Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols
Abstract
1. Introduction
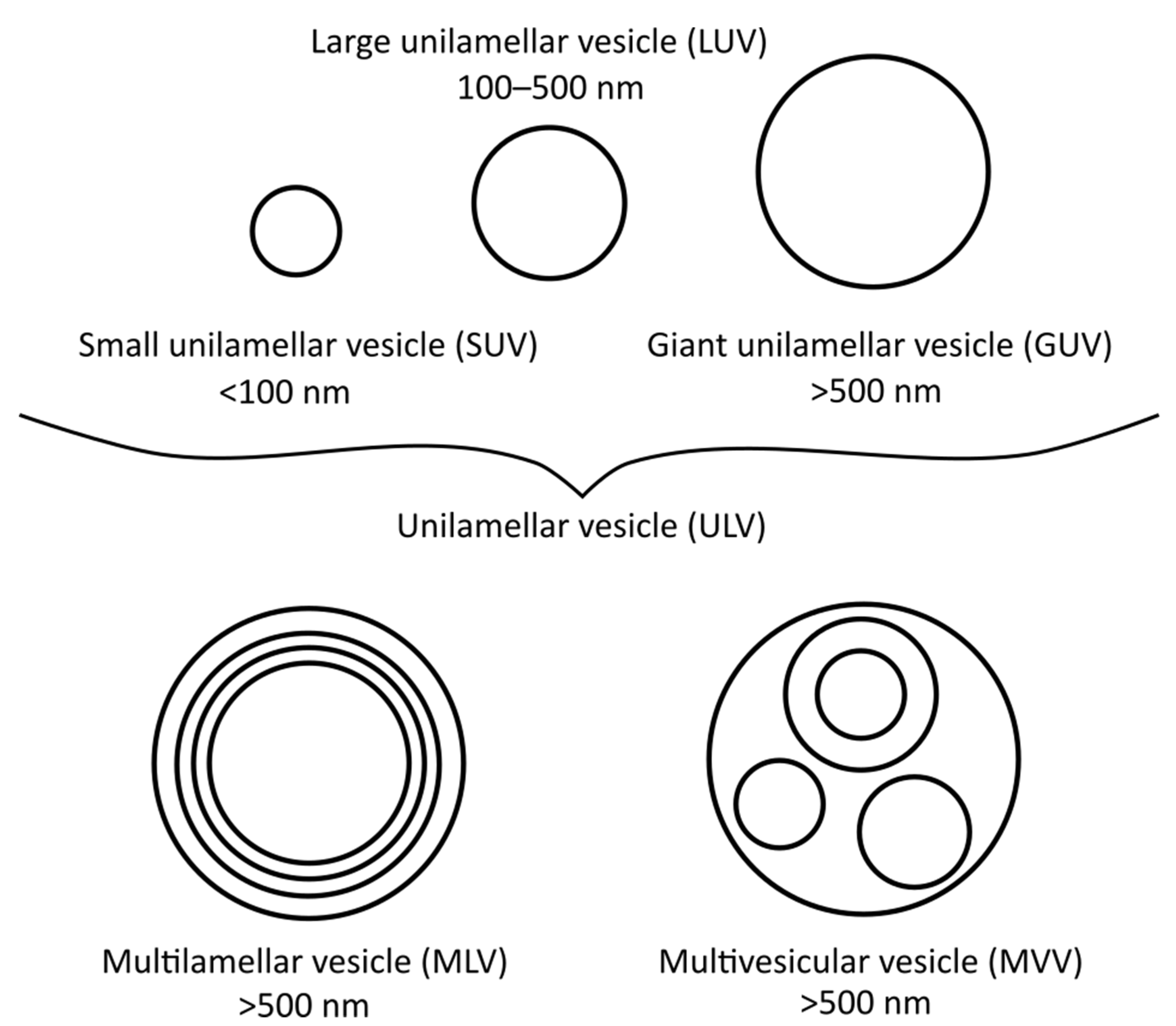
2. Preparation of Liposomes
2.1. Thin-Film Method
2.2. Proliposome Method
2.3. Injection Methods
2.3.1. Ethanol Injection
2.3.2. Ether Injection
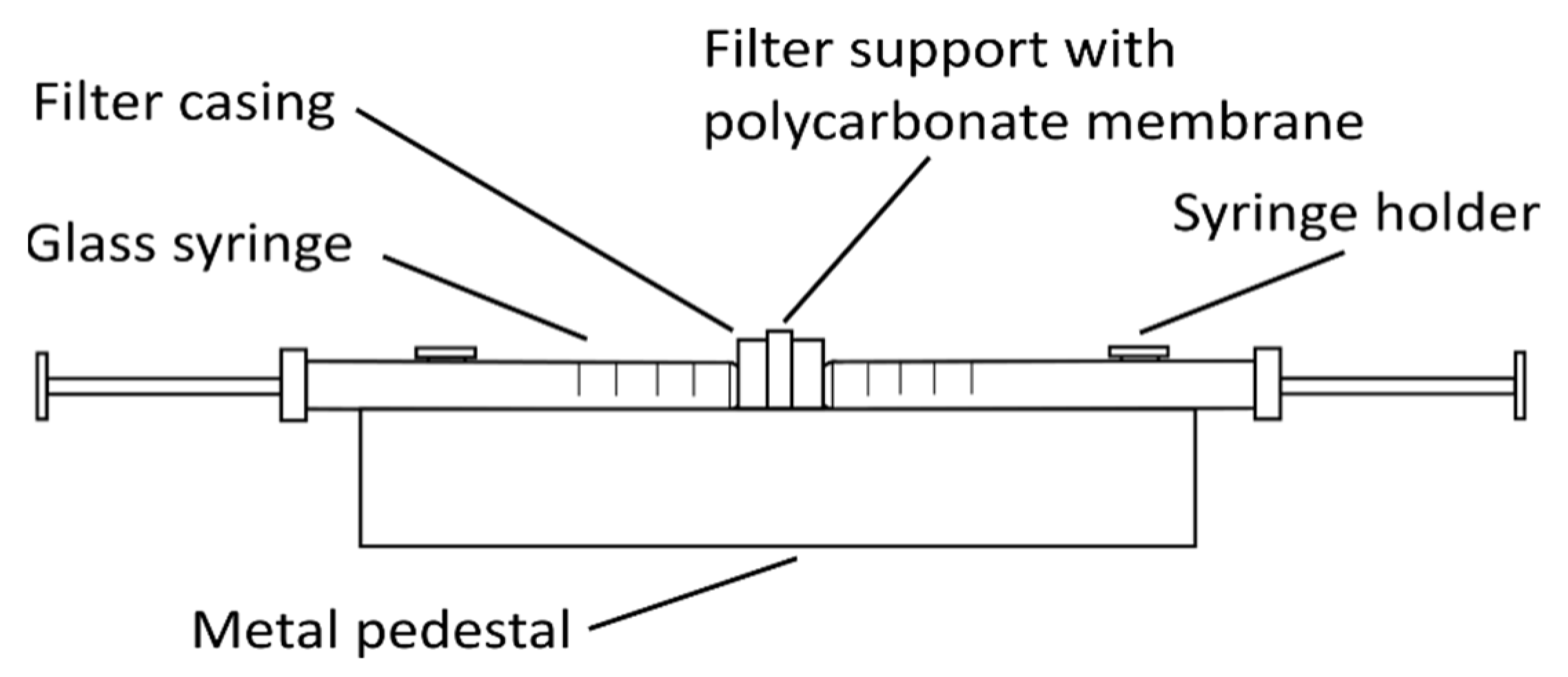
2.4. Emulsification Method
3. Spectroscopic Determination of the Position of the Compounds in the Bilayers
3.1. Polarisation and Anisotropy Measurements
3.2. Electron Paramagnetic Resonance Spectrometry for Determination of Compound’s Position
3.3. Nuclear Magnetic Resonance Spectrometry for Determination of Compound’s Position, Permeability and Its Effects on the Liposomes
4. Determination of the Effects of Compounds on the Liposomes
4.1. Determination of the Permeability of the Liposomal Bilayers: The Calcein Release Method
4.2. Spectroscopic Methods for Measuring Antioxidant Effects on the Model Lipid Membranes
4.2.1. Thiobarbituric Acid Reactive Substances
4.2.2. The BODIPY 581/591 C11 Method
4.2.3. Electron Paramagnetic Resonance Spectrometry for Determination of Antioxidant Activity
4.3. Surface Charge Measurements
- -
- Pipetting 20–50 µL of liposomal solution to the bottom of a specially designed zeta potential measuring cuvette, filled with filtered ultra-clean water (pore size, 0.45 µm)
- -
- Measuring the zeta-potential (in the case of aqueous liposomal suspensions, water is chosen as the solvent, the cuvette-type settings are selected according to the cuvette in use, and the number of measurements is defined by the machine software)
4.4. Size Measurements
4.4.1. Dynamic Light Scattering
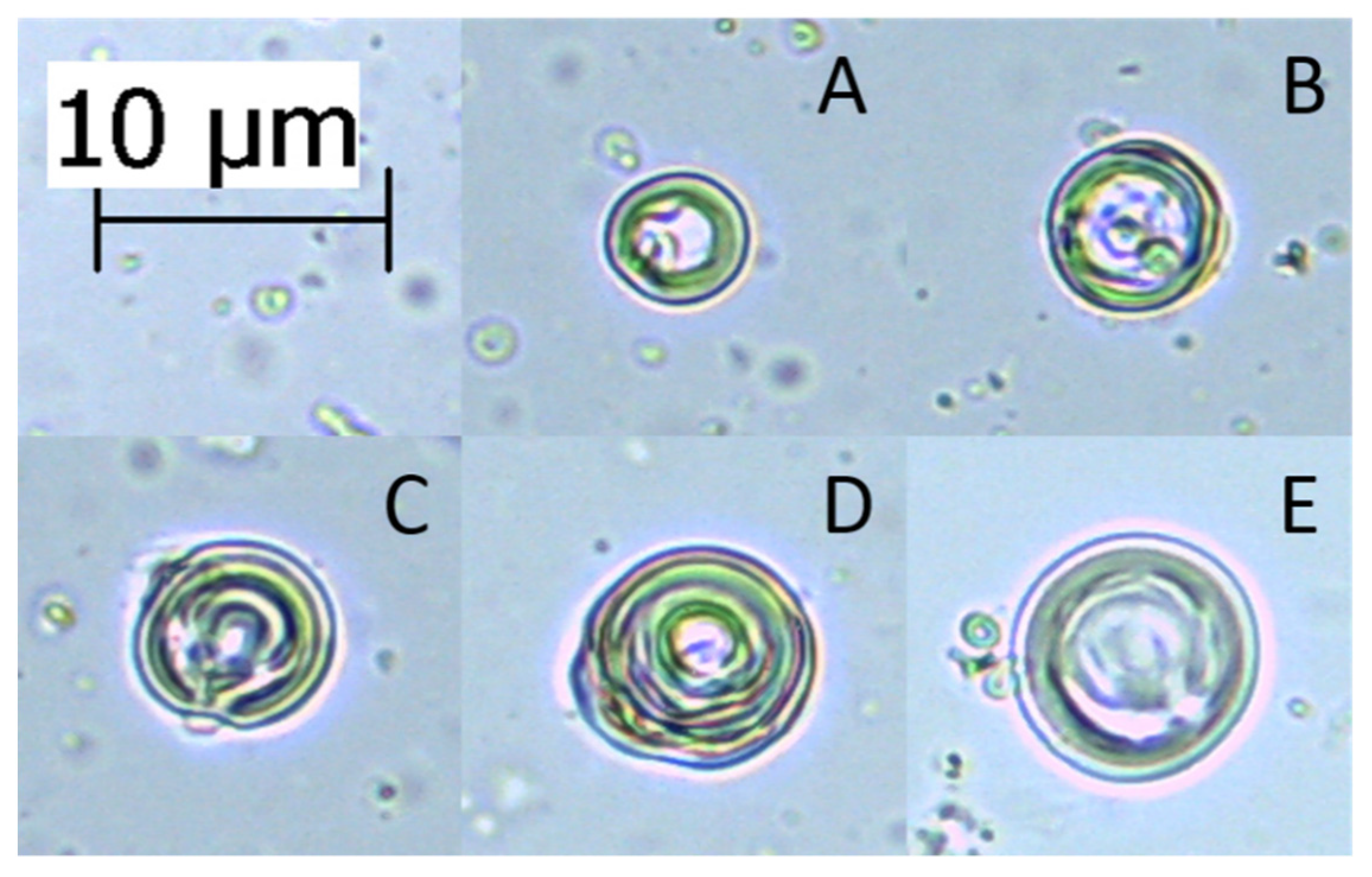
4.4.2. Light Microscopy
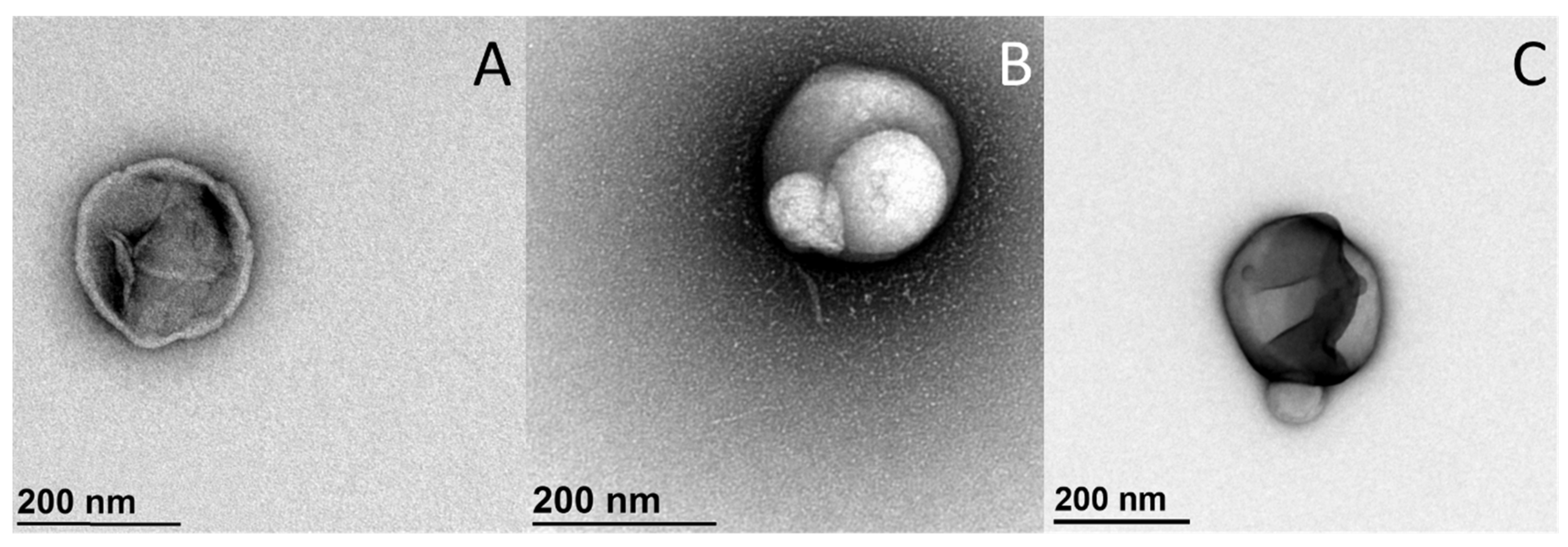
4.4.3. Transmission Electron Microscopy
4.5. Calorimetric Methods for Determination of the Phase Transition of the Membranes
4.5.1. Thermodynamic Profile of Thermotropic Phase Transition Using Differential Scanning Calorimetry
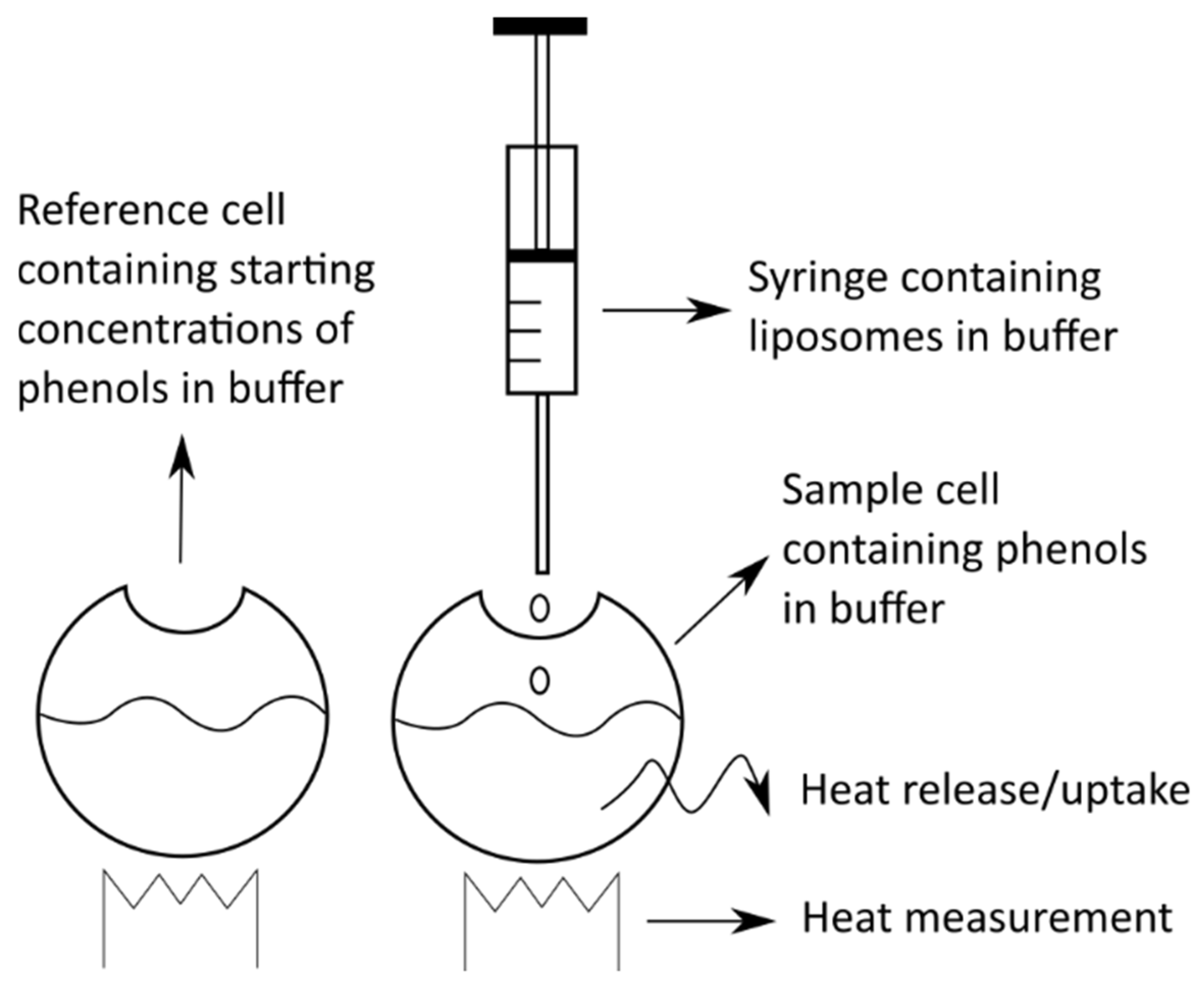
4.5.2. Isothermal Titration Calorimetry
5. Conclusions
Funding
Conflicts of Interest
References
- Lasic, D.D.; Papahadjopoulos, D. Medical Applications of Liposomes; Elsevier Science Publishers B. V.: Amsterdam, The Netherlands, 1998; p. 779. [Google Scholar]
- Bravo, L. Polyphenols: Chemistry, dietary sources, metabolism, and nutritional significance. Nutr. Rev. 1998, 56, 317–333. [Google Scholar] [CrossRef]
- Chebil, L.; Humeau, C.; Anthoni, J.; Dehez, F.; Engasser, J.-M.; Ghoul, M. Solubility of Flavonoids in Organic Solvents. J. Chem. Eng. Data 2007, 52, 1552–1556. [Google Scholar] [CrossRef]
- Scheidt, H.A.; Pampel, A.; Nissler, L.; Gebhardt, R.; Huster, D. Investigation of the membrane localization and distribution of flavonoids by high-resolution magic angle spinning NMR spectroscopy. BBA Biomembr. 2004, 1663, 97–107. [Google Scholar] [CrossRef]
- Tammela, P.; Laitinen, L.; Galkin, A.; Wennberg, T.; Heczko, R.; Vuorela, H.; Slotte, J.P.; Vuorela, P. Permeability characteristics and membrane affinity of flavonoids and alkyl gallates in Caco-2 cells and in phospholipid vesicles. Arch. Biochem. Biophys. 2004, 425, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Hendrich, A.B. Flavonoid-membrane interactions: Possible consequences for biological effects of some polyphenolic compounds. Acta Pharmacol. Sin. 2006, 27, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Amoros, M.; Simões, C.M.; Girre, L.; Sauvager, F.; Cormier, M. Synergistic effect of flavones and flavonols against herpes simplex virus type 1 in cell culture. Comparison with the antiviral activity of propolis. J. Nat. Prod. 1992, 55, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Siontorou, C.G.; Nikoleli, G.-P.; Nikolelis, D.P.; Karapetis, S.K. Artificial Lipid Membranes: Past, Present, and Future. Membranes 2017, 7, 38. [Google Scholar] [CrossRef]
- Negri, A.; Naponelli, V.; Rizzi, F.; Bettuzzi, S. Molecular Targets of Epigallocatechin—Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients 2018, 10, 1936. [Google Scholar] [CrossRef]
- Hoffman, J.F. Physiological characteristics of human red blood cell ghosts. J. Gen. Physiol. 1958, 42, 9–28. [Google Scholar] [CrossRef]
- Simons, T.J. The preparation of human red cell ghosts containing calcium buffers. J. Physiol. 1976, 256, 209–225. [Google Scholar] [CrossRef]
- Giess, F.; Friedrich, M.G.; Heberle, J.; Naumann, R.L.; Knoll, W. The protein-tethered lipid bilayer: A novel mimic of the biological membrane. Biophys. J. 2004, 87, 3213–3220. [Google Scholar] [CrossRef]
- Movileanu, L.; Neagoe, I.; Flonta, M.L. Interaction of the antioxidant flavonoid quercetin with planar lipid bilayers. Int. J. Pharmaceut. 2000, 205, 135–146. [Google Scholar] [CrossRef]
- Henn, F.A.; Thompson, T.E. Synthetic Lipid Bilayer Membranes. Annu. Rev. Biochem. 1969, 38, 241–262. [Google Scholar] [CrossRef]
- Peetla, C.; Stine, A.; Labhasetwar, V. Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery. Mol. Pharmaceut. 2009, 6, 1264–1276. [Google Scholar] [CrossRef]
- Bangham, A.D.; Hill, M.W.; Miller, N.G.A. Preparation and use of liposomes as models of biological membranes. In Methods in Membrane Biology; Korn, E.D., Ed.; Springer: Boston, MA, USA, 1974; Volume 1, pp. 1–68. [Google Scholar]
- Dua, J.S.; Rana, P.A.; Bhandari, D.K. Liposome: Methods of preparation and applications. Int. J. Pharm. Stud. Res. 2012, III, 14–20. [Google Scholar]
- Abram, V.; Berlec, B.; Ota, A.; Šentjurc, M.; Blatnik, P.; Ulrih, N.P. Effect of flavonoid structure on the fluidity of model lipid membranes. Food Chem. 2013, 139, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Weissig, V. Liposomes: Methods and Protocols: Pharmaceutical Nanocarriers; Humana Press (Springer Science+Business Media): New York, NY, USA, 2010; Volume 1, p. 564. [Google Scholar]
- Elhissi, A.M.; O’Neill, M.A.; Roberts, S.A.; Taylor, K.M. A calorimetric study of dimyristoylphosphatidylcholine phase transitions and steroid-liposome interactions for liposomes prepared by thin film and proliposome methods. Int. J. Pharmaceut. 2006, 320, 124–130. [Google Scholar] [CrossRef]
- Isailović, B.D.; Kostić, I.T.; Zvonar, A.; Đorđević, V.B.; Gašperlin, M.; Nedović, V.A.; Bugarski, B.M. Resveratrol loaded liposomes produced by different techniques. Innov. Food Sci. Emerg. 2013, 19, 181–189. [Google Scholar] [CrossRef]
- Jovanović, A.A.; Balanč, B.D.; Ota, A.; Ahlin Grabnar, P.; Djordjević, V.B.; Šavikin, K.P.; Bugarski, B.M.; Nedović, V.A.; Poklar Ulrih, N. Comparative Effects of Cholesterol and β-Sitosterol on the Liposome Membrane Characteristics. Eur. J. Lipid Sci. Tech. 2018, 120, 1800039. [Google Scholar] [CrossRef]
- Ota, A.; Abramovič, H.; Abram, V.; Poklar Ulrih, N. Interactions of p-coumaric, caffeic and ferulic acids and their styrenes with model lipid membranes. Food Chem. 2011, 125, 1256–1261. [Google Scholar] [CrossRef]
- Lasch, J.; Weissig, V.; Brandl, M. Preparation of liposomes. In Liposomes—A Practical Approach, 2nd ed.; Torchilin, V.P., Weissig, V., Eds.; Oxford University Press: New York, NY, USA, 2003; pp. 3–30. [Google Scholar]
- Lasic, D.D. Liposomes, from Physics to Application; Elsevier Science Publishers B. V.: Amsterdam, The Netherlands, 1993; p. 575. [Google Scholar]
- Ishikawa, H.; Shimoda, Y.; Matsumoto, K. Preparation of liposomal microcapsules by proliposome method with soybean lecithin. J. Fac. Agric. 2004, 49, 119–127. [Google Scholar]
- Elhissi, A.; Gill, H.; Ahmed, W.; Taylor, K. Vibrating-mesh nebulization of liposomes generated using an ethanol-based proliposome technology. J. Lipos. Res. 2011, 21, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Istenič, K.; Cerc Korošec, R.; Poklar Ulrih, N. Encapsulation of (-)-epigallocatechin gallate into liposomes and into alginate or chitosan microparticles reinforced with liposomes. J. Sci. Food Agric. 2016, 96, 4623–4632. [Google Scholar] [CrossRef] [PubMed]
- Perrett, S.; Golding, M.; Williams, W.P. A simple method for the preparation of liposomes for pharmaceutical applications: Characterization of the liposomes. J. Pharm. Pharmacol. 1991, 43, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Torchilin, V.P.; Weissig, V. Liposomes—A Practical Approach, 2nd ed.; Oxford University Press: Oxford, UK, 2003; p. 369. [Google Scholar]
- Jaafar-Maalej, C.; Diab, R.; Andrieu, V.; Elaissari, A.; Fessi, H. Ethanol injection method for hydrophilic and lipophilic drug-loaded liposome preparation. J. Lipos. Res. 2010, 20, 228–243. [Google Scholar] [CrossRef]
- Pons, M.; Foradada, M.; Estelrich, J. Liposomes obtained by the ethanol injection method. Int. J. Pharmaceut. 1993, 95, 51–56. [Google Scholar] [CrossRef]
- Charcosset, C.; Juban, A.; Valour, J.-P.; Urbaniak, S.; Fessi, H. Preparation of liposomes at large scale using the ethanol injection method: Effect of scale-up and injection devices. Chem. Eng. Res. Des. 2015, 94, 508–515. [Google Scholar] [CrossRef]
- Pham, H.L.; Shaw, P.N.; Davies, N.M. Preparation of immuno-stimulating complexes (ISCOMs) by ether injection. Int. J. Pharmaceut. 2006, 310, 196–202. [Google Scholar] [CrossRef]
- Chen, G.; Li, D.; Jin, Y.; Zhang, W.; Teng, L.; Bunt, C.; Wen, J. Deformable liposomes by reverse-phase evaporation method for an enhanced skin delivery of (+)-catechin. Drug Dev. Ind. Pharm. 2014, 40, 260–265. [Google Scholar] [CrossRef]
- Rojanapanthu, P.; Sarisuta, N.; Chaturon, K.; Kraisintu, K. Physicochemical properties of amphotericin B liposomes prepared by reverse-phase evaporation method. Drug Dev. Ind. Pharm. 2003, 29, 31–37. [Google Scholar] [CrossRef]
- Cortesi, R.; Esposito, E.; Gambarin, S.; Telloli, P.; Menegatti, E.; Nastruzzi, C. Preparation of liposomes by reverse-phase evaporation using alternative organic solvents. J. Microencapsul. 1999, 16, 251–256. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Science+Business Media, LLC: New York, NY, USA, 2006; p. 954. [Google Scholar]
- do Canto, A.M.T.M.; Robalo, J.R.; Santos, P.D.; Carvalho, A.J.P.; Ramalho, J.P.P.; Loura, L.M.S. Diphenylhexatriene membrane probes DPH and TMA-DPH: A comparative molecular dynamics simulation study. BBA Biomembr. 2016, 1858, 2647–2661. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H. Structure-dependent membrane interaction of flavonoids associated with their bioactivity. Food Chem. 2010, 120, 1089–1096. [Google Scholar] [CrossRef]
- Martinović, N.; Abramovič, H.; Poklar Ulrih, N. Inhibition of copper-induced lipid peroxidation by sinapic acid and its derivatives in correlation to their effect on the membrane structural properties. BBA Biomembr. 2019, 1861, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Reiner, G.N.; Delgado-Marín, L.; Olguín, N.; Sánchez-Redondo, S.; Sánchez-Borzone, M.; Rodríguez-Farré, E.; Suñol, C.; García, D.A. GABAergic pharmacological activity of propofol related compounds as possible enhancers of general anesthetics and interaction with membranes. Cell. Biochem. Biophys. 2013, 67, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Ulrih, N.P.; Maričić, M.; Ota, A.; Šentjurc, M.; Abram, V. Kaempferol and quercetin interactions with model lipid membranes. Food Res. Int. 2015, 71, 146–154. [Google Scholar] [CrossRef]
- Pottel, H.; van der Meer, W.; Herreman, W. Correlation between the order parameter and the steady-state fluorescence anisotropy of 1,6-diphenyl-1,3,5-hexatriene and an evaluation of membrane fluidity. BBA Biomembr. 1983, 730, 181–186. [Google Scholar] [CrossRef]
- Qin, P.Z.; Warncke, K. Methods in Enzymology: Electron Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions. Part A, 1st ed.; Academic Press: London, UK, 2015; Volume 563, p. 702. [Google Scholar]
- Balanč, B.D.; Ota, A.; Djordjević, V.B.; Šentjurc, M.; Nedović, V.A.; Bugarski, B.M.; Ulrih, N.P. Resveratrol-loaded liposomes: Interaction of resveratrol with phospholipids. Eur. J. Lipid Sci. Tech. 2015, 117, 1615–1626. [Google Scholar] [CrossRef]
- Tedeschi, A.; D’Errico, G.; Lauro, M.R.; Sansone, F.; Di Marino, S.; D’Ursi, A.M.; Aquino, R.P. Effect of flavonoids on the Aβ(25-35)-phospholipid bilayers interaction. Eur. J. Med. Chem. 2010, 45, 3998–4003. [Google Scholar] [CrossRef]
- Pawlikowska-Pawlęga, B.; Misiak, L.E.; Zarzyka, B.; Paduch, R.; Gawron, A.; Gruszecki, W.I. FTIR, 1H NMR and EPR spectroscopy studies on the interaction of flavone apigenin with dipalmitoylphosphatidylcholine liposomes. BBA Biomembr. 2013, 1828, 518–527. [Google Scholar] [CrossRef]
- Kates, M.; Manson, L.A. Biomembranes: Membrane Fluidity, 12th ed.; Plenum Press: New York, NY, USA, 1984. [Google Scholar]
- Skoog, D.A.; Holler, F.J.; Crouch, S.R. Principles of Instrumental Analysis, 6th ed.; Thomson Brooks/Cole: Belmont, CA, USA, 2007. [Google Scholar]
- Yu, X.; Chu, S.; Hagerman, A.E.; Lorigan, G.A. Probing the Interaction of Polyphenols with Lipid Bilayers by Solid-State NMR Spectroscopy. J. Agric. Food Chem. 2011, 59, 6783–6789. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.R.; De Kruyff, B.; Richards, R.E. Factors affecting the motion of the polar headgroup in phospholipid bilayers. A 31P NMR study of unsonicated phosphatidylcholine liposomes. BBA Biomembr. 1976, 426, 433–446. [Google Scholar] [CrossRef]
- Kajiya, K.; Kumazawa, S.; Naito, A.; Nakayama, T. Solid-state NMR analysis of the orientation and dynamics of epigallocatechin gallate, a green tea polyphenol, incorporated into lipid bilayers. Magn. Reson. Chem. 2008, 46, 174–177. [Google Scholar] [CrossRef]
- Uekusa, Y.; Kamihira, M.; Nakayama, T. Dynamic Behavior of Tea Catechins Interacting with Lipid Membranes As Determined by NMR Spectroscopy. J. Agric. Food Chem. 2007, 55, 9986–9992. [Google Scholar] [CrossRef]
- Gennis, R.B. Biomembranes. Molecular Structure and Function, 1st ed.; Springer-Verlag: New York, NY, USA, 1989; p. 533. [Google Scholar]
- Rumplecker, A.; Förster, S.; Zähres, M.; Mayer, C. Molecular exchange through vesicle membranes: A pulsed field gradient nuclear magnetic resonance study. J. Chem. Phys. 2004, 120, 8740–8747. [Google Scholar] [CrossRef]
- Cruciani, O.; Mannina, L.; Sobolev, A.P.; Cametti, C.; Segre, A. An improved NMR study of liposomes using 1-palmitoyl-2-oleoyl-sn-glycero-3-phospatidylcholine as model. Molecules 2006, 11, 334–344. [Google Scholar] [CrossRef]
- Katsu, T.; Imamura, T.; Komagoe, K.; Masuda, K.; Mizushima, T. Simultaneous Measurements of K+ and Calcein Release from Liposomes and the Determination of Pore Size Formed in a Membrane. Anal. Sci. 2007, 23, 517–522. [Google Scholar] [CrossRef]
- Shimanouchi, T.; Ishii, H.; Yoshimoto, N.; Umakoshi, H.; Kuboi, R. Calcein permeation across phosphatidylcholine bilayer membrane: Effects of membrane fluidity, liposome size, and immobilization. Colloid Surf. B 2009, 73, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-M.; Na, Y.-G.; Lee, H.-S.; Son, G.-H.; Jeon, S.-H.; Bang, K.-H.; Kim, S.-J.; Lee, H.-J.; Cho, C.-W. Improvement of cellular uptake of hydrophilic molecule, calcein, formulated by liposome. J. Pharm. Investig. 2018, 48, 595–601. [Google Scholar] [CrossRef]
- Tyagi, P.; Singh, M.; Kumari, H.; Kumari, A.; Mukhopadhyay, K. Bactericidal Activity of Curcumin I is Associated with Damaging of Bacterial Membrane. PLoS ONE 2015, 10, e0121313. [Google Scholar] [CrossRef] [PubMed]
- Yavlovich, A.; Singh, A.; Tarasov, S.; Capala, J.; Blumenthal, R.; Puri, A. Design of liposomes containing photopolymerizable phospholipids for triggered release of contents. J. Therm. Anal. Calorim. 2009, 98, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kuboi, R.; Shimanouchi, T.; Yoshimoto, M.; Umakoshi, H. Detection of protein conformation under stress conditions using liposomes as sensor materials. Sens. Mater. 2004, 16, 241–254. [Google Scholar]
- Maherani, B.; Arab-Tehrany, E.; Kheirolomoom, A.; Geny, D.; Linder, M. Calcein release behavior from liposomal bilayer; influence of physicochemical/mechanical/structural properties of lipids. Biochimie 2013, 95, 2018–2033. [Google Scholar] [CrossRef]
- Ghani, M.A.; Barril, C.; Bedgood, D.R.; Prenzler, P.D. Measurement of antioxidant activity with the thiobarbituric acid reactive substances assay. Food Chem. 2017, 230, 195–207. [Google Scholar] [CrossRef]
- Domínguez-Rebolledo, Á.; Martínez-Pastor, F.; Fernández-Santos, M.R.; Del Olmo, E.; Bisbal, A.; Ros-Santaella, J.L.; Garde, J.J. Comparison of the TBARS Assay and BODIPY C11 Probes for Assessing Lipid Peroxidation in Red Deer Spermatozoa. Reprod. Domest. Anim. 2010, 45, e360–e368. [Google Scholar] [CrossRef]
- Janero, D.R. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Radical Bio. Med. 1990, 9, 515–540. [Google Scholar] [CrossRef]
- Nogueira, A.O.d.M.; de Sousa, R.S.; Pereira, L.S.; Mallmann, C.; da Silva Ferreira, A.; Clementin, R.M.; de Lima, V.R. Physicochemical interactions among α-eleostearic acid-loaded liposomes applied to the development of drug delivery systems. J. Mol. Struct. 2018, 1154, 248–255. [Google Scholar] [CrossRef]
- Rael, L.T.; Thomas, G.W.; Craun, M.L.; Curtis, C.G.; Bar-Or, R.; Bar-Or, D. Lipid peroxidation and the thiobarbituric acid assay: Standardization of the assay when using saturated and unsaturated fatty acids. J. Biochem. Mol. Biol. 2004, 37, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Pelle, E.; Maes, D.; Padulo, G.A.; Kim, E.K.; Smith, W.P. An in vitro model to test relative antioxidant potential: Ultraviolet-induced lipid peroxidation in liposomes. Arch. Biochem. Biophys. 1990, 283, 234–240. [Google Scholar] [CrossRef]
- Marks, D.L.; Bittman, R.; Pagano, R.E. Use of Bodipy-labeled sphingolipid and cholesterol analogs to examine membrane microdomains in cells. Histochem. Cell Biol. 2008, 130, 819–832. [Google Scholar] [CrossRef]
- Thuwanut, P.; Axnér, E.; Johanisson, A.; Chatdarong, K. Detection of lipid peroxidation reaction in frozen-thawed epididymal cat spermatozoa using BODIPY(581/591) C11. Reprod. Domest. Anim. 2009, 44 (Suppl. 2), 373–376. [Google Scholar] [CrossRef] [PubMed]
- Jovanović, A.A.; Balanč, B.D.; Djordjević, V.B.; Ota, A.; Skrt, M.; Šavikin, K.P.; Bugarski, B.M.; Nedović, V.A.; Ulrih, N.P. Effect of gentisic acid on the structural-functional properties of liposomes incorporating β-sitosterol. Colloid Surf. B 2019, 183, 110422. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.C.; Valentim, I.B.; Silva, C.A.; Bechara, E.J.H.; Barros, M.P.d.; Mano, C.M.; Goulart, M.O.F. Total phenolic content and free radical scavenging activities of methanolic extract powders of tropical fruit residues. Food Chem. 2009, 115, 469–475. [Google Scholar] [CrossRef]
- Neunert, G.; Górnaś, P.; Dwiecki, K.; Siger, A.; Polewski, K. Synergistic and antagonistic effects between alpha-tocopherol and phenolic acids in liposome system: Spectroscopic study. Eur. Food Res. Technol. 2015, 241, 749–757. [Google Scholar] [CrossRef]
- Ota, A.; Šentjurc, M.; Bele, M.; Grabnar, P.A.; Ulrih, N.P. Impact of Carrier Systems on the Interactions of Coenzyme Q10 with Model Lipid Membranes. Food Biophys. 2016, 11, 60–70. [Google Scholar] [CrossRef]
- Frenzel, M.; Steffen-Heins, A. Impact of quercetin and fish oil encapsulation on bilayer membrane and oxidation stability of liposomes. Food Chem. 2015, 185, 48–57. [Google Scholar] [CrossRef]
- Nakarada, D.; Pejin, B.; Tommonaro, G.; Mojovic, M. Liposomal integration method for assessing antioxidative activity of water insoluble compounds towards biologically relevant free radicals: Example of avarol. J. Lipos. Res. 2020, 30, 218–226. [Google Scholar] [CrossRef]
- McLaughlin, S. Electrostatic Potentials at Membrane-Solution Interfaces. In Current Topics in Membranes and Transport; Bronner, F., Kleinzeller, A., Eds.; Academic Press: Cambridge, MA, USA, 1977; Volume 9, pp. 71–144. [Google Scholar]
- Ollila, F.; Halling, K.; Vuorela, P.; Vuorela, H.; Slotte, J.P. Characterization of flavonoid--biomembrane interactions. Arch. Biochem. Biophys. 2002, 399, 103–108. [Google Scholar] [CrossRef]
- van Dijk, C.; Driessen, A.J.; Recourt, K. The uncoupling efficiency and affinity of flavonoids for vesicles. Biochem. Pharmacol. 2000, 60, 1593–1600. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Zhang, T.; Wang, C.; Huang, Z.; Luo, X.; Deng, Y. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharm. Sci. 2015, 10, 81–98. [Google Scholar] [CrossRef]
- González-Ortega, R.; Šturm, L.; Skrt, M.; Di Mattia, C.D.; Pittia, P.; Poklar Ulrih, N. Liposomal Encapsulation of Oleuropein and an Olive Leaf Extract: Molecular Interactions, Antioxidant Effects and Applications in Model Food Systems. Food Biophys. 2020. [Google Scholar] [CrossRef]
- Forbes, N.; Hussain, M.T.; Briuglia, M.L.; Edwards, D.P.; Horst, J.H.t.; Szita, N.; Perrie, Y. Rapid and scale-independent microfluidic manufacture of liposomes entrapping protein incorporating in-line purification and at-line size monitoring. Int. J. Pharmaceut. 2019, 556, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Alund, S.J.; Hiorth, M.; Kjøniksen, A.-L.; Smistad, G. Studies on pectin coating of liposomes for drug delivery. Colloid Surf. B 2011, 88, 664–673. [Google Scholar] [CrossRef]
- Kavya, J.; Amsaveni, G.; Yasmin, H.; Murugesan, R.; Girigoswami, A. Gene expression profile induced by liposomal nanoformulation of anticancer agents: Insight onto cell death mechanism. Adv. Sci. Eng. Med. 2014, 6, 159–165. [Google Scholar] [CrossRef]
- Bhatia, A.; Kumar, R.; Katare, O.P. Tamoxifen in topical liposomes: Developement, characterisation and in-vitro evaluation. J. Pharm. Pharm. Sci. 2004, 7, 252–259. [Google Scholar] [PubMed]
- Baxa, U. Imaging of liposomes by transmission electron microscopy. In Characterisation of Nanoparticles Intended for Drug Delivery. Methods in Molecular Biology; McNeil, S., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1682, pp. 73–88. [Google Scholar]
- Sinha, R.; Joshi, A.; Joshi, U.J.; Srivastava, S.; Govil, G. Localization and interaction of hydroxyflavones with lipid bilayer model membranes: A study using DSC and multinuclear NMR. Eur. J. Med. Chem. 2014, 80, 285–294. [Google Scholar] [CrossRef]
- He, M.; Wu, T.; Pan, S.; Xu, X. Antimicrobial mechanism of flavonoids against Escherichia coli ATCC 25922 by model membrane study. Appl. Surf. Sci. 2014, 305, 515–521. [Google Scholar] [CrossRef]
- Bastos, M.; Aguas, A.C.P. Using DCS to Characterise Thermotropic Phase Transitions in Lipid Blayer Membranes; Aplication note (AN160630DSCLiposomeSamplePreparation); Malvern instruments Limited: Worcestershire, UK, 2016; p. 21. [Google Scholar]
- Gabbott, P. Principles and Applications of Thermal Analysis; Blackwell Publishing Ltd.: Oxford, UK, 2008; p. 480. [Google Scholar]
- Ladbury, J.E.; Chowdhry, B.Z. Biocalorimetry: Application of Calorimetry in the Biological Sciences; John Wiley & Sons Ltd.: New York, NY, USA, 1998; p. 364. [Google Scholar]
- Wu, G.; Lee, K.Y.C. Interaction of Poloxamers with Liposomes: An Isothermal Titration Calorimetry Study. J. Phys. Chem. B 2009, 113, 15522–15531. [Google Scholar] [CrossRef]
- Manrique-Moreno, M.; Howe, J.; Suwalsky, M.; Garidel, P.; Brandenburg, K. Physicochemical Interaction Study of Non-Steroidal Anti-Inflammatory Drugs with Dimyristoylphosphatidylethanolamine Liposomes. Lett. Drug Des. Discov. 2010, 7, 50–56. [Google Scholar] [CrossRef]
- Heerklotz, H.; Seelig, J. Titration calorimetry of surfactant–membrane partitioning and membrane solubilization. BBA Biomembr. 2000, 1508, 69–85. [Google Scholar] [CrossRef]
- Witzke, S.; Duelund, L.; Kongsted, J.; Petersen, M.; Mouritsen, O.G.; Khandelia, H. Inclusion of terpenoid plant extracts in lipid bilayers investigated by molecular dynamics simulations. J. Phys. Chem. B 2010, 114, 15825–15831. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šturm, L.; Poklar Ulrih, N. Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols. Int. J. Mol. Sci. 2021, 22, 6547. https://doi.org/10.3390/ijms22126547
Šturm L, Poklar Ulrih N. Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols. International Journal of Molecular Sciences. 2021; 22(12):6547. https://doi.org/10.3390/ijms22126547
Chicago/Turabian StyleŠturm, Luka, and Nataša Poklar Ulrih. 2021. "Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols" International Journal of Molecular Sciences 22, no. 12: 6547. https://doi.org/10.3390/ijms22126547
APA StyleŠturm, L., & Poklar Ulrih, N. (2021). Basic Methods for Preparation of Liposomes and Studying Their Interactions with Different Compounds, with the Emphasis on Polyphenols. International Journal of Molecular Sciences, 22(12), 6547. https://doi.org/10.3390/ijms22126547







