Culture Medium and Sex Drive Epigenetic Reprogramming in Preimplantation Bovine Embryos
Abstract
1. Introduction
2. Results
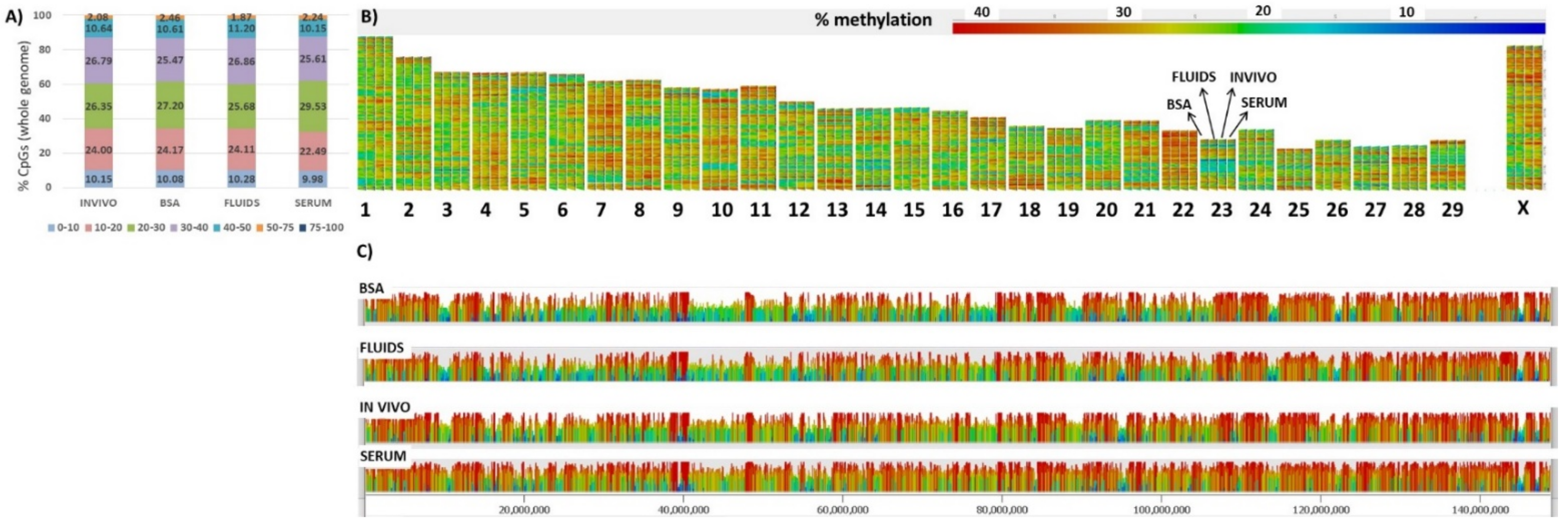
2.1. Global DNA Methylation Landscape in Single Bovine Blastocysts Obtained after In Vitro Production (IVP)
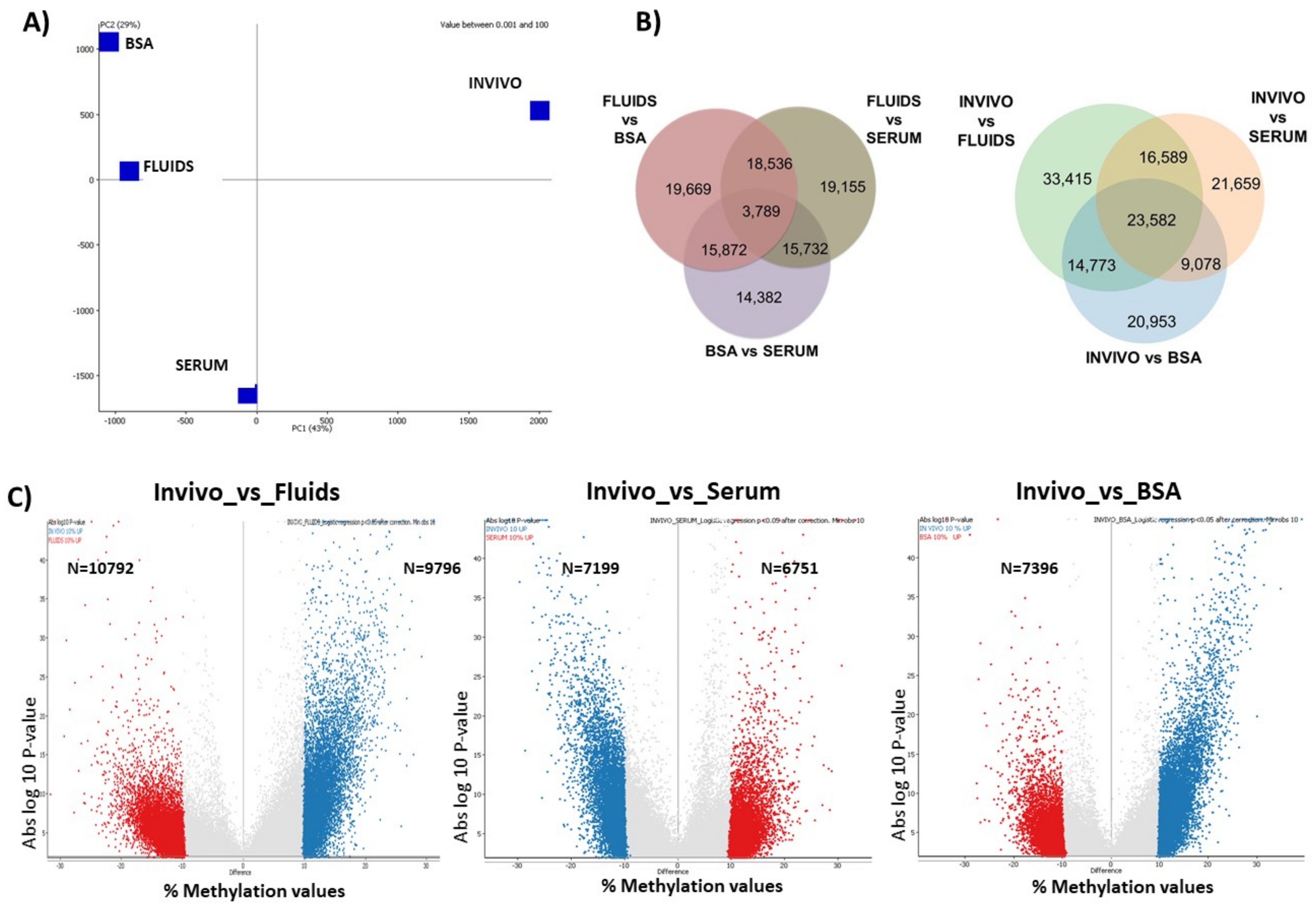
2.2. DNA Methylation Differences in Bovine Blastocysts under In Vitro vs. In Vivo Culture
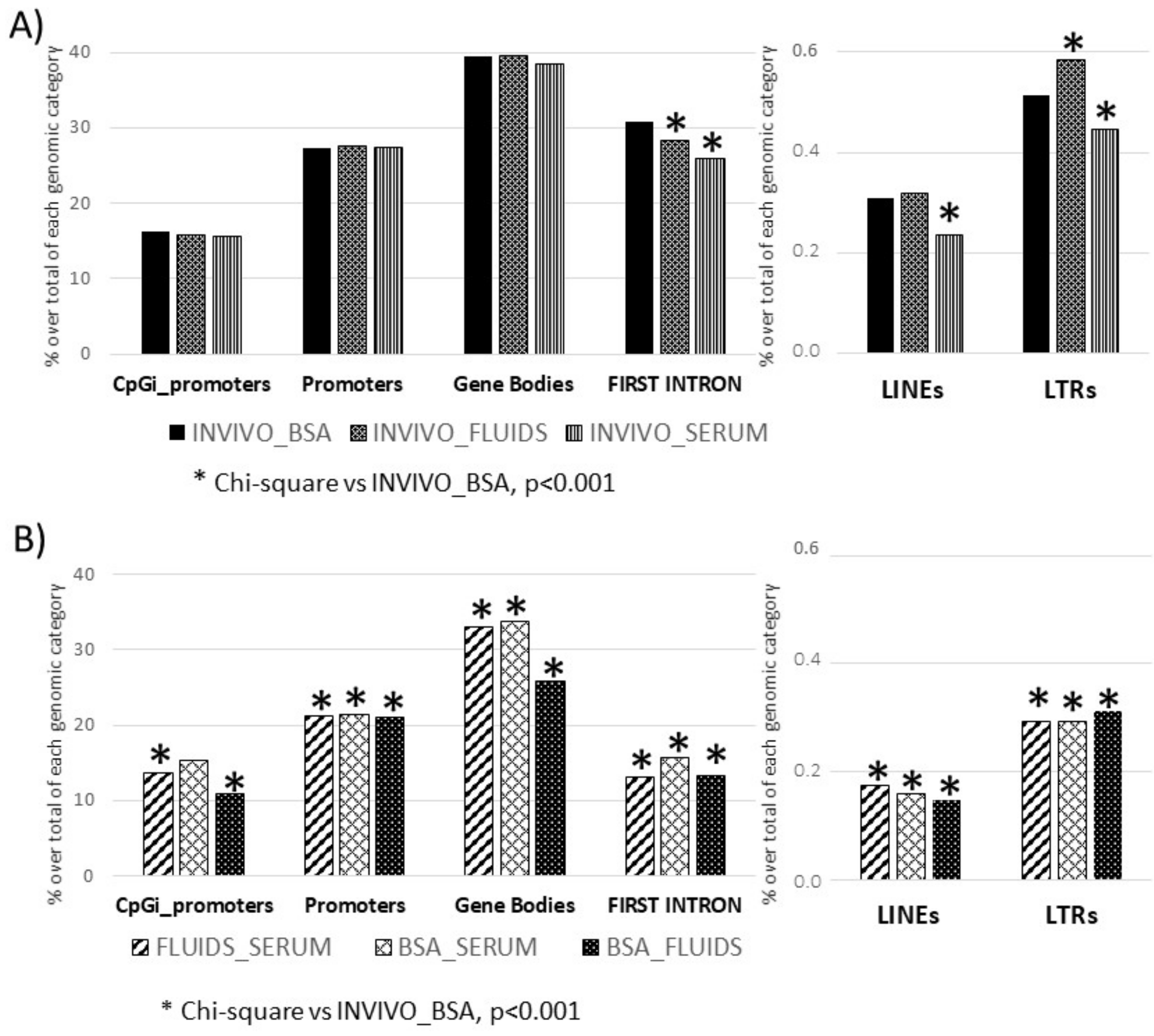
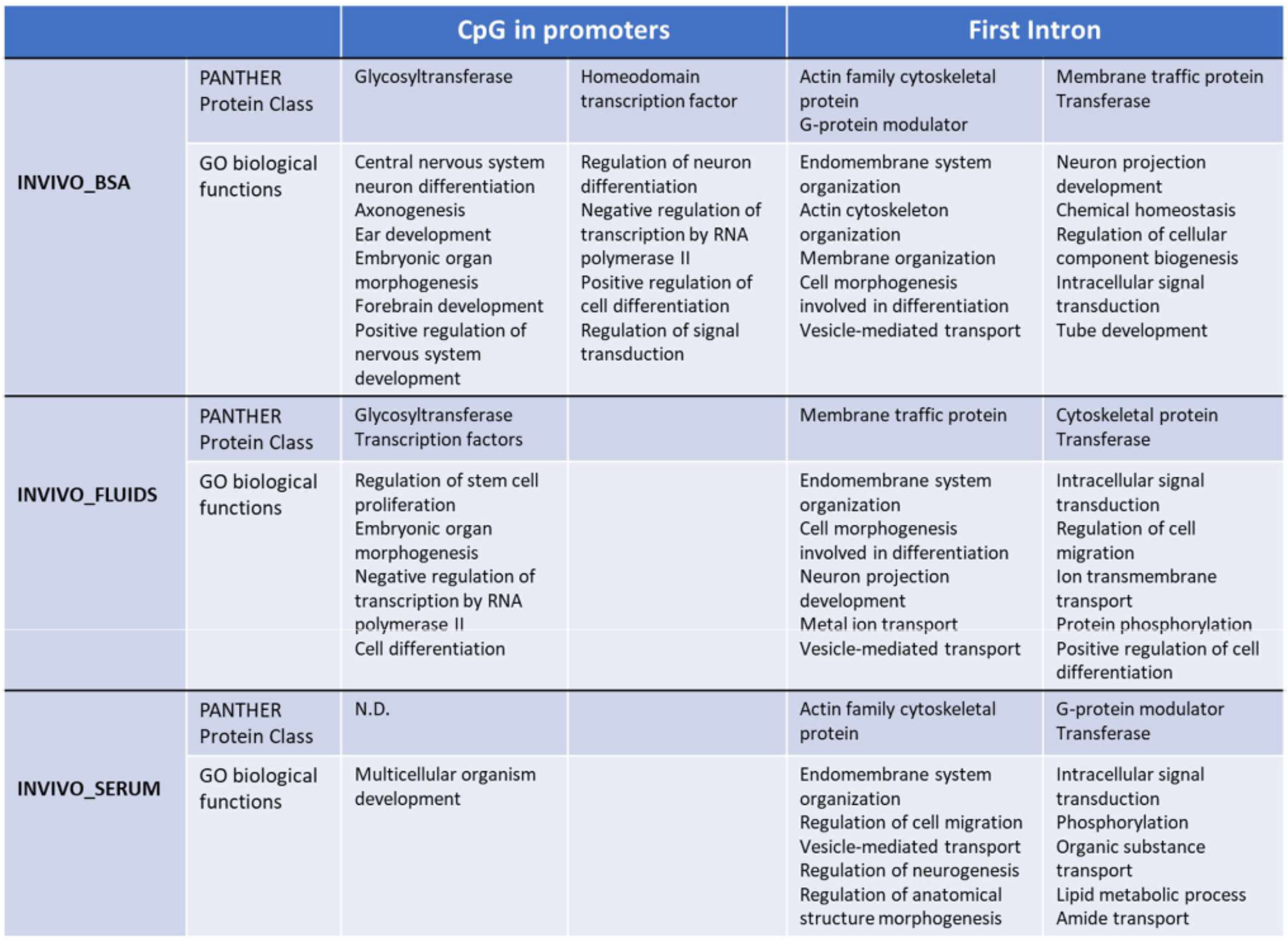
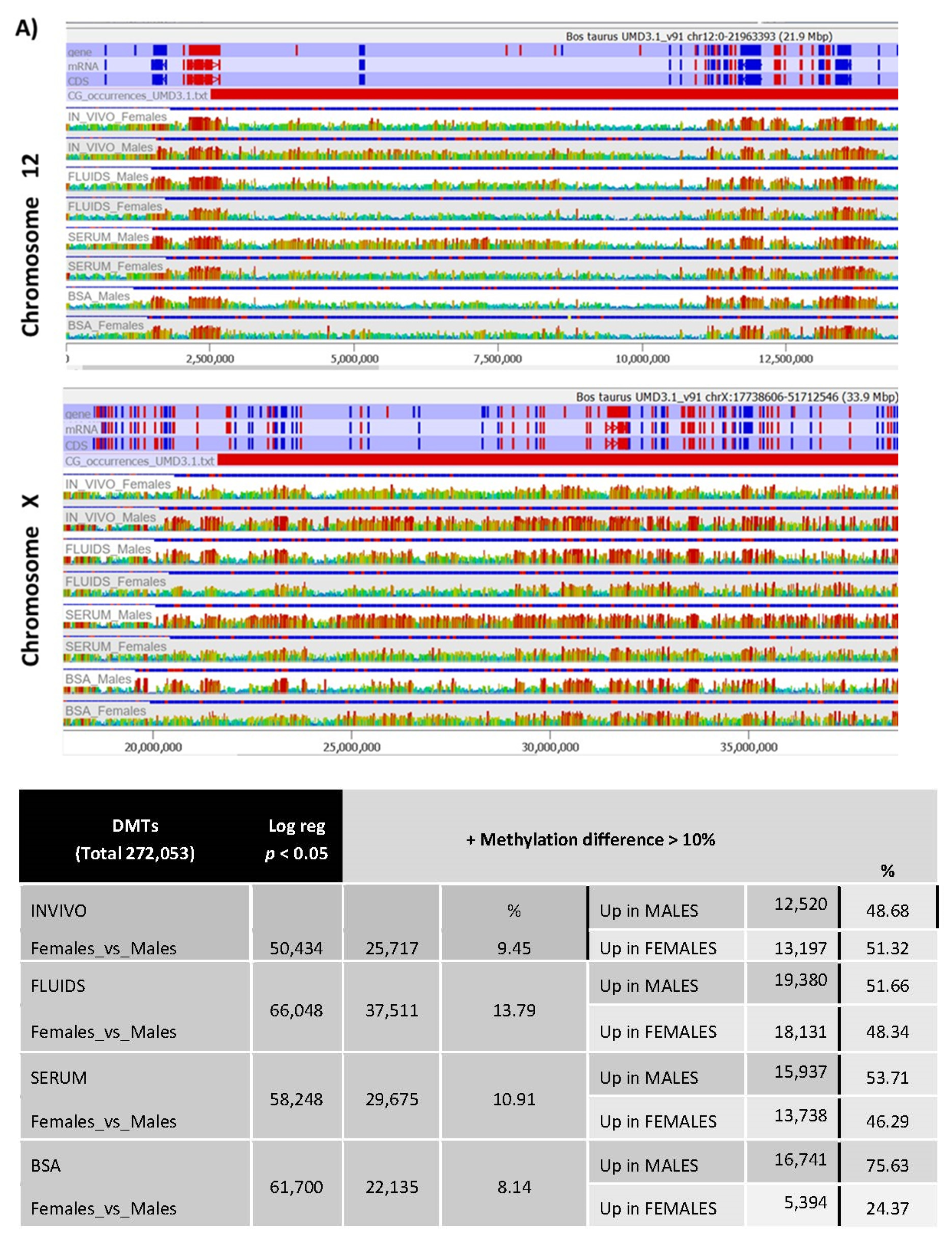
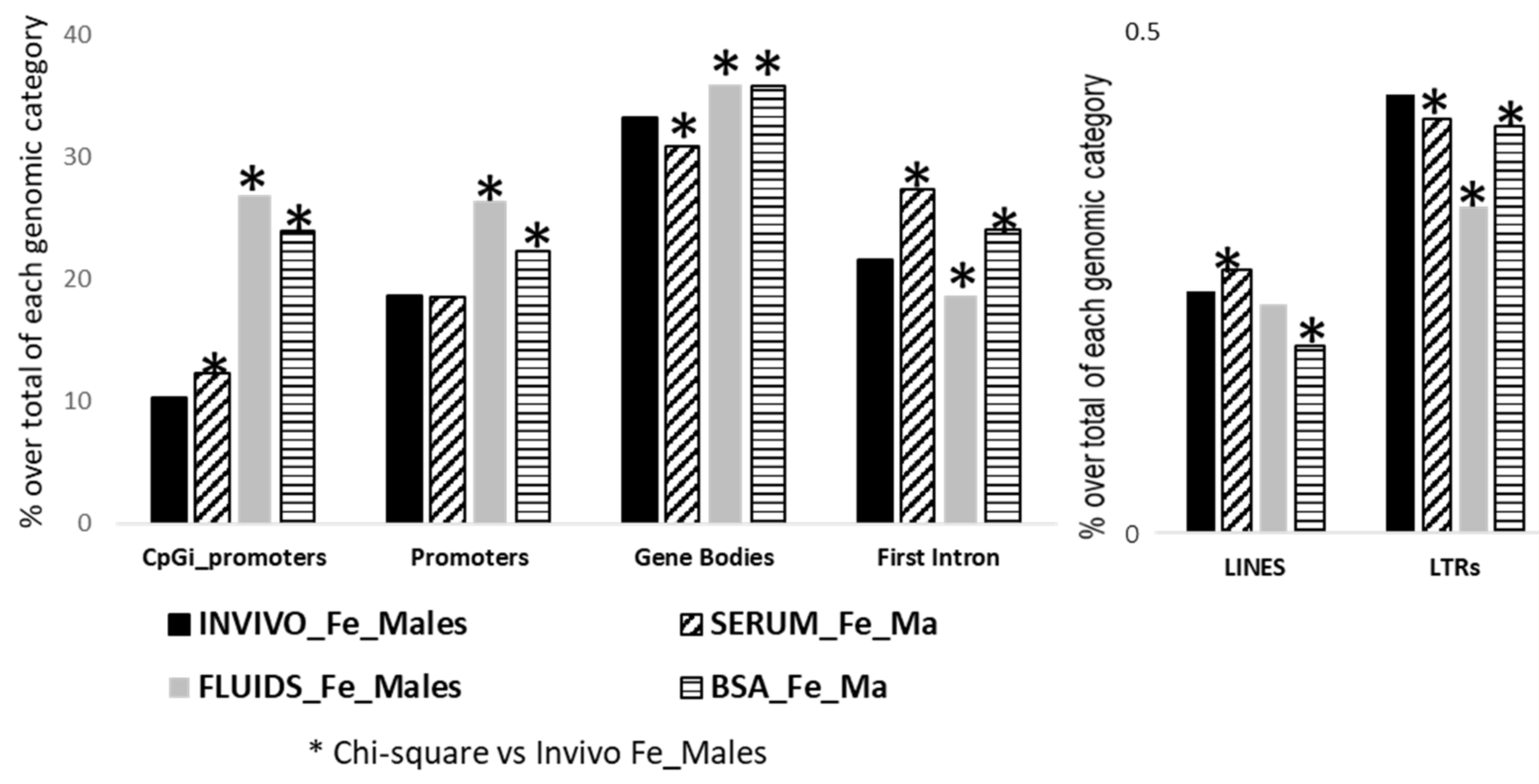
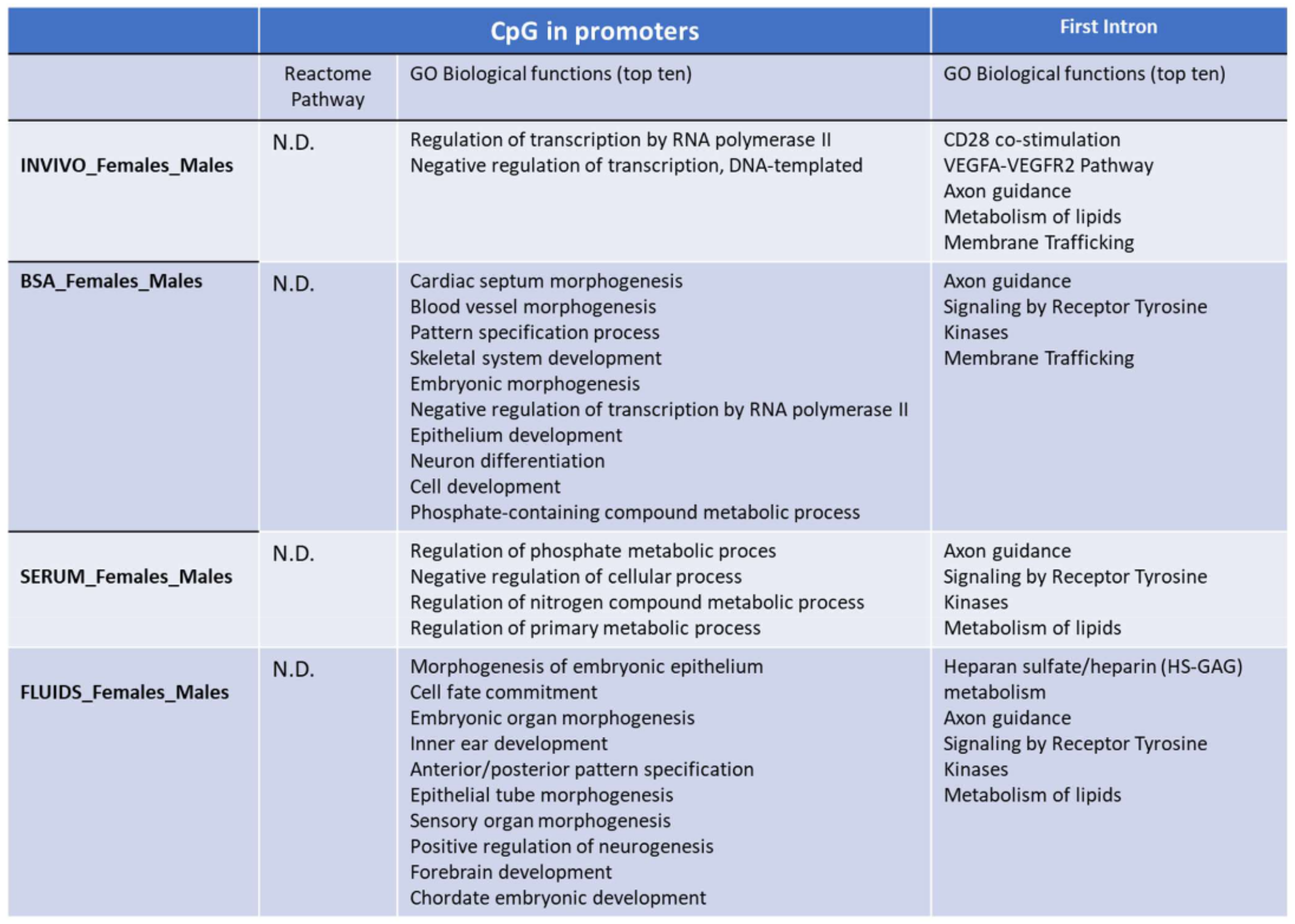
2.3. Targeted Analysis (Differentially Methylated Genomic Elements)
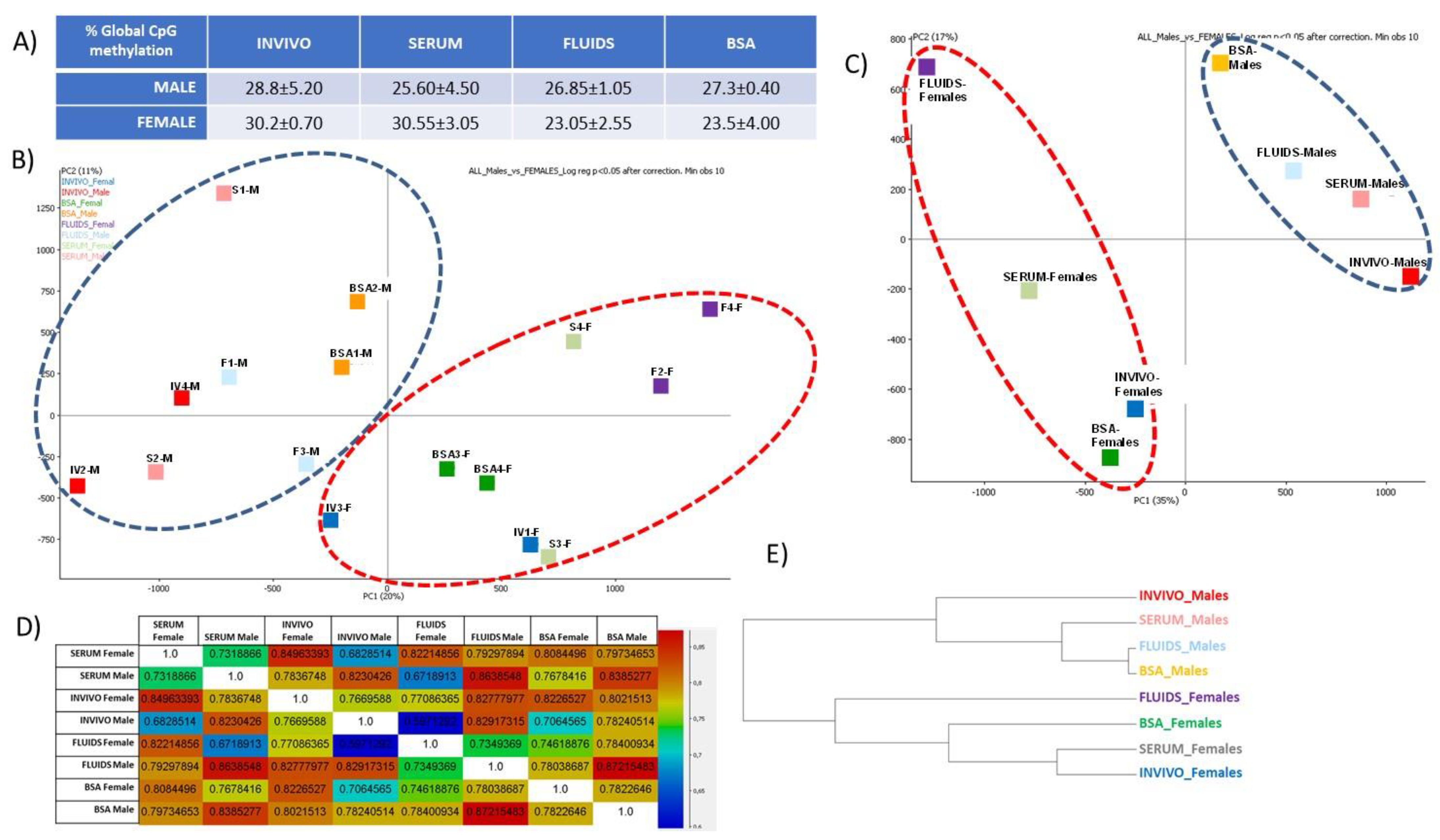
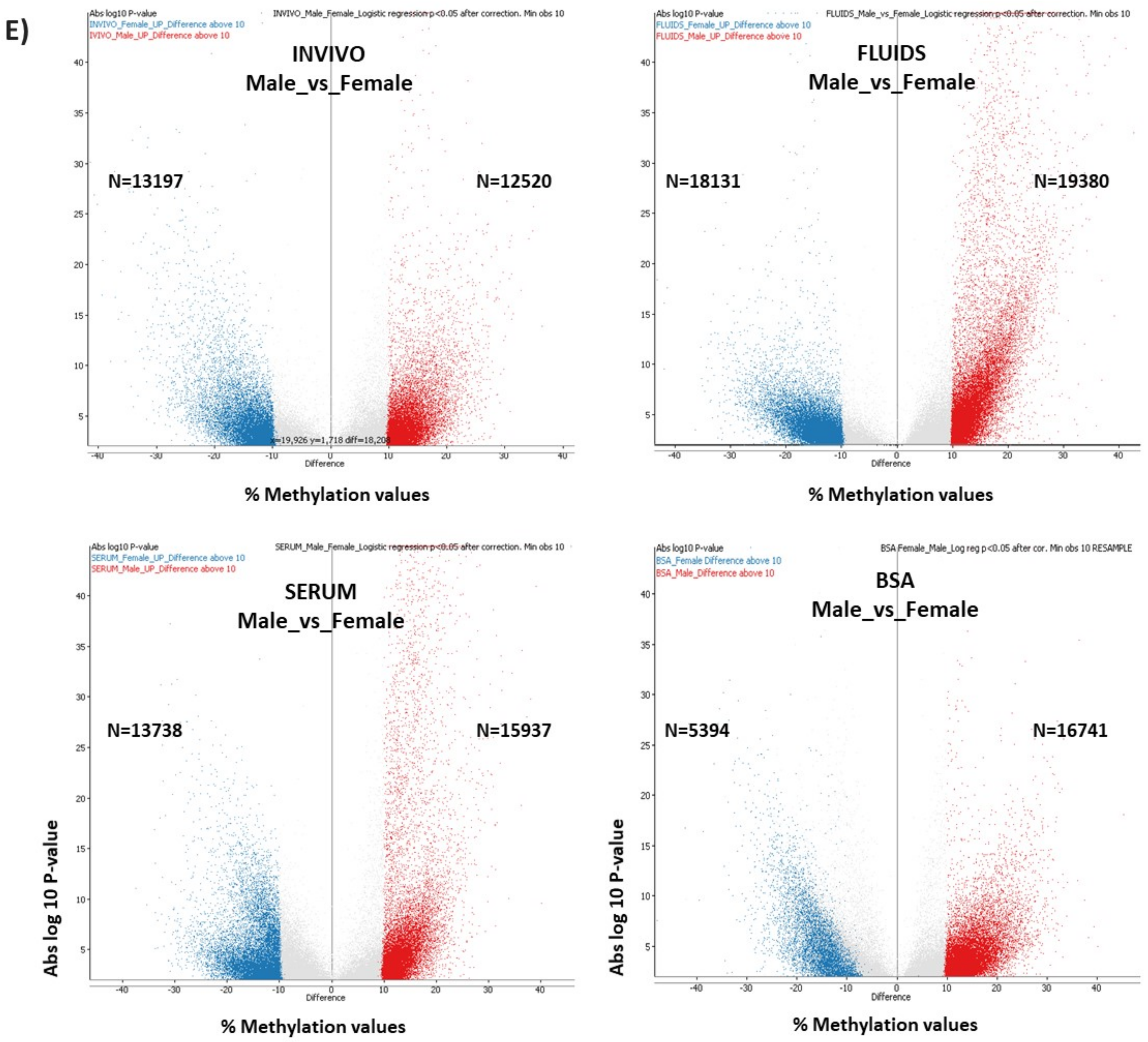
2.4. DNA Methylation Differences in Female versus Male Bovine In Vivo- or In Vitro-Derived Blastocysts
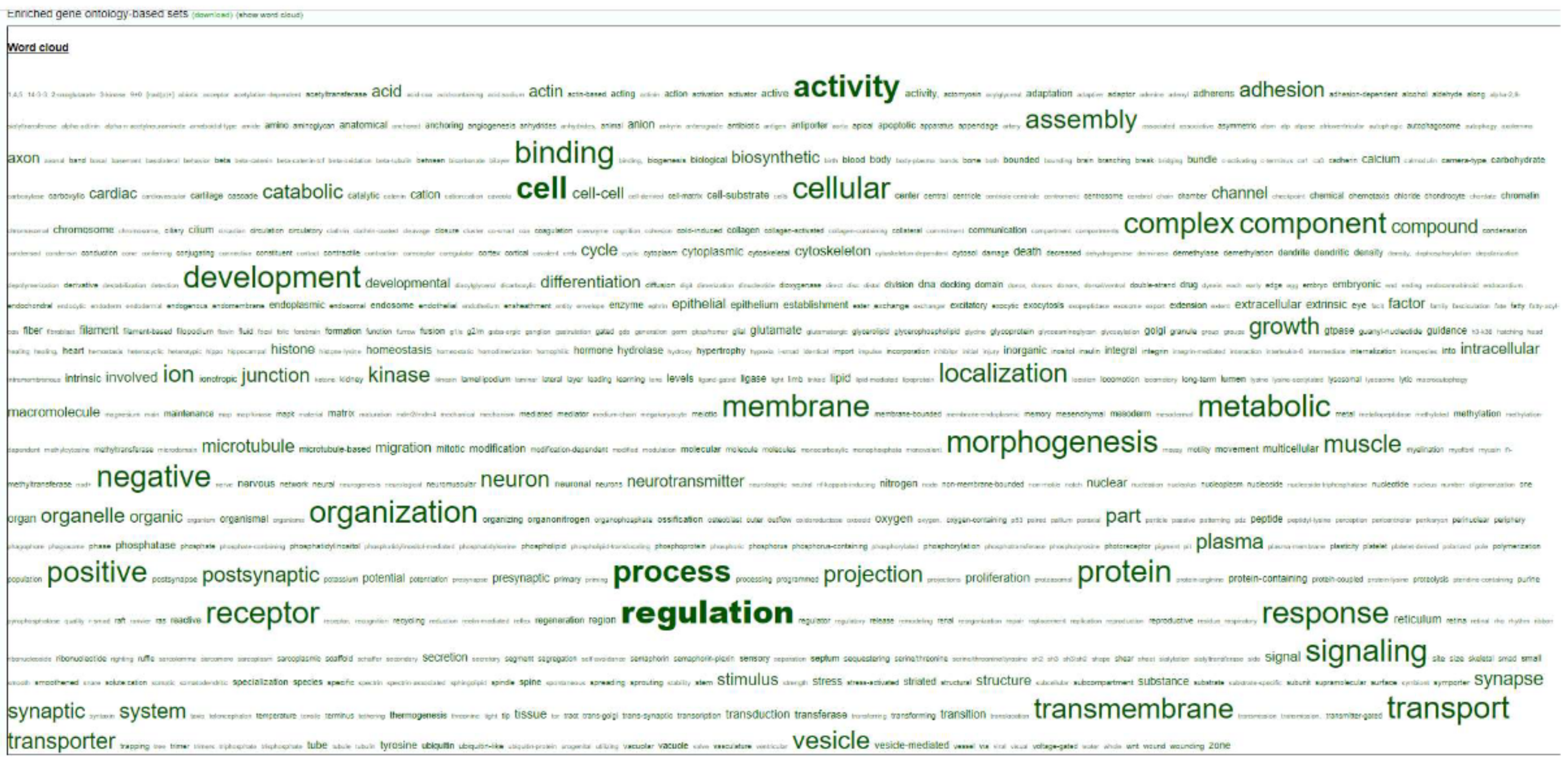
2.5. Unbiased Analysis
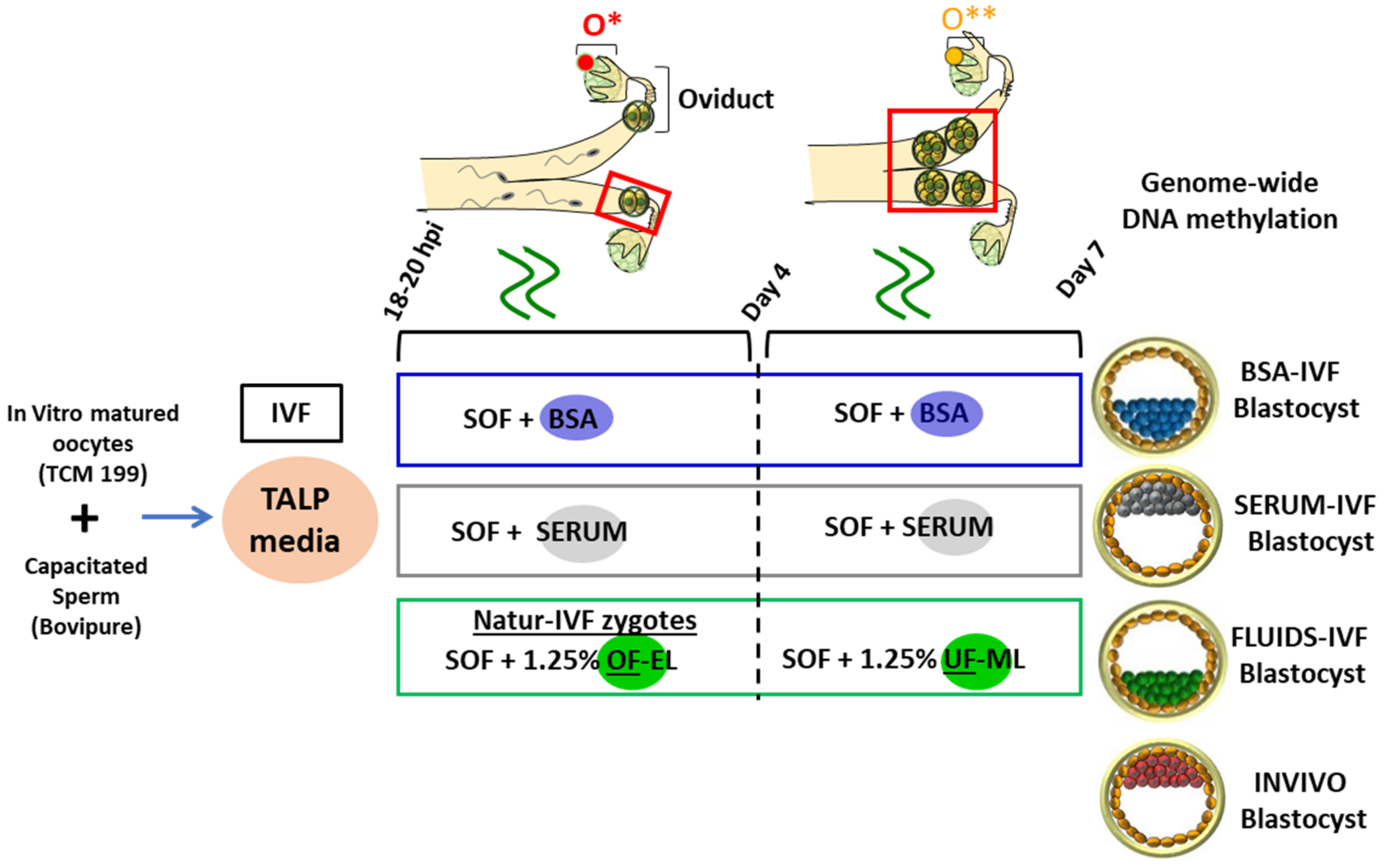
3. Materials and Methods
3.1. Oocyte Collection and IVP
3.2. In Vivo Collection of Embryos
3.3. DNA Library Preparation Based on Post-Bisulfite Adapter Tagging
3.4. Analysis of Methylation Data
3.5. Sexing of Embryos
3.6. Functional Analysis by PANTHER
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gluckman, P.D.; Hanson, M.A.; Beedle, A.S. Early life events and their consequences for later disease: A life history and evolutionary perspective. Am. J. Hum. Biol. 2007, 19, 1–19. [Google Scholar] [CrossRef]
- Baker, D.E.; Harrison, N.J.; Maltby, E.; Smith, K.; Moore, H.D.; Shaw, P.J.; Heath, P.R.; Holden, H.; Andrews, P.W. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat. Biotechnol. 2007, 25, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Boulet, S.L.; Kirby, R.S.; Reefhuis, J.; Zhang, Y.; Sunderam, S.; Cohen, B.; Bernson, D.; Copeland, G.; Bailey, M.A.; Jamieson, D.J.; et al. Assisted Reproductive Technology and Birth Defects Among Liveborn Infants in Florida, Massachusetts, and Michigan, 2000–2010. JAMA Pediatrics 2016, 170, e154934. [Google Scholar] [CrossRef] [PubMed]
- Kissin, D.M.; Jamieson, D.J.; Barfield, W.D. Monitoring health outcomes of assisted reproductive technology. N. Engl. J. Med. 2014, 371, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Choufani, S.; Turinsky, A.L.; Melamed, N.; Greenblatt, E.; Brudno, M.; Berard, A.; Fraser, W.D.; Weksberg, R.; Trasler, J.; Monnier, P. Impact of assisted reproduction, infertility, sex and paternal factors on the placental DNA methylome. Hum. Mol. Genet. 2019, 28, 372–385. [Google Scholar] [CrossRef]
- Chen, Z.; Hagen, D.E.; Elsik, C.G.; Ji, T.; Morris, C.J.; Moon, L.E.; Rivera, R.M. Characterization of global loss of imprinting in fetal overgrowth syndrome induced by assisted reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 4618–4623. [Google Scholar] [CrossRef]
- De Waal, E.; Mak, W.; Calhoun, S.; Stein, P.; Ord, T.; Krapp, C.; Coutifaris, C.; Schultz, R.M.; Bartolomei, M.S. In vitro culture increases the frequency of stochastic epigenetic errors at imprinted genes in placental tissues from mouse concepti produced through assisted reproductive technologies. Biol. Reprod. 2014, 90, 22. [Google Scholar] [CrossRef] [PubMed]
- De Waal, E.; Vrooman, L.A.; Fischer, E.; Ord, T.; Mainigi, M.A.; Coutifaris, C.; Schultz, R.M.; Bartolomei, M.S. The cumulative effect of assisted reproduction procedures on placental development and epigenetic perturbations in a mouse model. Hum. Mol. Genet. 2015, 24, 6975–6985. [Google Scholar] [CrossRef] [PubMed]
- Rivera, R.M.; Stein, P.; Weaver, J.R.; Mager, J.; Schultz, R.M.; Bartolomei, M.S. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum. Mol. Genet. 2008, 17, 1–14. [Google Scholar] [CrossRef]
- Fernandez-Gonzalez, R.; Moreira, P.; Bilbao, A.; Jimenez, A.; Perez-Crespo, M.; Ramirez, M.A.; Rodriguez De Fonseca, F.; Pintado, B.; Gutierrez-Adan, A. Long-term effect of in vitro culture of mouse embryos with serum on mRNA expression of imprinting genes, development, and behavior. Proc. Natl. Acad. Sci. USA 2004, 101, 5880–5885. [Google Scholar] [CrossRef]
- Robertson, S.A.; Chin, P.Y.; Femia, J.G.; Brown, H.M. Embryotoxic cytokines-Potential roles in embryo loss and fetal programming. J. Reprod. Immunol. 2018, 125, 80–88. [Google Scholar] [CrossRef]
- Ziebe, S.; Loft, A.; Povlsen, B.B.; Erb, K.; Agerholm, I.; Aasted, M.; Gabrielsen, A.; Hnida, C.; Zobel, D.P.; Munding, B.; et al. A randomized clinical trial to evaluate the effect of granulocyte-macrophage colony-stimulating factor (GM-CSF) in embryo culture medium for in vitro fertilization. Fertil. Steril. 2013, 99, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Sjoblom, C.; Wikland, M.; Robertson, S.A. Granulocyte-macrophage colony-stimulating factor promotes human blastocyst development in vitro. Hum. Reprod. 1999, 14, 3069–3076. [Google Scholar] [CrossRef]
- Sjoblom, C.; Roberts, C.T.; Wikland, M.; Robertson, S.A. Granulocyte-macrophage colony-stimulating factor alleviates adverse consequences of embryo culture on fetal growth trajectory and placental morphogenesis. Endocrinology 2005, 146, 2142–2153. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.S.; Jeung, S.H.; Biswas, D.; Jeon, Y.B.; Hyun, S.H. Effects of porcine granulocyte-macrophage colony-stimulating factor on porcine in vitro-fertilized embryos. Theriogenology 2012, 77, 1186–1197. [Google Scholar] [CrossRef]
- Denicol, A.C.; Block, J.; Kelley, D.E.; Pohler, K.G.; Dobbs, K.B.; Mortensen, C.J.; Ortega, M.S.; Hansen, P.J. The WNT signaling antagonist Dickkopf-1 directs lineage commitment and promotes survival of the preimplantation embryo. FASEB J. 2014, 28, 3975–3986. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, B.; Bonilla, L.; Block, J.; Fear, J.M.; Bonilla, A.Q.; Hansen, P.J. Colony-stimulating factor 2 (CSF-2) improves development and posttransfer survival of bovine embryos produced in vitro. Endocrinology 2009, 150, 5046–5054. [Google Scholar] [CrossRef]
- Dobbs, K.B.; Gagne, D.; Fournier, E.; Dufort, I.; Robert, C.; Block, J.; Sirard, M.A.; Bonilla, L.; Ealy, A.D.; Loureiro, B.; et al. Sexual dimorphism in developmental programming of the bovine preimplantation embryo caused by colony-stimulating factor 2. Biol. Reprod. 2014, 91, 80. [Google Scholar] [CrossRef] [PubMed]
- Canha-Gouveia, A.; Paradela, A.; Ramos-Fernandez, A.; Prieto-Sanchez, M.T.; Sanchez-Ferrer, M.L.; Corrales, F.; Coy, P. Which Low-Abundance Proteins are Present in the Human Milieu of Gamete/Embryo Maternal Interaction? Int. J. Mol. Sci. 2019, 20, 5305. [Google Scholar] [CrossRef]
- Canovas, S.; Ivanova, E.; Romar, R.; Garcia-Martinez, S.; Soriano-Ubeda, C.; Garcia-Vazquez, F.A.; Saadeh, H.; Andrews, S.; Kelsey, G.; Coy, P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. eLife 2017, 6. [Google Scholar] [CrossRef]
- Lopera-Vasquez, R.; Hamdi, M.; Maillo, V.; Lloreda, V.; Coy, P.; Gutierrez-Adan, A.; Bermejo-Alvarez, P.; Rizos, D. Effect of bovine oviductal fluid on development and quality of bovine embryos produced in vitro. Reprod. Fertil. Dev. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, M.; Lopera-Vasquez, R.; Maillo, V.; Sanchez-Calabuig, M.J.; Núnez, C.; Gutierrez-Adan, A.; Rizos, D. Bovine oviductal and uterine fluid support in vitro embryo development. Reprod. Fertil. Dev. 2018, 30, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Alvarez, P.; Rizos, D.; Rath, D.; Lonergan, P.; Gutierrez-Adan, A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc. Natl. Acad. Sci. USA 2010, 107, 3394–3399. [Google Scholar] [CrossRef]
- Hansen, P.J.; Dobbs, K.B.; Denicol, A.C.; Siqueira, L.G.B. Sex and the preimplantation embryo: Implications of sexual dimorphism in the preimplantation period for maternal programming of embryonic development. Cell Tissue Res. 2016, 363, 237–247. [Google Scholar] [CrossRef]
- Lowe, R.; Gemma, C.; Rakyan, V.K.; Holland, M.L. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genom. 2015, 16, 295. [Google Scholar] [CrossRef] [PubMed]
- Drake, A.J.; O’Shaughnessy, P.J.; Bhattacharya, S.; Monteiro, A.; Kerrigan, D.; Goetz, S.; Raab, A.; Rhind, S.M.; Sinclair, K.D.; Meharg, A.A.; et al. In utero exposure to cigarette chemicals induces sex-specific disruption of one-carbon metabolism and DNA methylation in the human fetal liver. BMC Med 2015, 13, 18. [Google Scholar] [CrossRef]
- Saenz-de-Juano, M.D.; Ivanova, E.; Billooye, K.; Herta, A.C.; Smitz, J.; Kelsey, G.; Anckaert, E. Genome-wide assessment of DNA methylation in mouse oocytes reveals effects associated with in vitro growth, superovulation, and sexual maturity. Clin. Epigenetics 2019, 11, 197. [Google Scholar] [CrossRef]
- Salilew-Wondim, D.; Saeed-Zidane, M.; Hoelker, M.; Gebremedhn, S.; Poirier, M.; Pandey, H.O.; Tholen, E.; Neuhoff, C.; Held, E.; Besenfelder, U.; et al. Genome-wide DNA methylation patterns of bovine blastocysts derived from in vivo embryos subjected to in vitro culture before, during or after embryonic genome activation. BMC Genom. 2018, 19, 424. [Google Scholar] [CrossRef]
- Kindsfather, A.J.; Czekalski, M.A.; Pressimone, C.A.; Erisman, M.P.; Mann, M.R.W. Perturbations in imprinted methylation from assisted reproductive technologies but not advanced maternal age in mouse preimplantation embryos. Clin. Epigenetics 2019, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Hattori, H.; Hiura, H.; Kitamura, A.; Miyauchi, N.; Kobayashi, N.; Takahashi, S.; Okae, H.; Kyono, K.; Kagami, M.; Ogata, T.; et al. Association of four imprinting disorders and ART. Clin. Epigenetics 2019, 11, 21. [Google Scholar] [CrossRef]
- Monteagudo-Sánchez, A.; Sánchez-Delgado, M.; Mora, J.R.H.; Santamaría, N.T.; Gratacós, E.; Esteller, M.; de Heredia, M.L.; Nunes, V.; Choux, C.; Fauque, P.; et al. Differences in expression rather than methylation at placenta-specific imprinted loci is associated with intrauterine growth restriction. Clin. Epigenetics 2019, 11, 35. [Google Scholar] [CrossRef]
- Anastasiadi, D.; Esteve-Codina, A.; Piferrer, F. Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 2018, 11, 37. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Alvarez, P.; Rizos, D.; Rath, D.; Lonergan, P.; Gutierrez-Adan, A. Epigenetic differences between male and female bovine blastocysts produced in vitro. Physiol. Genom. 2008, 32, 264–272. [Google Scholar] [CrossRef]
- Gebert, C.; Wrenzycki, C.; Herrmann, D.; Gröger, D.; Thiel, J.; Reinhardt, R.; Lehrach, H.; Hajkova, P.; Lucas-Hahn, A.; Carnwath, J.W.; et al. DNA methylation in the IGF2 intragenic DMR is re-established in a sex-specific manner in bovine blastocysts after somatic cloning. Genomics 2009, 94, 63–69. [Google Scholar] [CrossRef]
- Lopera-Vasquez, R.; Hamdi, M.; Maillo, V.; Gutierrez-Adan, A.; Bermejo-Alvarez, P.; Ramirez, M.A.; Yanez-Mo, M.; Rizos, D. Effect of bovine oviductal extracellular vesicles on embryo development and quality in vitro. Reproduction 2017, 153, 461–470. [Google Scholar] [CrossRef]
- Holm, P.; Booth, P.J.; Schmidt, M.H.; Greve, T.; Callesen, H. High bovine blastocyst development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myo-inositol with or without serum-proteins. Theriogenology 1999, 52, 683–700. [Google Scholar] [CrossRef]
- Stringfellow, D.; Givens, M. Manual of the International Embryo Transfer Society (IETS), 4th ed.; IETS: Champaign, IL, USA, 2010. [Google Scholar]
- Miura, F.; Enomoto, Y.; Dairiki, R.; Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 2012, 40, e136. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Canovas, S.; Garcia-Martínez, S.; Romar, R.; Lopes, J.S.; Rizos, D.; Sanchez-Calabuig, M.J.; Krueger, F.; Andrews, S.; Perez-Sanz, F.; et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 2020, 12, 64. [Google Scholar] [CrossRef]
- Peat, J.R.; Dean, W.; Clark, S.J.; Krueger, F.; Smallwood, S.A.; Ficz, G.; Kim, J.K.; Marioni, J.C.; Hore, T.A.; Reik, W. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 2014, 9, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, T.G.; Sinclair, K.D.; Young, L.E.; Wilmut, I.; Robinson, J.J. Large offspring syndrome and other consequences of ruminant embryo culture in vitro: Relevance to blastocyst culture in human ART. Hum. Fertil. 2000, 3, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Rizos, D.; Gutierrez-Adan, A.; Perez-Garnelo, S.; De La Fuente, J.; Boland, M.P.; Lonergan, P. Bovine embryo culture in the presence or absence of serum: Implications for blastocyst development, cryotolerance, and messenger RNA expression. Biol. Reprod. 2003, 68, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Cagnone, G.; Sirard, M.A. The impact of exposure to serum lipids during in vitro culture on the transcriptome of bovine blastocysts. Theriogenology 2014, 81, 712–722. [Google Scholar] [CrossRef]
- Heras, S.; De Coninck, D.I.; Van Poucke, M.; Goossens, K.; Bogado Pascottini, O.; Van Nieuwerburgh, F.; Deforce, D.; De Sutter, P.; Leroy, J.L.; Gutierrez-Adan, A.; et al. Suboptimal culture conditions induce more deviations in gene expression in male than female bovine blastocysts. BMC Genom. 2016, 17, 72. [Google Scholar] [CrossRef] [PubMed]
- Katz-Jaffe, M.G.; McCallie, B.R.; Preis, K.A.; Filipovits, J.; Gardner, D.K. Transcriptome analysis of in vivo and in vitro matured bovine MII oocytes. Theriogenology 2009, 71, 939–946. [Google Scholar] [CrossRef]
- Niemann, H.; Carnwath, J.W.; Herrmann, D.; Wieczorek, G.; Lemme, E.; Lucas-Hahn, A.; Olek, S. DNA methylation patterns reflect epigenetic reprogramming in bovine embryos. Cell Reprogram. 2010, 12, 33–42. [Google Scholar] [CrossRef]
- Deshmukh, R.S.; Ostrup, O.; Ostrup, E.; Vejlsted, M.; Niemann, H.; Lucas-Hahn, A.; Petersen, B.; Li, J.; Callesen, H.; Hyttel, P. DNA methylation in porcine preimplantation embryos developed in vivo and produced by in vitro fertilization, parthenogenetic activation and somatic cell nuclear transfer. Epigenetics 2011, 6, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Ealy, A.D.; Wooldridge, L.K.; McCoski, S.R. BOARD INVITED REVIEW: Post-transfer consequences of in vitro-produced embryos in cattle. J. Anim. Sci. 2019, 97, 2555–2568. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Zhang, K.X.; Shafiq, T.A.; Liao, W.W.; Hargan-Calvopina, J.; Chen, P.Y.; Clark, A.T. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell 2015, 161, 1425–1436. [Google Scholar] [CrossRef]
- Liang, X.W.; Cui, X.S.; Sun, S.C.; Jin, Y.X.; Heo, Y.T.; Namgoong, S.; Kim, N.H. Superovulation induces defective methylation in line-1 retrotransposon elements in blastocyst. Reprod. Biol. Endocrinol. 2013, 11, 69. [Google Scholar] [CrossRef]
- Ghosh, J.; Coutifaris, C.; Sapienza, C.; Mainigi, M. Global DNA methylation levels are altered by modifiable clinical manipulations in assisted reproductive technologies. Clin. Epigenetics 2017, 9, 14. [Google Scholar] [CrossRef]
- Zhu, J.; Li, M.; Chen, L.; Liu, P.; Qiao, J. The protein source in embryo culture media influences birthweight: A comparative study between G1 v5 and G1-PLUS v5. Hum. Reprod. 2014, 29, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Meintjes, M.; Chantilis, S.J.; Ward, D.C.; Douglas, J.D.; Rodriguez, A.J.; Guerami, A.R.; Bookout, D.M.; Barnett, B.D.; Madden, J.D. A randomized controlled study of human serum albumin and serum substitute supplement as protein supplements for IVF culture and the effect on live birth rates. Hum. Reprod. 2009, 24, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Engel, N. Sex Differences in Early Embryogenesis: Inter-Chromosomal Regulation Sets the Stage for Sex-Biased Gene Networks: The dialogue between the sex chromosomes and autosomes imposes sexual identity soon after fertilization. Bioessays 2018, 40, e1800073. [Google Scholar] [CrossRef]
- Siqueira, L.G.; Hansen, P.J. Sex differences in response of the bovine embryo to colony-stimulating factor 2. Reproduction 2016, 152, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Spate, L.D.; Green, M.P.; Roberts, R.M. Effects of D-glucose concentration, D-fructose, and inhibitors of enzymes of the pentose phosphate pathway on the development and sex ratio of bovine blastocysts. Mol. Reprod. Dev. 2005, 72, 201–207. [Google Scholar] [CrossRef]
- Pérez-Crespo, M.; Ramírez, M.A.; Fernández-González, R.; Rizos, D.; Lonergan, P.; Pintado, B.; Gutiérrez-Adán, A. Differential sensitivity of male and female mouse embryos to oxidative induced heat-stress is mediated by glucose-6-phosphate dehydrogenase gene expression. Mol. Reprod. Dev. 2005, 72, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Clement, K.; Huebner, A.J.; Webster, J.; Rose, C.M.; Brumbaugh, J.; Walsh, R.M.; Lee, S.; Savol, A.; Etchegaray, J.P.; et al. DUSP9 Modulates DNA Hypomethylation in Female Mouse Pluripotent Stem Cells. Cell Stem Cell 2017, 20, 706–719. [Google Scholar] [CrossRef] [PubMed]
- Milagre, I.; Stubbs, T.M.; King, M.R.; Spindel, J.; Santos, F.; Krueger, F.; Bachman, M.; Segonds-Pichon, A.; Balasubramanian, S.; Andrews, S.R.; et al. Gender Differences in Global but Not Targeted Demethylation in iPSC Reprogramming. Cell Rep. 2017, 18, 1079–1089. [Google Scholar] [CrossRef]
- Dobbs, K.B.; Rodriguez, M.; Sudano, M.J.; Ortega, M.S.; Hansen, P.J. Dynamics of DNA methylation during early development of the preimplantation bovine embryo. PLoS ONE 2013, 8, e66230. [Google Scholar] [CrossRef]
- Jenkins, K.J.; Correa, A.; Feinstein, J.A.; Botto, L.; Britt, A.E.; Daniels, S.R.; Elixson, M.; Warnes, C.A.; Webb, C.L.; Young, A.H.A.C.o.C.D.i.t. Noninherited risk factors and congenital cardiovascular defects: Current knowledge: A scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: Endorsed by the American Academy of Pediatrics. Circulation 2007, 115, 2995–3014. [Google Scholar] [CrossRef] [PubMed]
- Somerville, J. The Denolin Lecture: The woman with congenital heart disease. Eur. Heart J. 1998, 19, 1766–1775. [Google Scholar] [CrossRef]
- Sinclair, K.D.; Allegrucci, C.; Singh, R.; Gardner, D.S.; Sebastian, S.; Bispham, J.; Thurston, A.; Huntley, J.F.; Rees, W.D.; Maloney, C.A.; et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl. Acad. Sci. USA 2007, 104, 19351–19356. [Google Scholar] [CrossRef] [PubMed]
- Aiken, C.E.; Ozanne, S.E. Sex differences in developmental programming models. Reproduction 2013, 145, R1–R13. [Google Scholar] [CrossRef]
- Jiang, Z.; Lin, J.; Dong, H.; Zheng, X.; Marjani, S.L.; Duan, J.; Ouyang, Z.; Chen, J.; Tian, X.C. DNA methylomes of bovine gametes and in vivo produced preimplantation embryos. Biol. Reprod. 2018, 99, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Lengner, C.J.; Gimelbrant, A.A.; Erwin, J.A.; Cheng, A.W.; Guenther, M.G.; Welstead, G.G.; Alagappan, R.; Frampton, G.M.; Xu, P.; Muffat, J.; et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 2010, 141, 872–883. [Google Scholar] [CrossRef]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef]
- Nazor, K.L.; Altun, G.; Lynch, C.; Tran, H.; Harness, J.V.; Slavin, I.; Garitaonandia, I.; Müller, F.J.; Wang, Y.C.; Boscolo, F.S.; et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 2012, 10, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Tomoda, K.; Takahashi, K.; Leung, K.; Okada, A.; Narita, M.; Yamada, N.A.; Eilertson, K.E.; Tsang, P.; Baba, S.; White, M.P.; et al. Derivation conditions impact X-inactivation status in female human induced pluripotent stem cells. Cell Stem Cell 2012, 11, 91–99. [Google Scholar] [CrossRef]
- Carvalheira, L.R.; Tríbulo, P.; Borges, Á.; Hansen, P.J. Sex affects immunolabeling for histone 3 K27me3 in the trophectoderm of the bovine blastocyst but not labeling for histone 3 K18ac. PLoS ONE 2019, 14, e0223570. [Google Scholar] [CrossRef]
- Mathers, T.C.; Mugford, S.T.; Percival-Alwyn, L.; Chen, Y.; Kaithakottil, G.; Swarbreck, D.; Hogenhout, S.A.; van Oosterhout, C. Sex-specific changes in the aphid DNA methylation landscape. Mol. Ecol. 2019, 28, 4228–4241. [Google Scholar] [CrossRef] [PubMed]

| DMTs (100 CpG Tiles) | # Tiles (% over Total Titles) Differentially Methylated, p < 0.05 | # Tiles Differentially Methylated with > 10% Methylation dif (% over DMTs) | ||
|---|---|---|---|---|
| INVIVO vs. BSA | 68,386 (25.13%) | 15,930 (23.29%) | Up in INVIVO | 7,396 |
| Up in BSA | 8,534 | |||
| INVIVO vs. SERUM | 70,908 (26.06%) | 13,950 (19.67%) | Up in INVIVO | 7,199 |
| Up in SERUM | 6,751 | |||
| INVIVO vs. FLUIDS | 88,359 (32.48%) | 20,588 (23.30%) | Up in INVIVO | 9,796 |
| Up in FLUIDS | 10,792 | |||
| FLUIDS vs. BSA | 49,775 (18.30%) | 9,871 (19.83%) | Up in FLUIDS | 5,893 |
| Up in BSA | 3,978 | |||
| FLUIDS vs. SERUM | 57,866 (21.27%) | 9,333 (16.13%) | Up in FLUIDS | 5,364 |
| Up in SERUM | 3,969 | |||
| BSA vs. SERUM | 57,212 (21.03 %) | 8,652 (15.13 %) | Up in BSA | 4,235 |
| Up in SERUM | 4,417 | |||
| INVIVO | SERUM | FLUIDS | BSA | |
|---|---|---|---|---|
| DNMT1 |  females females |  females females |  females females | |
| DNMT3A |  females females | |||
| DNMT3B | ||||
| ZFP57 |  females females | |||
| TET1 |  females females | |||
| TET2 |  females females |  females females |  females females | |
| TET3 |  females females |  females females | ||
| UHRF2 |  females females |  females females |
| DMTs (Total 272053) | FEMALES | MALES | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p < 0.05 | + Methylation Difference > 10% | % | p < 0.05 | + Methylation Difference > 10% | % | |||||||||
| % Over Total | % Over Total | |||||||||||||
| INVIVO vs. FLUIDS | 78,979 | 47,969 | * | 17.63 | Up in INVIVO | 22,148 | 46.17 | 65,570 | 30,849 | * | 11.34 | Up in INVIVO | 15,122 | 49.02 |
| Up in FLUIDS | 25,821 | 53.83 | Up in FLUIDS | 15,727 | 50.98 | |||||||||
| INVIVO vs. BSA | 56,015 | 30,571 | 11.24 | Up in INVIVO | 15,776 | 51.60 | 75,793 | 40,885 | 15.03 | Up in INVIVO | 20,864 | 51.03 | ||
| Up in BSA | 14,795 | 48.40 | Up in BSA | 20,021 | 48.97 | |||||||||
| INVIVO vs. SERUM | 35,762 | 22,185 | * | 8.15 | Up in INVIVO | 12,999 | 58.59 | 77,265 | 33,389 | * | 12.27 | Up in INVIVO | 16,981 | 50.86 |
| Up in SERUM | 9,186 | 41.41 | Up in SERUM | 16,408 | 49.14 | |||||||||
| FLUIDS vs. BSA | 71,191 | 26,075 | * | 9.58 | Up in FLUIDS | 21,218 | 81.37 | 52,545 | 15,510 | * | 5.70 | Up in FLUIDS | 12,350 | 79.63 |
| Up in BSA | 4,857 | 18.63 | Up in BSA | 3,160 | 20.37 | |||||||||
| FLUIDS vs. SERUM | 49,662 | 27,937 | * | 10.27 | Up in FLUIDS | 13,010 | 46.57 | 67,148 | 27,412 | * | 10.08 | Up in FLUIDS | 14,487 | 52.85 |
| Up in SERUM | 14,927 | 53.43 | Up in SERUM | 12,925 | 47.15 | |||||||||
| SERUM vs. BSA | 48,251 | 26,563 | * | 9.76 | Up in SERUM | 12,434 | 46.81 | 71,791 | 32,638 | * | 12.00 | Up in SERUM | 15,368 | 47.09 |
| Up in BSA | 14,129 | 53.19 | Up in BSA | 17,270 | 52.91 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Canovas, S.; Ivanova, E.; Hamdi, M.; Perez-Sanz, F.; Rizos, D.; Kelsey, G.; Coy, P. Culture Medium and Sex Drive Epigenetic Reprogramming in Preimplantation Bovine Embryos. Int. J. Mol. Sci. 2021, 22, 6426. https://doi.org/10.3390/ijms22126426
Canovas S, Ivanova E, Hamdi M, Perez-Sanz F, Rizos D, Kelsey G, Coy P. Culture Medium and Sex Drive Epigenetic Reprogramming in Preimplantation Bovine Embryos. International Journal of Molecular Sciences. 2021; 22(12):6426. https://doi.org/10.3390/ijms22126426
Chicago/Turabian StyleCanovas, Sebastian, Elena Ivanova, Meriem Hamdi, Fernando Perez-Sanz, Dimitrios Rizos, Gavin Kelsey, and Pilar Coy. 2021. "Culture Medium and Sex Drive Epigenetic Reprogramming in Preimplantation Bovine Embryos" International Journal of Molecular Sciences 22, no. 12: 6426. https://doi.org/10.3390/ijms22126426
APA StyleCanovas, S., Ivanova, E., Hamdi, M., Perez-Sanz, F., Rizos, D., Kelsey, G., & Coy, P. (2021). Culture Medium and Sex Drive Epigenetic Reprogramming in Preimplantation Bovine Embryos. International Journal of Molecular Sciences, 22(12), 6426. https://doi.org/10.3390/ijms22126426






