RNA Induction and Inheritance of Epigenetic Cardiac Hypertrophy in the Mouse. Dev. Cell 2008, 14, 962–969. [Google Scholar] [CrossRef] [Green Version]
- Grandjean, V.; Fourré, S.; De Abreu, D.A.; Derieppe, M.A.; Remy, J.J.; Rassoulzadegan, M. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci. Rep. 2015, 5, 18193. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Wang, W.; Ali, M.A.; Wang, Y.; Zhang, Y.; Zhang, M.; Zhou, G.; Yang, J.D.; Zeng, C. Transcriptome-wide m6A profiling reveals mRNA post-transcriptional modification of boar sperm during cryopreservation. BMC Genom. 2021, 22, 588. [Google Scholar] [CrossRef]
- Oliva, R. Protamines and male infertility. Hum. Reprod. Update 2006, 12, 417–435. [Google Scholar] [CrossRef] [Green Version]
- Jodar, M. Sperm and Seminal Plasma RNAs: What Roles Do They Play beyond Fertilization? Reproduction 2019, 158, R113–R123. [Google Scholar] [CrossRef]
- Dai, C.; Zhang, Z.; Shan, G.; Chu, L.-T.; Huang, Z.; Moskovtsev, S.; Librach, C.; Jarvi, K.; Sun, Y. Advances in Sperm Analysis: Techniques, Discoveries and Applications. Nat. Rev. Urol. 2021, 18, 447–467. [Google Scholar] [CrossRef]
- D’Amico, F.; Zucchiatti, P.; Latella, K.; Pachetti, M.; Gessini, A.; Masciovecchio, C.; Vaccari, L.; Pascolo, L. Investigation of genomic DNA methylation by ultraviolet resonant Raman spectroscopy. J. Biophotonics 2020, 13, e202000150. [Google Scholar] [CrossRef]
- Rowe, P.J.; Comhaire, F.H.; Hargreave, T.B.; Mahmoud, A.M.; World Health Organization. WHO Manual for the Standardized Investigation, Diagnosis and Management of the Infertile Male; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Kobayashi, H.; Sato, A.; Otsu, E.; Hiura, H.; Tomatsu, C.; Utsunomiya, T.; Sasaki, H.; Yaegashi, N.; Arima, T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007, 16, 2542–2551. [Google Scholar] [CrossRef]
- You, C.; Dai, X.; Wang, Y. Position-Dependent Effects of Regioisomeric Methylated Adenine and Guanine Ribonucleosides on Translation. Nucleic Acids Res. 2017, 45, 9059–9067. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.W.; Lieu, D.K.; Huser, T.; Li, R.A. Label-Free Separation of Human Embryonic Stem Cells and Their Cardiac Derivatives Using Raman Spectroscopy. Anal. Chem. 2009, 81, 1324–1331. [Google Scholar] [CrossRef] [Green Version]
- Hobro, A.J.; Rouhi, M.; Blanch, E.W.; Conn, G.L. Raman and Raman Optical Activity (ROA) Analysis of RNA Structural Motifs in Domain I of the EMCV IRES. Nucleic Acids Res. 2007, 35, 1169–1177. [Google Scholar] [CrossRef]
- Kumamoto, Y.; Taguchi, A.; Smith, N.I.; Kawata, S. Deep ultraviolet resonant Raman imaging of a cell. J. Biomed. Opt. 2012, 17, 076001. [Google Scholar] [CrossRef] [Green Version]
- Morla-Folch, J.; Alvarez-Puebla, R.A.; Guerrini, L. Direct Quantification of DNA Base Composition by Surface-Enhanced Raman Scattering Spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3037–3041. [Google Scholar] [CrossRef]
- Pachetti, M.; D’Amico, F.; Pascolo, L.; Pucciarelli, S.; Gessini, A.; Parisse, P.; Vaccari, L.; Masciovecchio, C. UV Resonance Raman Explores Protein Structural Modification upon Fibrillation and Ligand Interaction. Biophys. J. 2021, 120, 4575–4589. [Google Scholar] [CrossRef]
- Jakubek, R.S.; Handen, J.; White, S.E.; Asher, S.A.; Lednev, I.K. Ultraviolet Resonance Raman Spectroscopic Markers for Protein Structure and Dynamics. Trends Analyt. Chem. 2018, 103, 223–229. [Google Scholar] [CrossRef]
- Oladepo, S.A.; Xiong, K.; Hong, Z.; Asher, S.A.; Handen, J.; Lednev, I.K. UV Resonance Raman Investigations of Peptide and Protein Structure and Dynamics. Chem. Rev. 2012, 112, 2604–2628. [Google Scholar] [CrossRef] [Green Version]
- Zhong, D.; Chen, M.; Zhang, L.; Chen, H.; Shi, D.; Liu, Q.; Li, H. Aberrant regulation of RNA methylation during spermatogenesis. Reprod. Domest. Anim. 2021, 56, 3–11. [Google Scholar] [CrossRef]
- Yao, L.; Yin, H.; Hong, M.; Wang, Y.; Yu, T.; Teng, Y.; Li, T.; Wu, Q. RNA Methylation in Hematological Malignancies and Its Interactions with Other Epigenetic Modifications. Leukemia 2021, 35, 1243–1257. [Google Scholar] [CrossRef]
- Nguyen, D.B.; Nguyen, T.D.; Kim, S.; Joo, S.-W. Vibrational Fingerprints of N6-Methyladenine and N6,N6-Dimethyladenine in Raman Spectra. Vib. Spectrosc. 2017, 90, 7–13. [Google Scholar] [CrossRef]
- Barhoumi, A.; Halas, N.J. Detecting Chemically Modified DNA Bases Using Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. Lett. 2011, 2, 3118–3123. [Google Scholar] [CrossRef] [Green Version]
- Oladepo, S.A.; Loppnow, G.R. Initial Excited-State Structural Dynamics of 9-Methyladenine from UV Resonance Raman Spectroscopy. J. Phys. Chem. B 2011, 115, 6149–6156. [Google Scholar] [CrossRef]
- Suen, W.; Spiro, T.G.; Sowers, L.C.; Fresco, J.R. Identification by UV Resonance Raman Spectroscopy of an Imino Tautomer of 5-Hydroxy-2′-Deoxycytidine, a Powerful Base Analog Transition Mutagen with a Much Higher Unfavored Tautomer Frequency than That of the Natural Residue 2′-Deoxycytidine. Proc. Natl. Acad. Sci. USA 1999, 96, 4500–4505. [Google Scholar] [CrossRef] [Green Version]
- D’Amico, F.; Cammisuli, F.; Addobbati, R.; Rizzardi, C.; Gessini, A.; Masciovecchio, C.; Rossi, B.; Pascolo, L. Oxidative Damage in DNA Bases Revealed by UV Resonant Raman Spectroscopy. Analyst 2015, 140, 1477–1485. [Google Scholar] [CrossRef]
- Benevides, J.M.; Overman, S.A.; Thomas, G.J., Jr. Raman, Polarized Raman and Ultraviolet Resonance Raman Spectroscopy of Nucleic Acids and Their Complexes. J. Raman Spectrosc. 2005, 36, 279–299. [Google Scholar] [CrossRef]
- D’Amico, F.; Saito, M.; Bencivenga, F.; Marsi, M.; Gessini, A.; Camisasca, G.; Principi, E.; Cucini, R.; Di Fonzo, S.; Battistoni, A.; et al. UV Resonant Raman Scattering Facility at Elettra. Nucl. Instrum. Methods Phys. Res. A 2013, 703, 33–37. [Google Scholar] [CrossRef]
| Patient | Concentration (106/mL) | Total Motility (%) | Progressive Motility (%) | WHO Classification |
|---|---|---|---|---|
| 1 | 9 | 40 | 30 | oligoasthenozoospermia |
| 2 | 60 | 50 | 20 | asthenozoospermia |
| 3 | 28 | 70 | 30 | asthenozoospermia |
| 4 | 15 | 45 | 25 | oligoasthenozoospermia |
| 5 | 3 | 35 | 10 | oligoasthenozoospermia |
| Samples | RNA Concentration (ng/µL) | Volume of Elution (µL) |
|---|---|---|
| 3T3 J2 cells | >4271 | 50 |
| VeroE6 cells | 1622.5 | 50 |
| 1 | 120.7 | 15 |
| 2 | 82.5 | 15 |
| 3 | 36.6 | 15 |
| 4 | 16.7 | 15 |
| 5 | 5.3 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pachetti, M.; D’Amico, F.; Zupin, L.; Luppi, S.; Martinelli, M.; Crovella, S.; Ricci, G.; Pascolo, L. Strategies and Perspectives for UV Resonance Raman Applicability in Clinical Analyses of Human Sperm RNA. Int. J. Mol. Sci. 2021, 22, 13134. https://doi.org/10.3390/ijms222313134
Pachetti M, D’Amico F, Zupin L, Luppi S, Martinelli M, Crovella S, Ricci G, Pascolo L. Strategies and Perspectives for UV Resonance Raman Applicability in Clinical Analyses of Human Sperm RNA. International Journal of Molecular Sciences. 2021; 22(23):13134. https://doi.org/10.3390/ijms222313134
Chicago/Turabian StylePachetti, Maria, Francesco D’Amico, Luisa Zupin, Stefania Luppi, Monica Martinelli, Sergio Crovella, Giuseppe Ricci, and Lorella Pascolo. 2021. "Strategies and Perspectives for UV Resonance Raman Applicability in Clinical Analyses of Human Sperm RNA" International Journal of Molecular Sciences 22, no. 23: 13134. https://doi.org/10.3390/ijms222313134
APA StylePachetti, M., D’Amico, F., Zupin, L., Luppi, S., Martinelli, M., Crovella, S., Ricci, G., & Pascolo, L. (2021). Strategies and Perspectives for UV Resonance Raman Applicability in Clinical Analyses of Human Sperm RNA. International Journal of Molecular Sciences, 22(23), 13134. https://doi.org/10.3390/ijms222313134









