An FYVE-Domain-Containing Protein, PsFP1, Is Involved in Vegetative Growth, Oxidative Stress Response and Virulence of Phytophthora sojae
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis of PsFP1 and PsFP2
2.2. Expression Analysis of PsFP1 and PsFP2
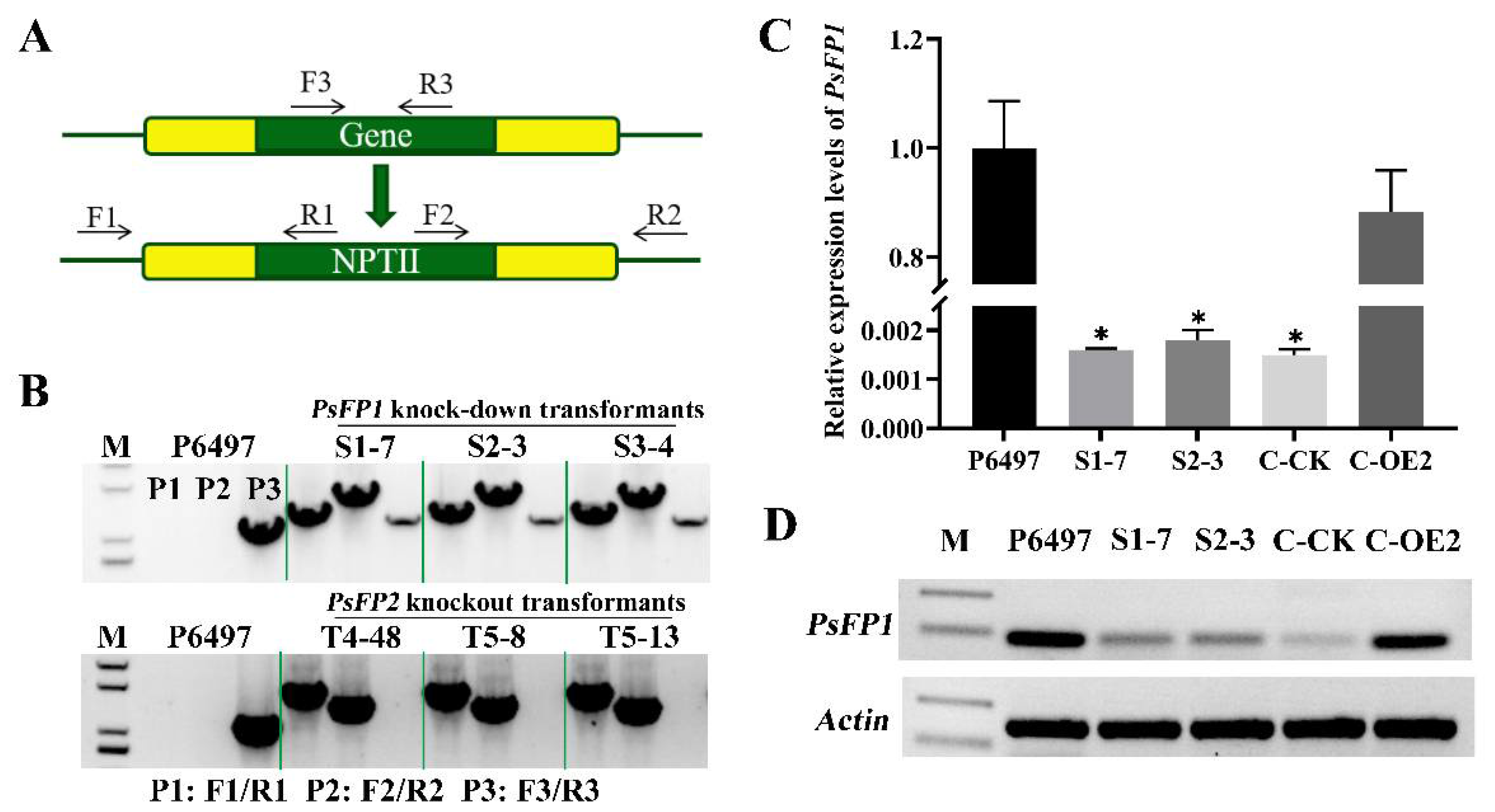
2.3. Generation of PsFP1 or PsFP2 Deletion Transformants
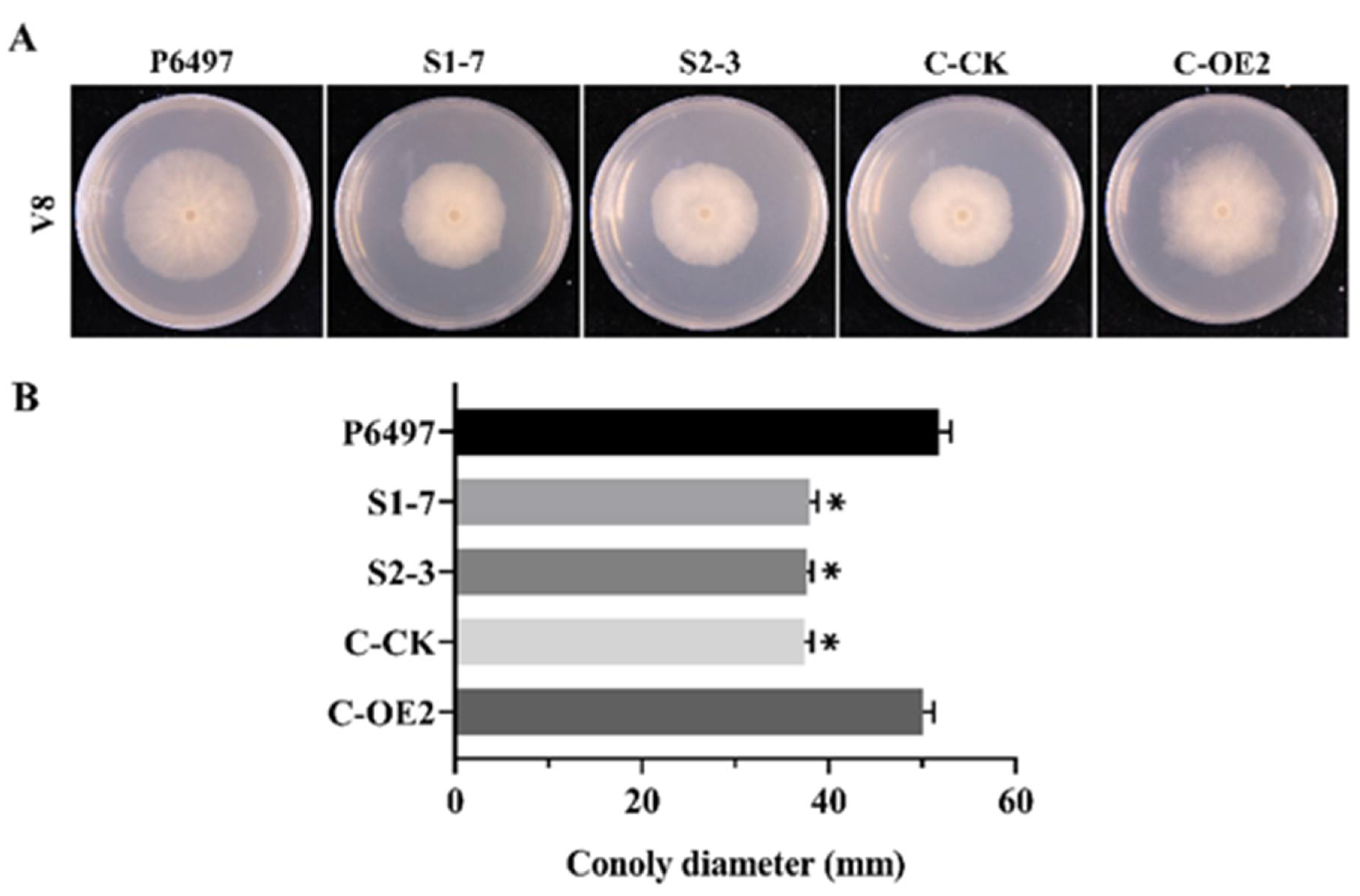
2.4. The Phenotypes of P. sojae Transformants
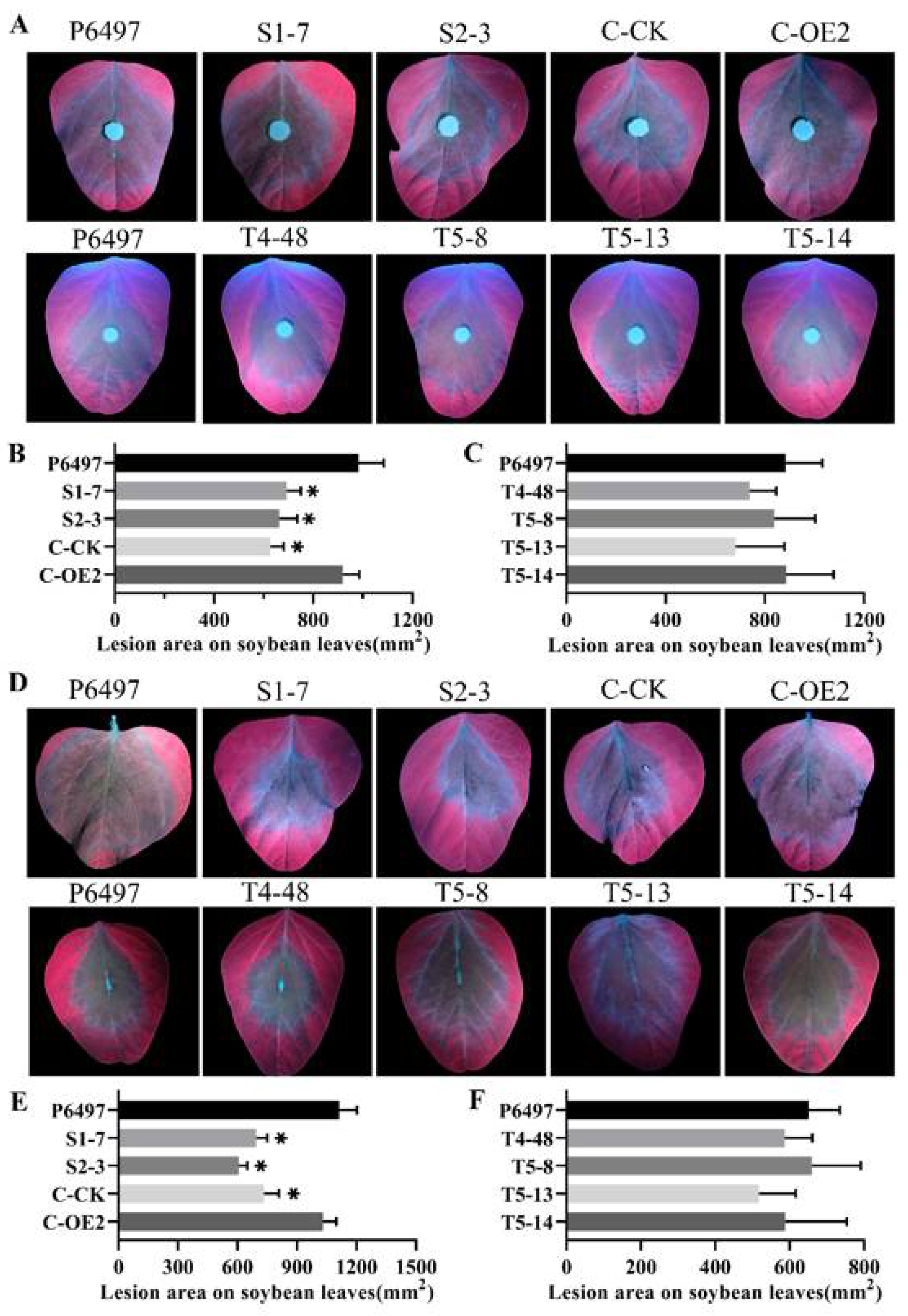
2.5. PsFP1 Is Required for Full Virulence of P. sojae
2.6. PsFP1 Is Important for P. sojae to Manage with Oxidative Stress
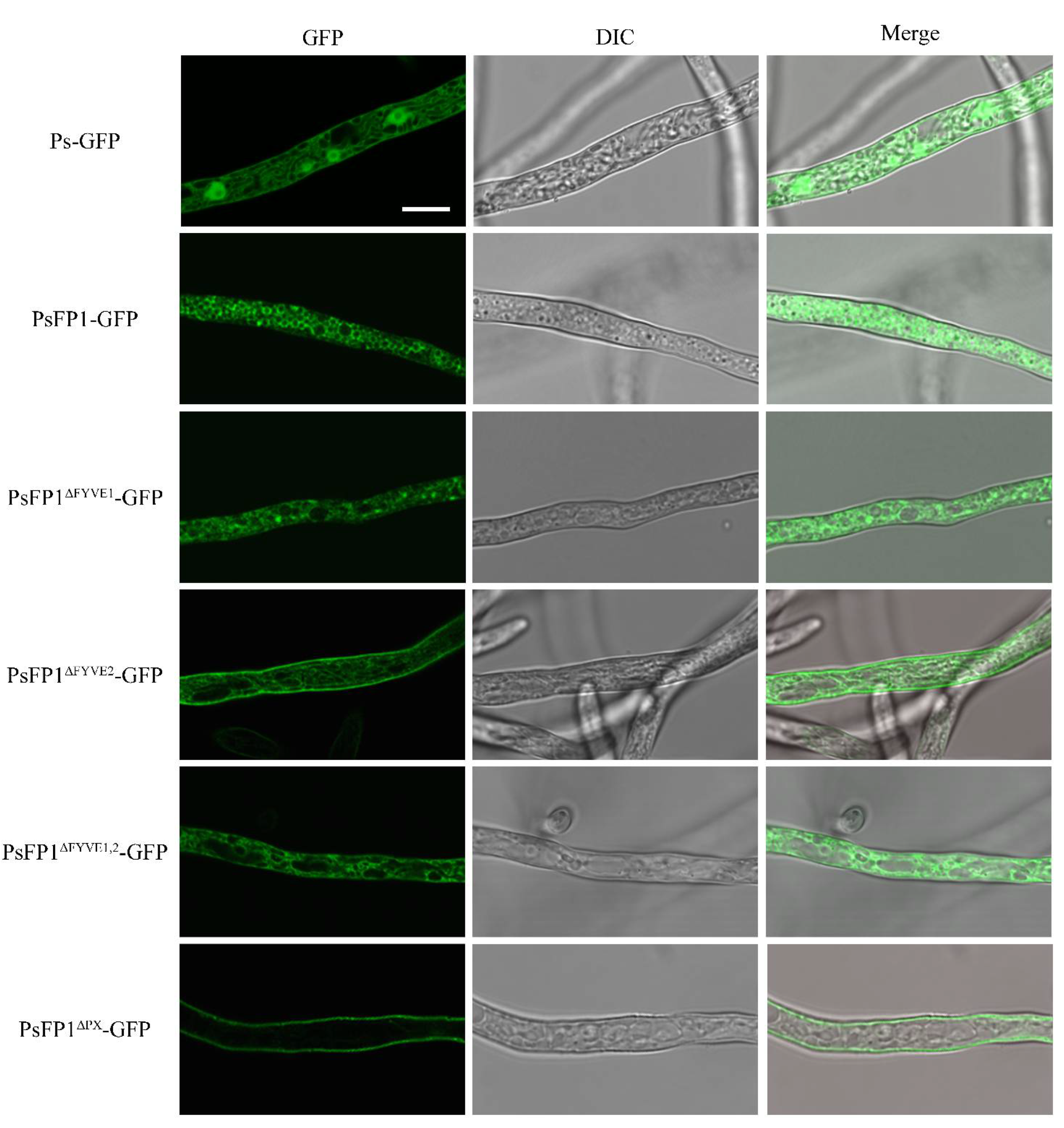
2.7. Subcellular Localization of PsFP1 is Determined by the FYVE and PX Domains
3. Discussion
4. Materials and Methods
4.1. P. sojae and Plant Cultivation
4.2. Construction of Plasmids
4.3. P. sojae Transformation
4.4. Phenotype Analysis of Transformants and P. sojae
4.5. Quantitative Reverse Transcription Polymerase Chain Reaction Analysis
4.6. Measurement of Hydrogen Peroxide Content in Mycelia
4.7. Determination of Superoxide Dismutase (SOD) Activity and Catalase (CAT) Activity
5. Conclusion and Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stenmark, H.; Aasland, R.; Driscoll, P.C. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002, 513, 77–84. [Google Scholar] [CrossRef]
- Gaullier, J.M.; Simonsen, A.; D’Arrigo, A.; Bremnes, B.; Stenmark, H.; Aasland, R. FYVE fingers bind PtdIns(3)P. Nature 1998, 394, 432–433. [Google Scholar] [CrossRef]
- Fritzius, T.; Burkard, G.; Haas, E.; Heinrich, J.; Schweneker, M.; Bosse, M.; Zimmermann, S.; Frey, A.D.; Caelers, A.; Bachmann, A.S.; et al. A WD-FYVE protein binds to the kinases Akt and PKCζ/λ. Biochem. J. 2006, 399, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H.; Aasland, R.; Toh, B.H.; D’Arrigo, A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 1996, 271, 24048–24054. [Google Scholar] [CrossRef] [PubMed]
- Gaullier, J.M.; Simonsen, A.; Arrigo, A.D.; Bremnes, B.; Monaghan, P.; Metcalfe, N.B. A functional PtdIns(3)P-binding motif. Nature 1998, 394, 433–434. [Google Scholar]
- Gillooly, D.J.; Simonsen, A.; Stenmark, H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001, 355, 249–258. [Google Scholar] [CrossRef]
- Gaullier, J.M.; Ronning, E.; Gillooly, D.J.; Stenmark, H. Interaction of the EEA1 FYVE finger with phosphatidylinositol 3-phosphate and early endosomes. Role of conserved residues. J. Biol. Chem. 2000, 275, 24595–24600. [Google Scholar] [CrossRef]
- Burd, C.G.; Emr, S.D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 1998, 2, 157–162. [Google Scholar] [CrossRef]
- Falasca, M.; Maffucci, T. Rethinking phosphatidylinositol 3-monophosphate. Biochim. Biophys. Acta 2009, 1793, 1795–1803. [Google Scholar] [CrossRef]
- Dae, H.K.; Eu, Y.J.; Cheol, M.Y.; Kim, Y.W.; Kyeong, T.P.; Jing, B.J.; Soo, J.K.; Stenmark, H.; Hwang, I. Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 2001, 13, 287–301. [Google Scholar] [CrossRef]
- Kale, S.D.; Gu, B.; Capelluto, D.G.S.; Dou, D.; Feldman, E.; Rumore, A.; Arredondo, F.D.; Hanlon, R.; Fudal, I.; Rouxel, T.; et al. External Lipid PI3P Mediates Entry of Eukaryotic Pathogen Effectors into Plant and Animal Host Cells. Cell 2010, 142, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Kale, S.D.; Wang, X.; Jiang, R.H.Y.; Bruce, N.A.; Arredondo, F.D.; Zhang, X.; Tyler, B.M. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell 2008, 20, 1930–1947. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, W.; Hou, Y.; Wu, Y.; Li, H.; Dou, D. Phosphatidylinositol 3-phosphate, an essential lipid in Phytophthora sojae, enriches in the haustoria during infection. Australas. Plant Pathol. 2016, 45, 435–441. [Google Scholar] [CrossRef]
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef]
- Tyler, B.M. Phytophthora sojae: Root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 2007, 8, 1–8. [Google Scholar] [CrossRef]
- Guetta, D.; Langou, K.; Grunwald, D.; Klein, G.; Aubry, L. FYVE-dependent endosomal targeting of an arrestin-related protein in amoeba. PLoS ONE 2010, 5, 15249. [Google Scholar] [CrossRef]
- Wywial, E.; Singh, S.M. Identification and structural characterization of FYVE domain-containing proteins of Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 157. [Google Scholar] [CrossRef]
- Banerjee, S.; Basu, S.; Sarkar, S. Comparative genomics reveals selective distribution and domain organization of FYVE and PX domain proteins across eukaryotic lineages. BMC Genom. 2010, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Ellson, C.D.; Andrews, S.; Stephens, L.R.; Hawkins, P.T. The PX domain: A new phosphoinositide-binding module. J. Cell Sci. 2002, 115, 1099–1105. [Google Scholar] [CrossRef]
- Kanai, F.; Liu, H.; Field, S.J.; Akbary, H.; Matsuo, T.; Brown, G.E.; Cantley, L.C.; Yaffe, M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001, 3, 675–678. [Google Scholar] [CrossRef]
- Ago, T.; Takeya, R.; Hiroaki, H.; Kuribayashi, F.; Ito, T.; Kohda, D.; Sumimoto, H. The PX domain as a novel phosphoinositide-binding module. Biochem. Biophys. Res. Commun. 2001, 287, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hortsman, H.; Seet, L.; Wong, S.H.; Hong, W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with Ptdlns(3)P. Nat. Cell Biol. 2001, 3, 658–666. [Google Scholar] [CrossRef]
- Ellson, C.D.; Gobert-Gosse, S.; Anderson, K.E.; Davidson, K.; Erdjument-Bromage, H.; Tempst, P.; Thuring, J.W.; Cooper, M.A.; Lim, Z.Y.; Holmes, A.B.; et al. Ptdlns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat. Cell Biol. 2001, 3, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.L.; Sato, T.K.; De Beer, T.; Kutateladze, T.G.; Emr, S.D.; Overduin, M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001, 3, 1–6. [Google Scholar] [CrossRef]
- Yu, J.W.; Lemmon, M.A. All Phox Homology (PX) Domains from Saccharomyces cerevisiae Specifically Recognize Phosphatidylinositol 3-Phosphate. J. Biol. Chem. 2001, 276, 44179–44184. [Google Scholar] [CrossRef] [PubMed]
- Whitley, P.; Hinz, S.; Doughty, J. Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol. 2009, 151, 1812–1822. [Google Scholar] [CrossRef]
- Hirano, T.; Sato, M.H. Arabidopsis FAB1A/B is possibly involved in the recycling of auxin transporters. Plant Signal. Behav. 2011, 6, 583–585. [Google Scholar] [CrossRef]
- Leevers, S.J.; Vanhaesebroeck, B.; Waterfield, M.D. Signalling through phosphoinositide 3-kinases: The lipids take centre stage. Cell Biol. 1998, 11, 219–225. [Google Scholar] [CrossRef]
- Corvera, S.; Czech, M.P. Direct targets of phosphoinositide 3-kinase products in membrane traffic and signal transduction. Trends Cell Biol. 1998, 8, 442–446. [Google Scholar] [CrossRef]
- Rameh, L.E.; Cantley, L.C. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 1999, 274, 8347–8350. [Google Scholar] [CrossRef]
- Lorenzo, Ó.; Urbé, S.; Clague, M.J. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J. Cell Sci. 2005, 118, 2005–2012. [Google Scholar] [CrossRef]
- Fang, Y.; Cui, L.; Gu, B.; Arredondo, F.; Tyler, B.M. Efficient genome editing in the oomycete phytophthora sojae using CRISPR/Cas9. Curr. Protoc. Microbiol. 2017, 44, 21A.1.1–21A.1.26. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef]
- Wu, X.; Scott, D.A.; Kriz, A.J.; Chiu, A.C.; Hsu, P.D.; Dadon, D.B.; Cheng, A.W.; Trevino, A.E.; Konermann, S.; Chen, S.; et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014, 32, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Yang, S.J.; Qiao, H.H.; Wang, X.; Hu, Q.; et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, W.; Zhang, X.; Yu, L.; Dong, W.; Pan, S.; Gao, S.; Huang, X.; Zhang, L. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016, 13, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Llavona, P.; Pinelli, M.; Mutarelli, M.; Marwah, V.S.; Schimpf-Linzenbold, S.; Thaler, S.; Yoeruek, E.; Vetter, J.; Kohl, S.; Wissinger, B. Allelic expression imbalance in the human retinal transcriptome and potential impact on inherited retinal diseases. Genes 2017, 8, 283. [Google Scholar] [CrossRef] [PubMed]
- Neve, B.; Froguel, P.; Corset, L.; Vaillant, E.; Vatin, V.; Boutin, P. Rapid SNP allele frequency determination in genomic DNA pools by PyrosequencingTM. Biotechniques 2002, 32, 1138–1142. [Google Scholar] [CrossRef]
- Chen, L.; Nordlander, C.; Behboudi, A.; Olsson, B.; Levan, K.K. Deriving evolutionary tree models of the oncogenesis of endometrial adenocarcinoma. Int. J. Cancer 2007, 120, 292–296. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, A.; Zheng, W.; Xu, H.; Shang, W.; Zheng, H.; Zhang, D.; Zhou, J.; Lu, G.; Li, G.; et al. Endosomal sorting complexes required for transport-0 is essential for fungal development and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2016, 18, 3742–3757. [Google Scholar] [CrossRef]
- Zheng, H.; Zheng, W.; Wu, C.; Yang, J.; Xi, Y.; Xie, Q.; Zhao, X.; Deng, X.; Lu, G.; Li, G.; et al. Rab GTPases are essential for membrane trafficking-dependent growth and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4580–4599. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Li, L.; Miao, P.; Wu, C.; Chen, X.; Yuan, M.; Fang, T.; Norvienyeku, J.; Li, G.; Zheng, W.; et al. FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3378–3392. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, D.J.; Stefan, C.J.; Babst, M.; Emr, S.D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003, 162, 413–423. [Google Scholar] [CrossRef]
- Hirano, T.; Matsuzawa, T.; Takegawa, K.; Sato, M.H. Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in arabidopsis. Plant Physiol. 2011, 155, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wu, S.; Chen, Y.; Han, X.; Gu, Q.; Yin, Y.; Ma, Z. The microtubule end-binding protein FgEB1 regulates polar growth and fungicide sensitivity via different interactors in Fusarium graminearum. Environ. Microbiol. 2017, 19, 1791–1807. [Google Scholar] [CrossRef]
- Cheng, W.; Lin, M.; Qiu, M.; Kong, L.; Xu, Y.; Li, Y.; Wang, Y.; Ye, W.; Dong, S.; He, S.; et al. Chitin synthase is involved in vegetative growth, asexual reproduction and pathogenesis of Phytophthora capsici and Phytophthora sojae. Environ. Microbiol. 2019, 21, 4537–4547. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Van Der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef]
- Fischer, R.L.; Bennett, A.B. Role of cell wall hydrolases in fruit ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 675–703. [Google Scholar] [CrossRef]
- Xu, H.; Mendgen, K. Targeted cell wall degradation at the penetration site of cowpea rust basidiosporelings. Mol. Plant Microbe Interact. 1997, 10, 87–94. [Google Scholar] [CrossRef][Green Version]
- Tyler, B.M. Entering and breaking: Virulence effector proteins of oomycete plant pathogens. Cell. Microbiol. 2009, 11, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Apostol, I.; Heinstein, P.F.; Low, P.S. Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells. Plant Physiol. 1989, 90, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus magnaporthe oryzae. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Molina, L.; Kahmann, R. An Ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell 2007, 19, 2293–2309. [Google Scholar] [CrossRef]
- Tanaka, A.; Christensen, M.J.; Takemoto, D.; Park, P.; Scott, B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell 2006, 18, 1052–1066. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.J.; Wang, Z.Y.; Jones, M.A.; Smirnoff, N.; Talbot, N.J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef]
- Segmüller, N.; Kokkelink, L.; Giesbert, S.; Odinius, D.; Van Kan, J.; Tudzynski, P. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant Microbe Interact. 2008, 21, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ye, W.; Wu, J.; Xuan, M.; Li, Y.; Gao, J.; Wang, Y.; Wang, Y.; Dong, S.; Wang, Y. The MADS-box transcription factor PsMAD1 is involved in zoosporogenesis and pathogenesis of phytophthora sojae. Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Ye, W.; Wang, X.; Tao, K.; Lu, Y.; Dai, T.; Dong, S.; Dou, D.; Gijzen, M.; Wang, Y. Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant Microbe Interact. 2011, 24, 1530–1539. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Du, X.; Zhou, X.; Jin, D.; Miao, J.; Liu, X. An FYVE-Domain-Containing Protein, PsFP1, Is Involved in Vegetative Growth, Oxidative Stress Response and Virulence of Phytophthora sojae. Int. J. Mol. Sci. 2021, 22, 6601. https://doi.org/10.3390/ijms22126601
Zhang J, Du X, Zhou X, Jin D, Miao J, Liu X. An FYVE-Domain-Containing Protein, PsFP1, Is Involved in Vegetative Growth, Oxidative Stress Response and Virulence of Phytophthora sojae. International Journal of Molecular Sciences. 2021; 22(12):6601. https://doi.org/10.3390/ijms22126601
Chicago/Turabian StyleZhang, Jinhui, Xiaoran Du, Xin Zhou, Duo Jin, Jianqiang Miao, and Xili Liu. 2021. "An FYVE-Domain-Containing Protein, PsFP1, Is Involved in Vegetative Growth, Oxidative Stress Response and Virulence of Phytophthora sojae" International Journal of Molecular Sciences 22, no. 12: 6601. https://doi.org/10.3390/ijms22126601
APA StyleZhang, J., Du, X., Zhou, X., Jin, D., Miao, J., & Liu, X. (2021). An FYVE-Domain-Containing Protein, PsFP1, Is Involved in Vegetative Growth, Oxidative Stress Response and Virulence of Phytophthora sojae. International Journal of Molecular Sciences, 22(12), 6601. https://doi.org/10.3390/ijms22126601





