MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation
Abstract
1. Introduction
2. Results
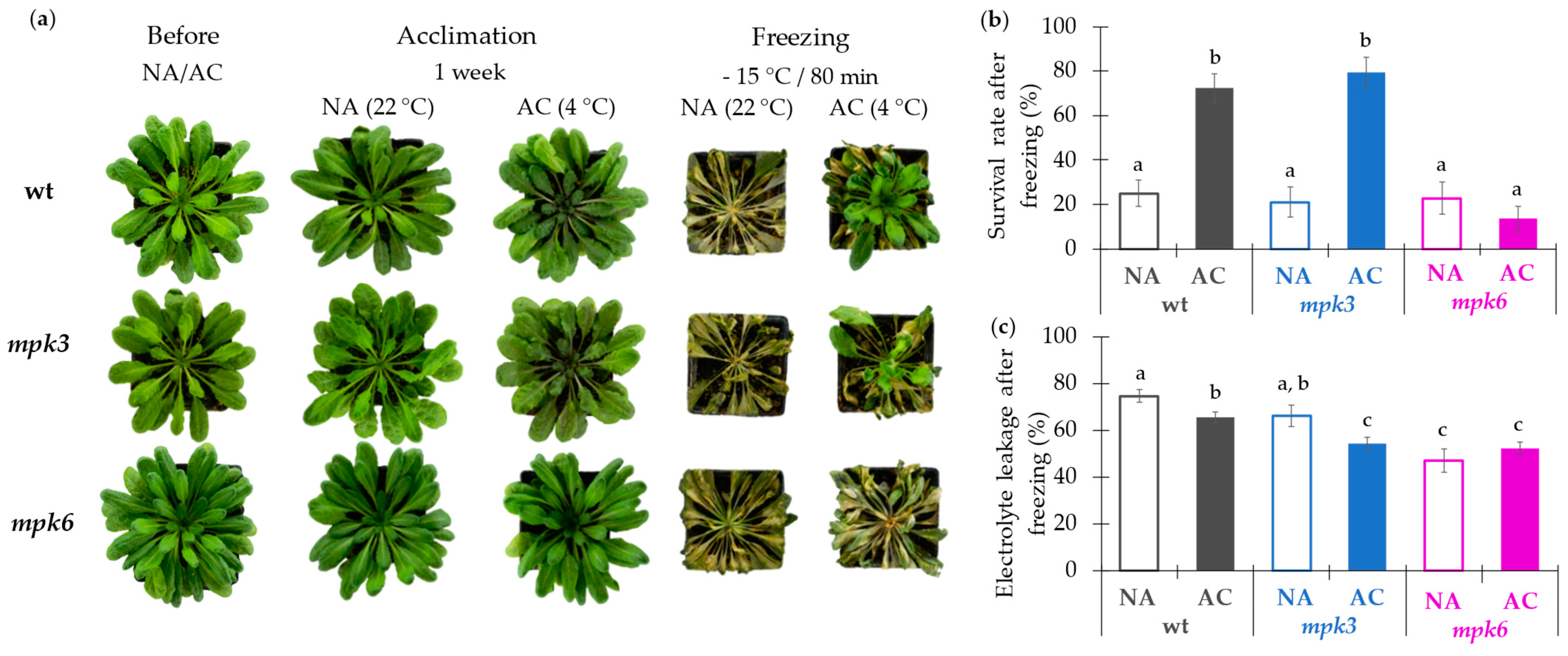
2.1. MPK6 Is Crucial for Freezing Tolerance
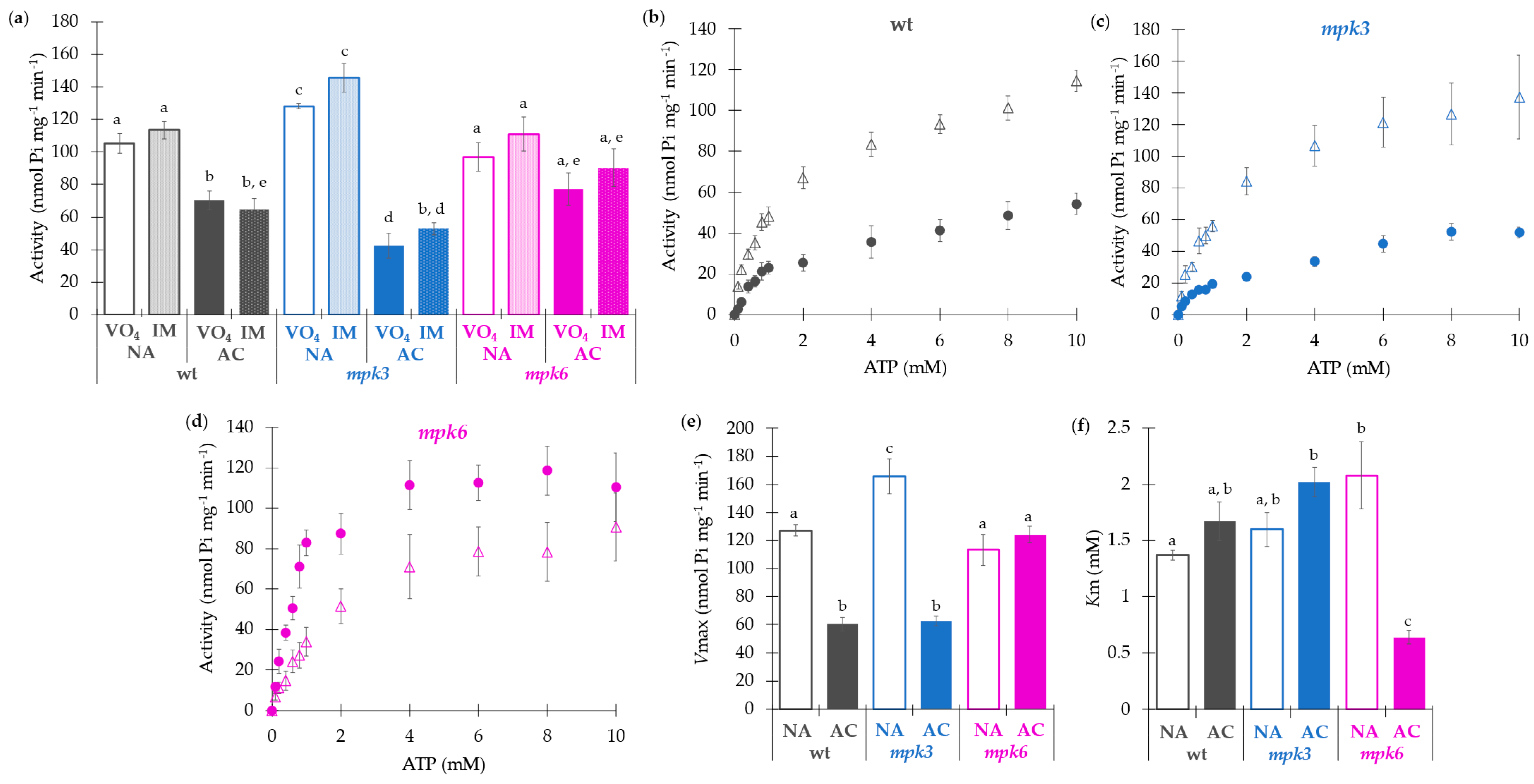
2.2. Negative Regulation of the Plasma Membrane H+-ATPase Activity Is Exerted by MPK3 in Non-Acclimated Conditions and by MPK6 in Acclimated Conditions
2.3. Kinetic Characterization of the Plasma Membrane H+-ATPase Activity Is Coincident with the Negative Regulatory Roles of MPK3 and MPK6 and Reveals Different Features of the Enzyme
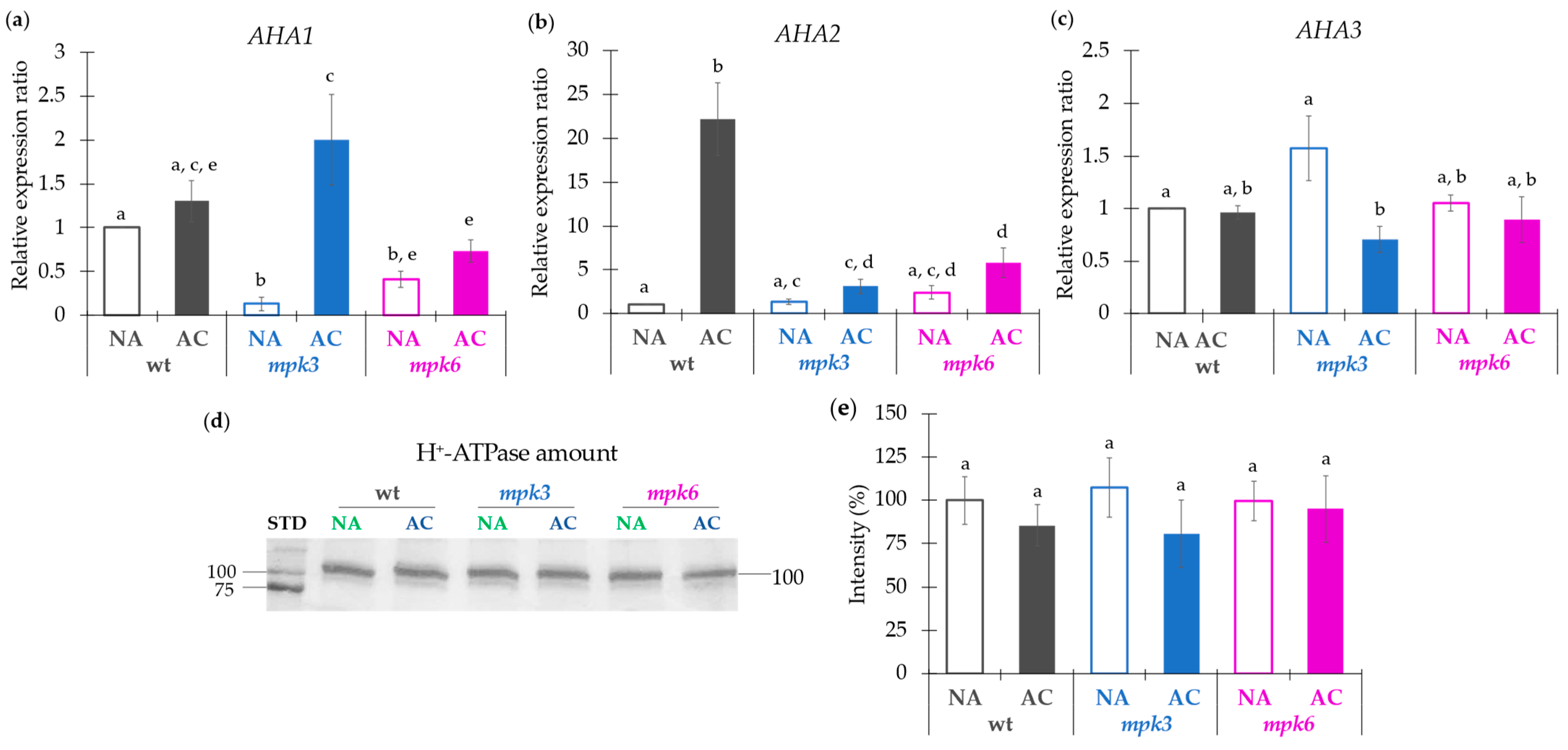
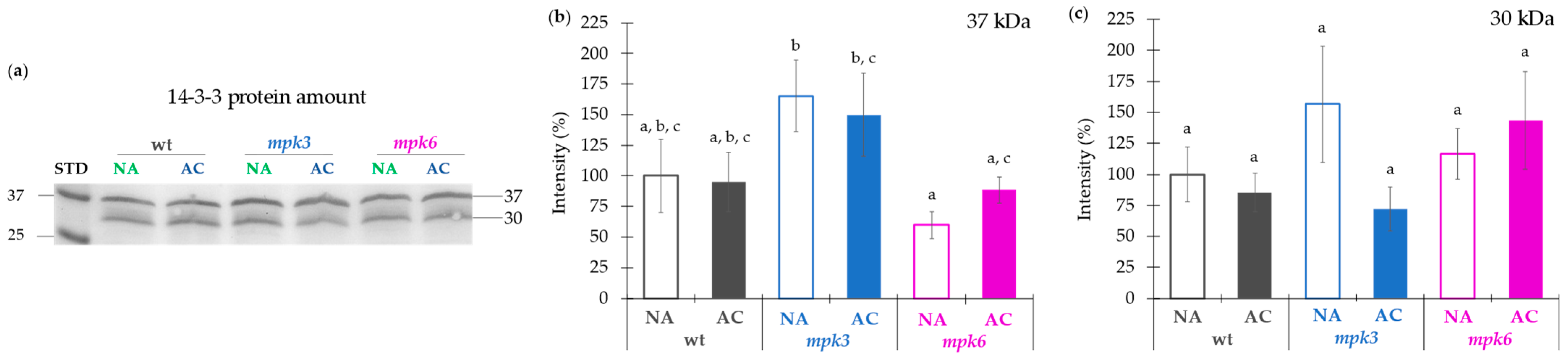
2.4. Contribution of Transcriptional or Translational Effects or Association of Regulatory Proteins to the Negative Control of MPK6 on the Plasma Membrane H+-ATPase Activity in Cold Acclimation
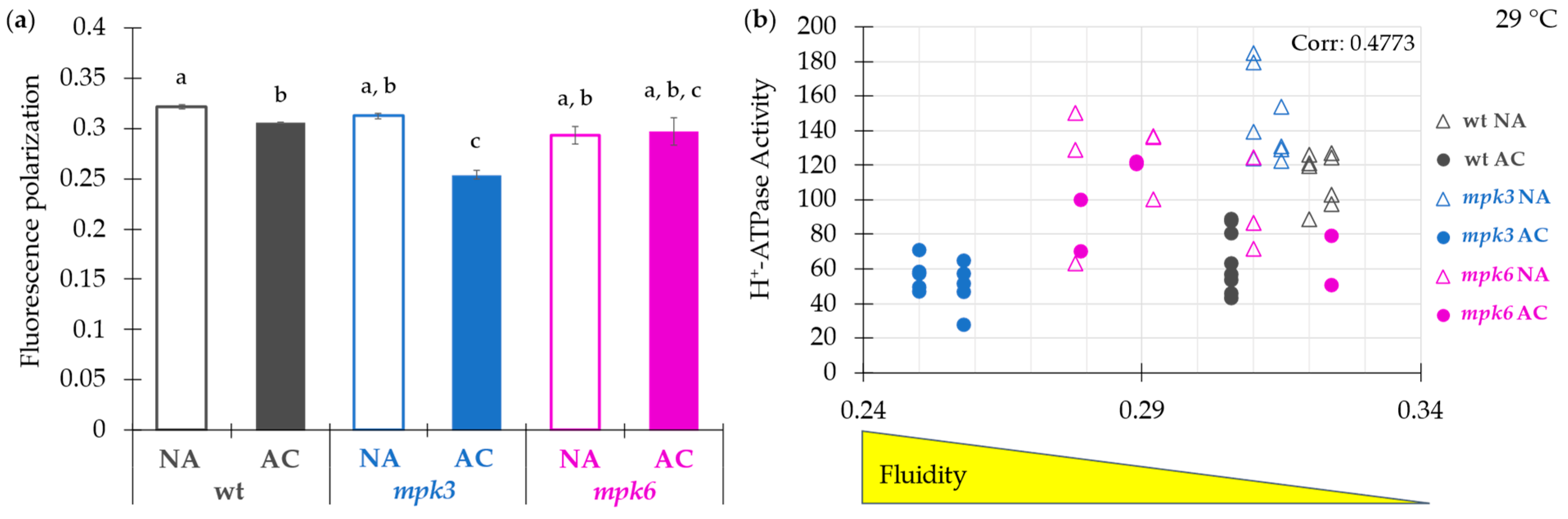
2.5. Influence of the Plasma Membrane Fluidity on the Negative Control of MPK6 Exerted on the H+-ATPase Activity under Cold Acclimation
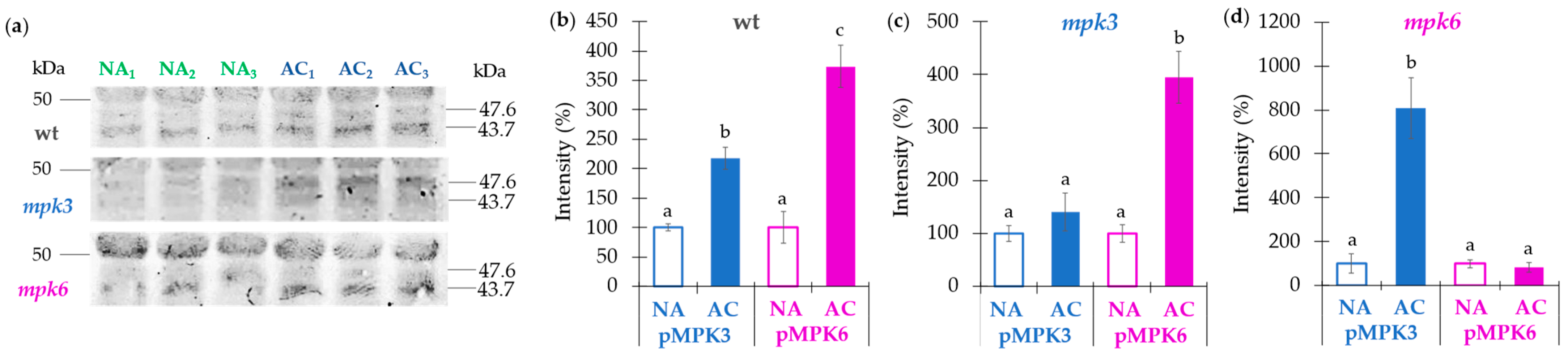
2.6. MPK3 and MPK6 Remain Activated after One Week of Cold Acclimation
3. Discussion
3.1. MPK6 Is a Positive Regulator of the Cold Acclimation Leading to Freezing Tolerance
3.2. MPK6 Influences Membrane Properties Involved in the Freezing Tolerance
3.3. Plasma Membrane H+-ATPase Is Negatively Regulated by MPK3 under Non-Acclimation and by MPK6 under Cold Acclimation
3.4. The Expression of Plasma Membrane H+-ATPase Isogenes Is Regulated by MPK3 and MPK6 and Depends on the AHA Isogene, the MPK, the NA or AC Condition but Is Unrelated to the H+-ATPase Activity Extent
3.5. Plasma Membrane H+-ATPase Activity Regulated by MPK3 or MPK6 in Non-Acclimation or Cold Acclimation Is Independent on the Amount of Protein Levels or Association of 14-3-3 Proteins
3.6. Plasma Membrane H+-ATPase Activity Regulated by MPK6 Is Associated to the Membrane Fluidity
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Physiological Analysis
4.3. Subcellular Fractionation
4.4. ATPase Activity and Kinetic Assays
4.5. Plasma Membrane Fluidity Assay
4.6. RNA Extraction and qPCR Analysis
4.7. Protein Determination
4.8. Immunoblotting
4.9. Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jenks, M.; Hasegawa, P. Plant Abiotic Stress, 2nd ed.; Wiley-Blackwell: New York, NY, USA, 2014. [Google Scholar]
- Vats, S. Biotic and Abiotic Stress Tolerance in Plants; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Yadav, S.K. Cold stress tolerance mechanisms in plants. A review. Agron. Sustain. Dev. 2010, 30, 515–527. [Google Scholar] [CrossRef]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant. 2016, 9, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Fuglsang, A.T.; Visconti, S.; Drumm, K.; Jahn, T.; Stensballe, A.; Mattei, B.; Jensen, O.N.; Aducci, P.; Palmgren, M.G. Binding of 14-3-3 Protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 1999, 274, 36774–36780. [Google Scholar] [CrossRef] [PubMed]
- Camoni, L.; Iori, V.; Marra, M.; Aducci, P. Phosphorylation-dependent interaction between plant plasma membrane H+-ATPase and 14-3-3 Proteins. J. Biol. Chem. 2000, 275, 9919–9923. [Google Scholar] [CrossRef] [PubMed]
- Kanczewska, J.; Marco, S.; Vandermeeren, C.; Maudoux, O.; Rigaud, J.-L.; Boutry, M. Activation of the plant plasma membrane H+-ATPase by phosphorylation and binding of 14-3-3 proteins converts a dimer into a hexamer. Proc. Natl. Acad. Sci. USA 2005, 102, 11675–11680. [Google Scholar] [CrossRef] [PubMed]
- Morales-Cedillo, F.; González-Solís, A.; Gutiérrez-Angoa, L.; Cano-Ramírez, D.; Gavilanes-Ruiz, M. Plant lipid environment and membrane enzymes: The case of the plasma membrane H+-ATPase. Plant Cell Rep. 2015, 34, 617–629. [Google Scholar] [CrossRef]
- Ahn, S.J.; Im, Y.J.; Chung, G.C.; Cho, B.H. Inducible expression of plasma membrane H+-ATPase in the roots of figleaf gourd plants under chilling root temperature. Physiol. Plant. 1999, 106, 35–40. [Google Scholar] [CrossRef]
- Chelysheva, V.V.; Smolenskaya, I.N.; Trofimova, M.C.; Babakov, A.V.; Muromtsev, G.S. Role of the 14-3-3 proteins in the regulation of H+-ATPase activity in the plasma membrane of suspension-cultured sugar beet cells under cold stress. FEBS Lett. 1999, 456, 22–26. [Google Scholar] [CrossRef]
- Ahn, S.-J.; Im, Y.-J.; Chung, G.-C.; Seong, K.-Y.; Cho, B.-H. Sensitivity of plasma membrane H+-ATPase of cucumber root system in response to low root temperature. Plant Cell Rep. 2000, 19, 831–835. [Google Scholar] [CrossRef]
- Kim, H.S.; Oh, J.M.; Luan, S.; Carlson, J.E.; Ahn, S.J. Cold stress causes rapid but differential changes in properties of plasma membrane H(+)-ATPase of camelina and rapeseed. J. Plant Physiol. 2013, 170, 828–837. [Google Scholar] [CrossRef]
- Sadura, I.; Libik-Konieczny, M.; Jurczyk, B.; Gruszka, D.; Janeczko, A. Plasma membrane ATPase and the aquaporin HvPIP1 in barley brassinosteroid mutants acclimated to high and low temperature. J. Plant Physiol. 2020, 244, 153090. [Google Scholar] [CrossRef]
- Janicka-Russak, M.; Kabała, K.; Wdowikowska, A.; Kłobus, G. Response of plasma membrane H+-ATPase to low temperature in cucumber roots. J. Plant Res. 2012, 125, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Pan, X.; Qu, H.; Underhill, S.J.R. Low temperature alters plasma membrane lipid composition and ATPase activity of pineapple fruit during blackheart development. J. Bioenerg. Biomembr. 2014, 46, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Muzi, C.; Camoni, L.; Visconti, S.; Aducci, P. Cold stress affects H+-ATPase and phospholipase D activity in Arabidopsis. Plant Physiol. Biochem. 2016, 108, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 2000, 24, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Furuya, T.; Matsuoka, D.; Nanmori, T. Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J. Plant Res. 2013, 126, 833–840. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.Y.; Chung, W.S. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627. [Google Scholar] [CrossRef]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell. 2017, 43, 1–13. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, P.; Si, T.; Hsu, C.-C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev. Cell. 2017, 43, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Tena, G.; Asai, T.; Chiu, W.-L.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Mishra, N.S.; Tuteja, R.; Tuteja, N. Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 2006, 452, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-García, M.; Gavilanes-Ruíz, M.; Arce-Cervantes, O. Long-chain bases, phosphatidic acid, MAPKs, and reactive oxygen species as nodal signal transducers in stress responses in Arabidopsis. Front. Plant Sci. 2015, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. Ann. Plant Rev. 2018, 33, 100–136. [Google Scholar] [CrossRef]
- Pitzschke, A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 2015, 20, 49–55. [Google Scholar] [CrossRef]
- Rodríguez, M.; Petersen, M.; Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2007, 61, 621–649. [Google Scholar] [CrossRef]
- Andreasson, E.; Ellis, B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010, 15, 106–113. [Google Scholar] [CrossRef]
- Yang, T.; Shad Ali, G.; Yang, L.; Du, L.; Reddy, A.S.N.; Poovaiah, B.W. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Benhassaine-Kesri, G.; Demandre, C.; Rézé, N.; Launay, A.; Pelletier, S.; Renou, J.P.; Zachowski, A.; Baudouin, E.; Guillas, I. Phytosphingosine-phosphate is a signal for AtMPK6 activation and Arabidopsis response to chilling. New Phytol. 2012, 194, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Furuya, T.; Matsuoka, D.; Nanmori, T. Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 2014, 588, 2025–2030. [Google Scholar] [CrossRef] [PubMed]
- Zuther, E.; Schulz, E.; Childs, L.H.; Hincha, D.K. Clinal variation in the non-acclimated and cold-acclimated freezing tolerance of Arabidopsis thaliana accessions. Plant Cell Environ. 2012, 35, 1860–1878. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Christensen, G. Functional comparisons between plant plasma membrane H(+)-ATPase isoforms expressed in yeast. J. Biol. Chem. 1994, 269, 3027–3033. [Google Scholar] [CrossRef]
- Alsterfjord, M.; Sehnke, P.C.; Arkell, A.; Larsson, H.; Svennelid, F.; Rosenquist, M.; Ferl, R.J.; Sommarin, M.; Larsson, C. Plasma membrane H(+)-ATPase and 14-3-3 isoforms of Arabidopsis leaves: Evidence for isoform specificity in the 14-3-3/H+-ATPase interaction. Plant Cell Physiol. 2004, 45, 1202–1210. [Google Scholar] [CrossRef]
- DeLille, J.M.; Sehnke, P.C.; Ferl, R.J. The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001, 126, 35–38. [Google Scholar] [CrossRef]
- Shinitzky, M.; Barenholz, Y. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim. Biophys. Acta 1978, 515, 367–394. [Google Scholar] [CrossRef]
- López-Bucio, J.S.; Dubrovsky, J.G.; Raya-González, J.; Ugartechea-Chirino, Y.; López-Bucio, J.; de Luna-Valdez, L.A.; Ramos-Vega, M.; León, P.; Guevara-García, A.A. Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J. Exp. Bot. 2014, 65, 169–183. [Google Scholar] [CrossRef]
- Sethi, V.; Raghuram, B.; Sinha, A.K.; Chattopadhyay, S. A mitogen-activated protein kinase cascade module, MKK3-MPK6 and MYC2, is involved in blue light-mediated seedling development in Arabidopsis. Plant Cell. 2014, 26, 3343–3357. [Google Scholar] [CrossRef]
- Smékalová, V.; Luptovčiak, I.; Komis, G.; Šamajová, O.; Ovečka, M.; Doskočilová, A.; Takáč, T.; Vadovič, P.; Novák, O.; Pechan, T.; et al. Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 2014, 203, 1175–1193. [Google Scholar] [CrossRef] [PubMed]
- Plieth, C.; Hansen, U.P.; Knight, H.; Knight, M.R. Temperature sensing by plants: The primary characteristics of signal perception and calcium response. Plant J. 1999, 18, 491–497. [Google Scholar] [CrossRef]
- Smallwood, M.; Bowles, D.J. Plants in a cold climate. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 831–847. [Google Scholar] [CrossRef] [PubMed]
- Titov, A.F.; Shibaeva, T.G.; Ikkonen, E.N.; Sherudilo, E.G. Plant responses to a daily short-term temperature drop: Phenomenology and mechanisms. Russ. J. Plant Physiol. 2020, 67, 1003–1017. [Google Scholar] [CrossRef]
- Leuendorf, J.E.; Frank, M.; Schmülling, T. Acclimation, priming and memory in the response of Arabidopsis thaliana seedlings to cold stress. Sci. Rep. 2020, 10, 689. [Google Scholar] [CrossRef]
- Hatsugai, N.; Katagiri, F. Quantification of plant cell death by electrolyte leakage assay. Bio-Protocol 2018, 8, e2758. [Google Scholar] [CrossRef]
- Orvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef]
- Mikami, K.; Murata, N. Membrane fluidity and the perception of environmental signals in Cyanobacteria and plants. Prog. Lipid Res. 2003, 42, 527–543. [Google Scholar] [CrossRef]
- Arora, R. Mechanism of freeze-thaw injury and recovery: A cool retrospective and warming up to new ideas. Plant Sci. 2018, 270, 301–313. [Google Scholar] [CrossRef]
- Gupta, R.K. A study of photosynthesis and leakage of solutes in relation to desiccation of bryophytes. Can. J. Bot. 1977, 55, 1186–1194. [Google Scholar] [CrossRef]
- Leopold, A.C.; Musgrave, M.E.; Williams, K.M. Solute leakage resulting from leaf desiccation. Plant Physiol. 1981, 68, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Palta, J.P.; Levitt, J.; Stadelmann, E.J. Freezing injury in onion bulb cells: I. Evaluation of the conductivity method and analysis of ion and sugar efflux from injured cells. Plant Physiol. 1977, 60, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Palta, J.P. Stress interactions at the cellular and membrane levels. Hortscience 1990, 25, 1377–1381. [Google Scholar] [CrossRef]
- Demidchik, V. ROS-activated ion channels in plants: Biophysical characteristics, physiological functions and molecular nature. Int. J. Mol. Sci. 2018, 19, 1263. [Google Scholar] [CrossRef]
- Miquel, M.; James, D., Jr.; Dooner, H.; Browse, J. Arabidopsis requires polyunsaturated lipids for low-temperature survival. Proc. Natl. Acad. Sci. USA 1993, 90, 6208–6212. [Google Scholar] [CrossRef]
- Chen, M.; Markham, J.E.; Cahoon, E.B. Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781. [Google Scholar] [CrossRef]
- Chen, M.; Thelen, J.J. ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. Plant Cell. 2013, 25, 1430–1444. [Google Scholar] [CrossRef]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef]
- Suri, S.S.; Dhindsa, R.S. A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ. 2008, 31, 218–226. [Google Scholar] [CrossRef]
- Wielandt, A.G.; Pedersen, J.T.; Falhof, J.; Kemmer, G.C.; Lund, A.; Ekberg, K.; Fuglsang, A.T.; Pomorski, T.G.; Buch-Pedersen, M.J.; Palmgren, M. Specific activation of the plant P-type plasma membrane H+-ATPase by lysophospholipids depends on the autoinhibitory N- and C-terminal domains. J. Biol. Chem. 2015, 290, 16281–16291. [Google Scholar] [CrossRef]
- Harper, J.F.; Surowy, T.K.; Sussman, M.R. Molecular cloning and sequence of cDNA encoding the plasma membrane proton pump (H+-ATPase) of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1989, 86, 1234–1238. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Burch, H.L.; Nelson, R.B.; Barrett-Wilt, G.; Kline, K.G.; Mohsin, S.B.; Young, J.C.; Otegui, M.S.; Sussman, M.R. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010, 285, 17918–17929. [Google Scholar] [CrossRef] [PubMed]
- Młodzińska, E.; Kłobus, G.; Christensen, M.D.; Fuglsang, A.T. The plasma membrane H+-ATPase AHA2 contributes to the root architecture in response to different nitrogen supply. Physiol. Plant. 2015, 154, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.D.; Olsen, L.I.; Ezike, C.V.; Pedersen, J.T.; Manstretta, R.; López-Marqués, R.L.; Palmgren, M. Roles of plasma membrane proton ATPases AHA2 and AHA7 in normal growth of roots and root hairs in Arabidopsis thaliana. Physiol. Plant 2019, 166, 848–861. [Google Scholar] [CrossRef] [PubMed]
- Lang, V.; Pertl-Obermeyer, H.; Safiarian, M.J.; Obermeyer, G. Pump up the volume—A central role for the plasma membrane H+ pump in pollen germination and tube growth. Protoplasma 2014, 251, 477–488. [Google Scholar] [CrossRef]
- Hoffmann, R.D.; Portes, M.T.; Olsen, L.I.; Damineli, D.; Hayashi, M.; Nunes, C.O.; Pedersen, J.T.; Lima, P.T.; Campos, C.; Feijó, J.A.; et al. Plasma membrane H+-ATPases sustain pollen tube growth and fertilization. Nat. Commun. 2020, 11, 2395. [Google Scholar] [CrossRef]
- Ferl, R.J. 14-3-3 Proteins and signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 49–73. [Google Scholar] [CrossRef]
- Jahn, T.; Fuglsang, A.T.; Olsson, A.; Brüntrup, I.M.; Collinge, D.B.; Volkmann, D.; Sommarin, M.; Palmgren, M.G.; Larsson, C. The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell 1997, 9, 1805–1814. [Google Scholar] [CrossRef]
- Bagnat, M.; Chang, A.; Simons, K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 2001, 12, 4129–4138. [Google Scholar] [CrossRef]
- Borner, G.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; Macaskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef]
- Carmona-Salazar, L.; Cahoon, R.E.; Gasca-Pineda, J.; González-Solís, A.; Vera-Estrella, R.; Treviño, V.; Cahoon, E.B.; Gavilanes-Ruiz, M. Plasma and vacuolar membrane sphingolipidomes: Composition and insights on the role of main molecular species. Plant Physiol. 2021, 186, 624–636. [Google Scholar] [CrossRef]
- Grennan, A.K. Lipid rafts in plants. Plant Physiol. 2007, 143, 1083–1085. [Google Scholar] [CrossRef]
- Uemura, M.; Tominaga, Y.; Nakagawara, C.; Shigematsu, S.; Minami, A.; Kawamura, Y. Responses of the plasma membrane to low temperatures. Physiol Plant. 2006, 126, 81–89. [Google Scholar] [CrossRef]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Ellis, B.E. Arabidopsis MAPK phosphatase 2 (MKP2) positively regulates oxidative stress tolerance and inactivates the MPK3 and MPK6 MAPKs. J. Biol. Chem. 2007, 282, 25020–25029. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; Kim, K.E.; Kim, K.C.; Nguyen, X.C.; Han, H.J.; Jung, M.S.; Kim, H.S.; Kim, S.H.; Park, H.C.; Yun, D.J.; et al. Cadmium activates Arabidopsis MPK3 and MPK6 via accumulation of reactive oxygen species. Phytochemistry 2010, 71, 614–618. [Google Scholar] [CrossRef]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 2014, 343, 408–411. [Google Scholar] [CrossRef] [PubMed]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S.; et al. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Kristensen, A.; Cuin, T.A.; Schulze, W.X.; Persson, J.; Thuesen, K.H.; Ytting, C.K.; Oehlenschlæger, C.B.; Mahmood, K.; Sondergaard, T.E.; et al. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. Plant J. 2014, 80, 951–964. [Google Scholar] [CrossRef]
- Jones, J.B. Hydroponics: Its history and use in plant nutrition studies. J. Plant Nutr. 1982, 5, 1003–1030. [Google Scholar] [CrossRef]
- Zavafer, A.; González-Solís, A.; Palacios-Bahena, S.; Saucedo-García, M.; Tapia de Aquino, C.; Vázquez-Santana, S.; King-Díaz, B.; Gavilanes-Ruiz, M. Organized disassembly of photosynthesis during programmed cell death mediated by long chain bases. Sci. Rep. 2020, 10, 10360. [Google Scholar] [CrossRef]
- Carmona-Salazar, L.; El Hafidi, M.; Enríquez-Arredondo, C.; Vázquez-Vázquez, C.; González de la Vara, L.E.; Gavilanes-Ruíz, M. Isolation of detergent-resistant membranes from plant photosynthetic and non-photosynthetic tissues. Anal. Biochem. 2011, 417, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Nieto, S.; García-Rubio, O.; Pacheco-Moisés, F.; Carballo, A.; Rodríguez-Sotres, R.; Gavilanes-Ruíz, M. Purification of plasma membranes from dry maize embryos. Physiol. Plant. 1997, 101, 157–164. [Google Scholar] [CrossRef]
- González-Romo, P.; Sánchez-Nieto, S.; Gavilanes-Ruíz, M. A modified colorimetric method for the determination of orthophosphate in the presence of high ATP concentrations. Anal. Biochem. 1992, 200, 235–238. [Google Scholar] [CrossRef]
- Koetsier, G.; Cantor, E. A Practical Guide to Analyzing Nucleic Acid Concentration and Purity with Microvolume Spectrophotometers; New England BioLabs, Inc.: Ipswich, MA, USA, 2019. [Google Scholar]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.-K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Peterson, G.L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 1977, 83, 346–356. [Google Scholar] [CrossRef]
- Schägger, H.; von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponce-Pineda, I.G.; Carmona-Salazar, L.; Saucedo-García, M.; Cano-Ramírez, D.; Morales-Cedillo, F.; Peña-Moral, A.; Guevara-García, Á.A.; Sánchez-Nieto, S.; Gavilanes-Ruíz, M. MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation. Int. J. Mol. Sci. 2021, 22, 6338. https://doi.org/10.3390/ijms22126338
Ponce-Pineda IG, Carmona-Salazar L, Saucedo-García M, Cano-Ramírez D, Morales-Cedillo F, Peña-Moral A, Guevara-García ÁA, Sánchez-Nieto S, Gavilanes-Ruíz M. MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation. International Journal of Molecular Sciences. 2021; 22(12):6338. https://doi.org/10.3390/ijms22126338
Chicago/Turabian StylePonce-Pineda, Ilian Giordano, Laura Carmona-Salazar, Mariana Saucedo-García, Dora Cano-Ramírez, Francisco Morales-Cedillo, Araceli Peña-Moral, Ángel Arturo Guevara-García, Sobeida Sánchez-Nieto, and Marina Gavilanes-Ruíz. 2021. "MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation" International Journal of Molecular Sciences 22, no. 12: 6338. https://doi.org/10.3390/ijms22126338
APA StylePonce-Pineda, I. G., Carmona-Salazar, L., Saucedo-García, M., Cano-Ramírez, D., Morales-Cedillo, F., Peña-Moral, A., Guevara-García, Á. A., Sánchez-Nieto, S., & Gavilanes-Ruíz, M. (2021). MPK6 Kinase Regulates Plasma Membrane H+-ATPase Activity in Cold Acclimation. International Journal of Molecular Sciences, 22(12), 6338. https://doi.org/10.3390/ijms22126338






