MicroRNAs: The Link between the Metabolic Syndrome and Oncogenesis
Abstract
1. Introduction
- (“neoplasm invasiveness/etiology”[MeSH Terms]) AND specific microRNA,
- ((“obesity”[MeSH Terms]) OR (“insulin resistance”[MeSH Terms])) OR (“metabolic syndrome”[MeSH Terms]) AND specific microRNA.
2. MicroRNAs Link the Metabolic Syndrome and Cancer
2.1. The Role of miRNAs in Cancer by Modulating Macrophage Phenotypes
2.2. The Role of CTBP1 in Cancer by Modulating microRNAs Expression
2.3. The Role of Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) in Obesity and Cancer
2.4. PI3K/AKT—Common Pathway in MetS and Cancer
2.5. Caveolin-1 (CAV1) Signaling
2.6. Wnt/β-catenin Signaling
2.7. PTEN Signaling
2.8. The miRNA-Processing Enzyme DICER
2.9. PPARα -FOXP4- NOTCH Pathway
2.10. SRC/SOX2/c-MYC Pathway
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Final Report. Circulation 2002, 106, 3143. [CrossRef]
- Grundy, S.M. Metabolic Syndrome Update. Trends Cardiovasc. Med. 2016, 26, 364–373. [Google Scholar] [CrossRef]
- Piotrowski, I.; Kulcenty, K.; Suchorska, W. Interplay between Inflammation and Cancer. Rep. Pr. Oncol. Radiother. 2020, 25, 422–427. [Google Scholar] [CrossRef]
- Heyn, G.S.; Corrêa, L.H.; Magalhães, K.G. The Impact of Adipose Tissue–Derived MiRNAs in Metabolic Syndrome, Obesity, and Cancer. Front. Endocrinol. 2020, 11, 563816. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The Impact of MicroRNAs on Protein Output. Nat. Cell Biol. 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, R.; Tanaka, C.; Sato, M.; Nagasaki, H.; Sugimura, K.; Okumura, K.; Nakagawa, Y.; Aoki, N. Adipocyte-Derived Microvesicles Contain RNA That Is Transported into Macrophages and Might Be Secreted into Blood Circulation. Biochem. Biophys. Res. Commun. 2010, 398, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal MiRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [PubMed]
- De Visser, K.E.; Eichten, A.; Coussens, L.M. Paradoxical Roles of the Immune System During Cancer Development. Nat. Rev. Cancer 2006, 6, 24–37. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, Y. Tumor-associated macrophages: From basic research to clinical application. J. Hematol. Oncol. 2017, 10, 58. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage Plasticity and Polarization: In Vivo Veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Baer, C.; Squadrito, M.L.; Laoui, D.; Thompson, D.; Hansen, S.K.; Kiialainen, A.; Hoves, S.; Ries, C.H.; Ooi, C.H.; De Palma, M. Suppression of MicroRNA Activity Amplifies IFN-γ-Induced Macrophage Activation and Promotes Anti-Tumour Immunity. Nat. Cell Biol. 2016, 18, 790–802. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Pucci, F.; Magri, L.; Moi, D.; Gilfillan, G.; Ranghetti, A.; Casazza, A.; Mazzone, M.; Lyle, R.; Naldini, L.; et al. MiR-511-3p Modulates Genetic Programs of Tumor-Associated Macrophages. Cell Rep. 2012, 1, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Mühlberg, L.; Kühnemuth, B.; Costello, E.; Shaw, V.; Sipos, B.; Huber, M.; Griesmann, H.; Krug, S.; Schober, M.; Gress, T.M.; et al. MiRNA Dynamics in Tumor-Infiltrating Myeloid Cells Modulating Tumor Progression in Pancreatic Cancer. OncoImmunology 2016, 5, e1160181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, L.; Hu, Y.; Huang, Y.; Zhang, Y.; Zheng, X.; Wang, S.; Wang, Y.; Yujuan, Z.; Zhang, M.; et al. MiRNA Let-7b Modulates Macrophage Polarization and Enhances Tumor-Associated Macrophages to Promote Angiogenesis and Mobility in Prostate Cancer. Sci. Rep. 2016, 6, 25602. [Google Scholar] [CrossRef]
- Ying, X.; Wu, Q.; Wu, X.; Zhu, Q.; Wang, X.; Jiang, L.; Chen, X.; Wang, X. Epithelial Ovarian Cancer-Secreted Exosomal MiR-222-3p Induces Polarization of Tumor-Associated Macrophages. Oncotarget 2016, 7, 43076–43087. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ying, X.; Wang, X.; Wu, X.; Zhu, Q.; Wang, X. Exosomes Derived from Hypoxic Epithelial Ovarian Cancer Deliver MicroRNA-940 to Induce Macrophage M2 Polarization. Oncol. Rep. 2017, 38, 522–528. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated Polarization of Tumor-Associated Macrophages by MiR-145 via Colorectal Cancer–Derived Extracellular Vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Jian, S.-F.; Lin, Y.-S.; Pan, Y.-C.; Wu, C.-Y.; Kuo, P.-L. Hypoxic Lung-Cancer-Derived Extracellular Vesicle MicroRNA-103a Increases the Oncogenic Effects of Macrophages by Targeting PTEN. Mol. Ther. 2018, 26, 568–581. [Google Scholar] [CrossRef]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef]
- Zonari, E.; Pucci, F.; Saini, M.; Mazzieri, R.; Politi, L.S.; Gentner, B.; Naldini, L. A Role for MiR-155 in Enabling Tumor-Infiltrating Innate Immune Cells to Mount Effective Antitumor Responses in Mice. Blood 2013, 122, 243–252. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Z.; Chen, C.; Liu, Y.; Si, Q.; Chuang, T.-H.; Li, N.; Gomezcabrero, A.; A Reisfeld, R.; Xiang, R.; et al. MicroRNA-19a-3p Inhibits Breast Cancer Progression and Metastasis by Inducing Macrophage Polarization through Downregulated Expression of Fra-1 Proto-Oncogene. Oncogene 2014, 33, 3014–3023. [Google Scholar] [CrossRef]
- Xu, S.; Wei, J.; Wang, F.; Kong, L.-Y.; Ling, X.-Y.; Nduom, E.; Gabrusiewicz, K.; Doucette, T.; Yang, Y.; Yaghi, N.K.; et al. Effect of MiR-142-3p on the M2 Macrophage and Therapeutic Efficacy Against Murine Glioblastoma. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef]
- Dong, L.; Xia, S.; Luo, Y.; Diao, H.; Zhang, J.; Chen, J.; Zhang, J. Targeting Delivery Oligonucleotide into Macrophages by Cationic Polysaccharide from Bletilla Striata Successfully Inhibited the Expression of TNF-α. J. Control. Release 2009, 134, 214–220. [Google Scholar] [CrossRef]
- Huang, Z.; Gan, J.; Long, Z.; Guo, G.; Shi, X.; Wang, C.; Zang, Y.; Ding, Z.; Chen, J.; Zhang, J.; et al. Targeted Delivery of Let-7b to Reprogramme Tumor-Associated Macrophages and Tumor Infiltrating Dendritic Cells for Tumor Rejection. Biomaterials 2016, 90, 72–84. [Google Scholar] [CrossRef]
- Teng, Y.; Mu, J.; Hu, X.; Samykutty, A.; Zhuang, X.; Deng, Z.; Zhang, L.; Cao, P.; Yan, J.; Miller, D.; et al. Grapefruit-Derived Nanovectors Deliver MiR-18a for Treatment of Liver Metastasis of Colon Cancer by Induction of M1 Macrophages. Oncotarget 2016, 7, 25683–25697. [Google Scholar] [CrossRef] [PubMed]
- Blevins, M.A.; Huang, M.; Zhao, R. The Role of CtBP1 in Oncogenic Processes and Its Potential As a Therapeutic Target. Mol. Cancer Ther. 2017, 16, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-L.; Huang, C.-X.; Zhang, J.; Inoue, A.; Zeng, S.-E.; Xiao, S.-J. CtBP1 Is Involved in Epithelial-Mesenchymal Transition and Is a Potential Therapeutic Target for Hepatocellular Carcinoma. Oncol. Rep. 2013, 30, 809–814. [Google Scholar] [CrossRef]
- De Luca, P.; Dalton, G.N.; Scalise, G.D.; Moiola, C.P.; Porretti, J.; Massillo, C.; Kordon, E.; Gardner, K.; Zalazar, F.; Flumian, C.; et al. CtBP1 Associates Metabolic Syndrome and Breast Carcinogenesis Targeting Multiple MiRNAs. Oncotarget 2016, 7, 18798–18811. [Google Scholar] [CrossRef] [PubMed]
- Dalton, G.N.; Massillo, C.; Scalise, G.D.; Duca, R.; Porretti, J.; Farré, P.L.; Gardner, K.; Paez, A.; Gueron, G.; De Luca, P.; et al. CTBP1 Depletion on Prostate Tumors Deregulates miRNA/MRNA Expression and Impairs Cancer Progression in Metabolic Syndrome Mice. Cell Death Dis. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Farré, P.L.; Scalise, G.D.; Duca, R.B.; Dalton, G.N.; Massillo, C.; Porretti, J.; Graña, K.; Gardner, K.; De Luca, P.; De Siervi, A. CTBP1 and Metabolic Syndrome Induce an MRNA and MiRNA Expression Profile Critical for Breast Cancer Progression and Metastasis. Oncotarget 2018, 9, 13848–13858. [Google Scholar] [CrossRef]
- Panigrahy, D.; Singer, S.; Shen, L.Q.; Butterfield, C.E.; Freedman, D.A.; Chen, E.J.; Moses, M.A.; Kilroy, S.; Duensing, S.; Fletcher, C.; et al. PPARγ Ligands Inhibit Primary Tumor Growth and Metastasis by Inhibiting Angiogenesis. J. Clin. Investig. 2002, 110, 923–932. [Google Scholar] [CrossRef]
- Weng, J.-R.; Chen, C.-Y.; Pinzone, J.J.; Ringel, M.D. Beyond Peroxisome Proliferator-Activated Receptor γ Signaling: The Multi-Facets of the Antitumor Effect of Thiazolidinediones. Endocr. Relat. Cancer 2006, 13, 401–413. [Google Scholar] [CrossRef]
- Seiri, P.; Abi, A.; Soukhtanloo, M. PPAR-γ: Its Ligand and Its Regulation by MicroRNAs. J. Cell. Biochem. 2019, 120, 10893–10908. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A Role OfmiR-27in the Regulation of Adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Lee, M.J.; Abdelmohsen, K.; Kim, W.; Kim, M.M.; Srikantan, S.; Martindale, J.L.; Hutchinson, E.R.; Kim, H.H.; Marasa, B.S.; et al. MiR-130 Suppresses Adipogenesis by Inhibiting PPAR{gamma} expression. Mol. Cell. Biol. 2010, 31, 626–638. [Google Scholar] [CrossRef]
- Yang, Z.; Bian, C.; Zhou, H.; Huang, S.; Wang, S.; Liao, L.; Zhao, R.C. MicroRNA Hsa-MiR-138 Inhibits Adipogenic Differentiation of Human Adipose Tissue-Derived Mesenchymal Stem Cells Through Adenovirus EID-1. Stem Cells Dev. 2011, 20, 259–267. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shaker, O.G.; Ismail, M.F.; Sayed, N.H. Peroxisome Proliferator-Activated Receptor Gamma in Obesity and Colorectal Cancer: The Role of Epigenetics. Sci. Rep. 2017, 7, 10714. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Hu, L.; Li, X.; Geng, J.; Dai, M.; Bai, X. MicroRNA-130b Promotes Lung Cancer Progression via PPARγ/VEGF-A/BCL-2-Mediated Suppression of Apoptosis. J. Exp. Clin. Cancer Res. 2016, 35, 1–15. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, F.; Mao, X.; Huang, J.; Yang, J.; Yin, X.; Wu, L.; Zheng, L.; Wang, Q. Elevation of MiR-27b by HPV16 E7 Inhibits PPARγ Expression and Promotes Proliferation and Invasion in Cervical Carcinoma Cells. Int. J. Oncol. 2015, 47, 1759–1766. [Google Scholar] [CrossRef]
- Winkler, I.; Bitter, C.; Winkler, S.; Weichenhan, D.; Thavamani, A.; Hengstler, J.G.; Borkham-Kamphorst, E.; Kohlbacher, O.; Plass, C.; Geffers, R.; et al. Identification of Pparγ-Modulated MiRNA Hubs That Target the Fibrotic Tumor Microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 454–463. [Google Scholar] [CrossRef]
- Chakraborty, C.; Doss, C.G.P.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of MiRNA in Insulin Signaling Pathway and Insulin Resistance: Micro-Molecules With a Major Role in Type-2 Diabetes. Wiley Interdiscip. Rev. RNA 2014, 5, 697–712. [Google Scholar] [CrossRef]
- Huang, F.; Chen, J.; Wang, J.; Zhu, P.; Lin, W. Palmitic Acid Induces MicroRNA-221 Expression to Decrease Glucose Uptake in HepG2 Cells via the PI3K/AKT/GLUT4 Pathway. BioMed Res. Int. 2019, 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Wu, Y.-Y.; Wei, L.-Q. MiR-221 Affects the Proliferation and Apoptosis of Laryngeal Cancer Cells through the PI3K/AKT Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1258–1263. [Google Scholar] [PubMed]
- Wu, X.; Huang, J.; Yang, Z.; Zhu, Y.; Zhang, Y.; Wang, J.; Yao, W. MicroRNA-221-3p Is Related to Survival and Promotes Tumour Progression in Pancreatic Cancer: A Comprehensive Study on Functions and Clinicopathological Value. Cancer Cell Int. 2020, 20, 1–25. [Google Scholar] [CrossRef]
- Pan, X.; Hong, X.; Lai, J.; Cheng, L.; Cheng, Y.; Yao, M.; Wang, R.; Hu, N. Exosomal MicroRNA-221-3p Confers Adriamycin Resistance in Breast Cancer Cells by Targeting PIK3R1. Front. Oncol. 2020, 10, 441. [Google Scholar] [CrossRef] [PubMed]
- Massillo, C.; Duca, R.B.; Lacunza, E.; Dalton, G.N.; Farré, P.L.; Taha, N.; Piccioni, F.; Scalise, G.D.; Gardner, K.; De Siervi, A. Adipose Tissue from Metabolic Syndrome Mice Induces an Aberrant MiRNA Signature Highly Relevant in Prostate Cancer Development. Mol. Oncol. 2020, 14, 2868–2883. [Google Scholar] [CrossRef] [PubMed]
- Ahonen, M.A.; Asghar, M.Y.; Parviainen, S.J.; Liebisch, G.; Höring, M.; Leidenius, M.; Fischer-Posovszky, P.; Wabitsch, M.; Mikkola, T.S.; Törnquist, K.; et al. Human Adipocyte Differentiation and Composition of Disease-Relevant Lipids Are Regulated by MiR-221-3p. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1, 1866. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Hua, Y.; Deng, F.; Wang, D.; Wu, Y.; Zhang, W.; Tang, J. MiR-145 in Cancer Therapy Resistance and Sensitivity: A Comprehensive Review. Cancer Sci. 2020, 111, 3122–3131. [Google Scholar] [CrossRef] [PubMed]
- Tait, S.; Baldassarre, A.; Masotti, A.; Calura, E.; Martini, P.; Varì, R.; Scazzocchio, B.; Gessani, S.; Del Corno, M. Integrated Transcriptome Analysis of Human Visceral Adipocytes Unravels Dysregulated MicroRNA-Long Non-Coding RNA-MRNA Networks in Obesity and Colorectal Cancer. Front. Oncol. 2020, 10, 1089. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 Regulate Insulin Sensitivity. Nat. Cell Biol. 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Li, M.; Pan, S.; Qiu, A. Roles of MicroRNA-221/222 in Type 2 Diabetic Patients With Post-Menopausal Breast Cancer. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.L.; Wahyono, A.; Aryandono, T.; Haryono, S.J. Caveolin-1 in Breast Cancer: Single Molecule Regulation of Multiple Key Signaling Pathways. Asian Pac. J. Cancer Prev. 2015, 16, 6803–6812. [Google Scholar] [CrossRef]
- Pucci, M.; Bravatà, V.; Forte, G.I.; Cammarata, F.P.; Messa, C.; Gilardi, M.C.; Minafra, L. Caveolin-1, Breast Cancer and Ionizing Radiation. Cancer Genom. Proteom. 2015, 12, 143–152. [Google Scholar]
- Shi, X.-Y.; Xiong, L.-X.; Xiao, L.; Meng, C.; Qi, G.-Y.; Li, W.-L. Downregulation of Caveolin-1 Upregulates the Expression of Growth Factors and Regulators in Co-Culture of Fibroblasts With Cancer Cells. Mol. Med. Rep. 2016, 13, 744–752. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, H.; Holly, J.; Bahl, A.; Perks, C. Inhibition of FASN and ERα Signalling During Hyperglycaemia-Induced Matrix-Specific EMT Promotes Breast Cancer Cell Invasion via a Caveolin-1-Dependent Mechanism. Cancer Lett. 2018, 419, 187–202. [Google Scholar] [CrossRef]
- Pandey, V.; Vijayakumar, M.V.; Ajay, A.K.; Malvi, P.; Bhat, M.K. Diet-Induced Obesity Increases Melanoma Progression: Involvement of Cav-1 and FASN. Int. J. Cancer 2011, 130, 497–508. [Google Scholar] [CrossRef]
- Karczewska-Kupczewska, M.; Stefanowicz, M.; Matulewicz, N.; Nikołajuk, A.; Straczkowski, M. Wnt Signaling Genes in Adipose Tissue and Skeletal Muscle of Humans With Different Degree of Insulin Sensitivity. J. Clin. Endocrinol. Metab. 2016, 101, 3079–3087. [Google Scholar] [CrossRef]
- Koni, M.; Pinnarò, V.; Brizzi, M.F. The Wnt Signalling Pathway: A Tailored Target in Cancer. Int. J. Mol. Sci. 2020, 21, 7697. [Google Scholar] [CrossRef]
- Debebe, A.; Medina, V.; Chen, C.-Y.; Mahajan, I.M.; Jia, C.; Fu, D.; He, L.; Zeng, N.; Stiles, B.W.; Chen, C.-L.; et al. Wnt/β-Catenin Activation and Macrophage Induction During Liver Cancer Development Following Steatosis. Oncogene 2017, 36, 6020–6029. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, F.; Zhao, Y.; He, K.; Zheng, X.; Pan, Y.; Shao, D.; Shang, P.; Yang, Y.; Zhang, D.; et al. Obesity-Associated MiR-27a Upregulation Promotes Hepatocellular Carcinoma Metastasis through Suppressing SFRP1. OncoTargets Ther. 2018, 11, 3281–3292. [Google Scholar] [CrossRef] [PubMed]
- Bulger, D.; Conley, J.; Conner, S.H.; Majumdar, G.; Solomon, S.S. Role of PTEN in TNFα Induced Insulin Resistance. Biochem. Biophys. Res. Commun. 2015, 461, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Naizabekov, S.; Chen, Z.; Tokay, T. Power of PTEN/AKT: Molecular Switch between Tumor Suppressors and Oncogenes. Oncol. Lett. 2016, 12, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Chen, M.; Pandolfi, P.P. The Functions and Regulation of the PTEN Tumour Suppressor: New Modes and Prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Brito, M.B.; Goulielmaki, E.; Papakonstanti, E.A. Focus on PTEN Regulation. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.-C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Patel, I.; Kumar, A.; Donovan, V.; Levenson, A.S. Grape Powder Supplementation Attenuates Prostate Neoplasia Associated With Pten Haploinsufficiency in Mice Fed High-Fat Diet. Mol. Nutr. Food Res. 2020, 64, e2000326. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in Metabolism and Metabolic Disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating miRNAs Regulate Gene Expression in Other Tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Sahasrabuddhe, N.A.; Huang, T.-C.; Kumar, P.; Yang, Y.; Ghosh, B.; Leach, S.D.; Chaerkady, R.; Pandey, A. Ablation of Dicer Leads to Widespread Perturbation of Signaling Pathways. Biochem. Biophys. Res. Commun. 2015, 463, 389–394. [Google Scholar] [CrossRef]
- Shan, W.; Sun, C.; Zhou, B.; Guo, E.; Lu, H.; Xia, M.; Li, K.; Weng, D.; Lin, X.; Meng, L.; et al. Role of Dicer As a Prognostic Predictor for Survival in Cancer Patients: A Systematic Review With a Meta-Analysis. Oncotarget 2016, 7, 72672–72684. [Google Scholar] [CrossRef]
- Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T.; et al. A MicroRNA Targeting Dicer for Metastasis Control. Cell 2010, 141, 1195–1207. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Cittelly, D.M.; Howe, E.N.; Spoelstra, N.S.; McKinsey, E.L.; LaPara, K.; Elias, A.; Yee, D.; Richer, J.K. MicroRNAs Link Estrogen Receptor Alpha Status and Dicer Levels in Breast Cancer. Horm. Cancer 2010, 1, 306–319. [Google Scholar] [CrossRef]
- Geng, L.; Sun, B.; Gao, B.; Wang, Z.; Quan, C.; Wei, F.; Fang, X.-D. MicroRNA-103 Promotes Colorectal Cancer by Targeting Tumor Suppressor DICER and PTEN. Int. J. Mol. Sci. 2014, 15, 8458–8472. [Google Scholar] [CrossRef]
- Su, Y.; Hsu, T.; Chen, H.; Su, C.; Huang, M.; Chuang, T.; Su, J.L.; Hsieh, C.; Chiu, C. ERK-mediated Transcriptional Activation of Dicer Is Involved in Gemcitabine Resistance of Pancreatic Cancer. J. Cell. Physiol. 2021, 236, 4420–4434. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Kulyté, A. MicroRNA Regulatory Networks in Human Adipose Tissue and Obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Huang, Y.; Zeng, H.; Hu, B.; Guan, L.; Zhang, H.; Yu, A.-M.; Johnson, C.; Gonzalez, F.J.; et al. PPARα Regulates Tumor Cell Proliferation and Senescence via a Novel Target Gene Carnitine Palmitoyltransferase 1C. Carcinogenesis 2017, 38, 474–483. [Google Scholar] [CrossRef]
- Rajarajan, D.; Selvarajan, S.; Raja, M.R.C.; Mahapatra, S.K.; Kasiappan, R. Genome-wide Analysis Reveals miR-3184-5p and miR-181c-3p As a Critical Regulator for adipocytes-associated Breast Cancer. J. Cell. Physiol. 2019, 234, 17959–17974. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, H.; Thakur, A.; Chen, T.; Xue, J.; Li, D.; Chen, M. FOXP4 Modulates Tumor Growth and Independently Associates With MiR-138 in Non-Small Cell Lung Cancer Cells. Tumor Biol. 2015, 36, 8185–8191. [Google Scholar] [CrossRef]
- Andersson, E.; Sandberg, R.; Lendahl, U. Notch Signaling: Simplicity in Design, Versatility in Function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Sharma, A.; Balaji, S.A.; Gowda, M.C.; Dighe, R.R.; Kumar, R.V.; Rangarajan, A. Coordinate Hyperactivation of Notch1 and Ras/MAPK Pathways Correlates With Poor Patient Survival: Novel Therapeutic Strategy for Aggressive Breast Cancers. Mol. Cancer Ther. 2014, 13, 3198–3209. [Google Scholar] [CrossRef]
- Bi, P.; Yue, F.; Karki, A.; Castro, B.; Wirbisky, S.E.; Wang, C.; Durkes, A.; Elzey, B.D.; Andrisani, O.M.; Bidwell, C.A.; et al. Notch Activation Drives Adipocyte Dedifferentiation and Tumorigenic Transformation in Mice. J. Exp. Med. 2016, 213, 2019–2037. [Google Scholar] [CrossRef] [PubMed]
- Battle, M.; Gillespie, C.; Quarshie, A.; Lanier, V.; Harmon, T.; Wilson, K.; Torroella-Kouri, M.; Gonzalez-Perez, R.R. Obesity Induced a Leptin-Notch Signaling Axis in Breast Cancer. Int. J. Cancer 2014, 134, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Picon-Ruiz, M.; Pan, C.; Drews-Elger, K.; Jang, K.; Besser, A.H.; Zhao, D.; Tarifa, C.M.; Kim, M.; Ince, T.A.; Azzam, D.J.; et al. Interactions between Adipocytes and Breast Cancer Cells Stimulate Cytokine Production and Drive Src/Sox2/MiR-302b–Mediated Malignant Progression. Cancer Res. 2016, 76, 491–504. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Hirsch, H.A.; Struhl, K. An Epigenetic Switch Involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 Links Inflammation to Cell Transformation. Cell 2009, 139, 693–706. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, L.; Zhang, L.; Li, R.; Liang, J.; Yu, W.; Sun, L.; Yang, X.; Wang, Y.; Zhang, Y.; et al. The Molecular Mechanism Governing the Oncogenic Potential of SOX2 in Breast Cancer. J. Biol. Chem. 2008, 283, 17969–17978. [Google Scholar] [CrossRef]
- Volinia, S.; Nuovo, G.; Drusco, A.; Costinean, S.; Abujarour, R.; Desponts, C.; Garofalo, M.; Baffa, R.; Aeqilan, R.; Maharry, K.; et al. Pluripotent Stem Cell MiRNAs and Metastasis in Invasive Breast Cancer. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef][Green Version]
- Seyhan, A.A.; Lopez, Y.N.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-Enriched MiRNAs Are Altered in the Circulation of Subjects With Diabetes: A Pilot Cross-Sectional Study. Sci. Rep. 2016, 6, 31479. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, S.; Mahdavi, R.; Alipoor, B.; Panahi, G.; Esfahani, E.N.; Razi, F.; Taghikhani, M.; Meshkani, R. Decreased Serum MicroRNA-21 Level Is Associated With Obesity in Healthy and Type 2 Diabetic Subjects. Arch. Physiol. Biochem. 2018, 124, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, L.-Z.; Yang, D.-G.; Zhang, Q.-Y.; Deng, Z.-N.; Wang, K.; Mao, X.-J. MiR-21 Antagomir Improves Insulin Resistance and Lipid Metabolism Disorder in Streptozotocin-Induced Type 2 Diabetes Mellitus Rats. Ann. Palliat. Med. 2020, 9, 394–404. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Zhang, W.; Zheng, X.; Chen, X. Decreased Insulin Resistance by Myo-Inositol Is Associated With Suppressed Interleukin 6/Phospho-STAT3 Signaling in a Rat Polycystic Ovary Syndrome Model. J. Med. Food 2020, 23, 375–387. [Google Scholar] [CrossRef]
- Ling, H.-Y.; Hu, B.; Hu, X.-B.; Zhong, J.; Feng, S.-D.; Qin, L.; Liu, G.; Wen, G.-B.; Liao, D.-F. MiRNA-21 Reverses High Glucose and High Insulin Induced Insulin Resistance in 3T3-L1 Adipocytes through Targeting Phosphatase and Tensin Homologue. Exp. Clin. Endocrinol. Diabetes 2012, 120, 553–559. [Google Scholar] [CrossRef]
- Mazloom, H.; Alizadeh, S.; Esfahani, E.N.; Razi, F.; Meshkani, R. Decreased Expression of MicroRNA-21 Is Associated With Increased Cytokine Production in Peripheral Blood Mononuclear Cells (PBMCs) of Obese Type 2 Diabetic and Non-Diabetic Subjects. Mol. Cell. Biochem. 2016, 419, 11–17. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, S.H.; Cho, H.H.; Shin, K.K.; Bae, Y.C.; Jung, J.S. MicroRNA 21 Regulates the Proliferation of Human Adipose Tissue-Derived Mesenchymal Stem Cells and High-Fat Diet-Induced Obesity Alters MicroRNA 21 Expression in White Adipose Tissues. J. Cell. Physiol. 2011, 227, 183–193. [Google Scholar] [CrossRef]
- Seeger, T.; Fischer, A.; Muhly-Reinholz, M.; Zeiher, A.M.; Dimmeler, S. Long-Term Inhibition of MiR-21 Leads to Reduction of Obesity in db/Db Mice. Obesity 2014, 22, 2352–2360. [Google Scholar] [CrossRef]
- He, Q.F.; Wang, L.X.; Zhong, J.M.; Hu, R.Y.; Fang, L.; Wang, H.; Gong, W.W.; Zhang, J.; Pan, J.; Yu, M. Circulating MicroRNA-21 Is Downregulated in Patients With Metabolic Syndrome. Biomed. Environ. Sci. 2016, 29, 385–389. [Google Scholar]
- Connolly, E.C.; Van Doorslaer, K.; Rogler, L.E.; Rogler, C.E. Overexpression of MiR-21 Promotes an In Vitro Metastatic Phenotype by Targeting the Tumor Suppressor RHOB. Mol. Cancer Res. 2010, 8, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Jajoo, S.; Mukherjea, D.; Kaur, T.; Sheehan, K.E.; Sheth, S.; Borse, V.; Rybak, L.P.; Ramkumar, V. Essential Role of NADPH Oxidase-Dependent Reactive Oxygen Species Generation in Regulating MicroRNA-21 Expression and Function in Prostate Cancer. Antioxid. Redox Signal 2013, 19, 1863–1876. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of Circulating MicroRNAs and Prediction of Aggressive Prostate Cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Amankwah, E.K.; Anegbe, E.; Park, H.; Pow-Sang, J.; Hakam, A.; Park, J.Y. MiR-21, MiR-221 and MiR-222 Expression and Prostate Cancer Recurrence Among Obese and Non-Obese Cases. Asian J. Androl. 2013, 15, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, Y.; Ang, L.; Li, X.; Liu, C.; Sun, B.; Lin, X.; Peng, Z.; Zhang, X.; Zhang, Q.; et al. Extracellular Vesicles-Derived OncomiRs Mediate Communication between Cancer Cells and Cancer-Associated Hepatic Stellate Cells in Hepatocellular Carcinoma Microenvironment. Carcinogenesis 2019, 41, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Ziyan, W.; Shuhua, Y.; Xiufang, W.; Xiaoyun, L. MicroRNA-21 Is Involved in Osteosarcoma Cell Invasion and Migration. Med. Oncol. 2011, 28, 1469–1474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fu, C.; Xu, Q.; Wei, X. Long Non-Coding RNA CASC7 Inhibits the Proliferation and Migration of Colon Cancer Cells via Inhibiting MicroRNA-21. Biomed. Pharmacother. 2017, 95, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, C.; Wang, Y.; Xu, B.; Zhang, W. LINC00312 Represses Proliferation and Metastasis of Colorectal Cancer Cells by Regulation of miR-21. J. Cell. Mol. Med. 2018, 22, 5565–5572. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Tang, Q.; Qiu, M.; Lang, N.; Li, M.; Zheng, Y.; Bi, F. MiR-21 Targets the Tumor Suppressor RhoB and Regulates Proliferation, Invasion and Apoptosis in Colorectal Cancer Cells. FEBS Lett. 2011, 585, 2998–3005. [Google Scholar] [CrossRef]
- Li, C.; Zhao, L.; Chen, Y.; He, T.; Chen, X.; Mao, J.; Li, C.; Lyu, J.; Meng, Q.H. MicroRNA-21 Promotes Proliferation, Migration, and Invasion of Colorectal Cancer, and Tumor Growth Associated With down-Regulation of sec23a Expression. BMC Cancer 2016, 16, 1–11. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, K.; Li, T.; Fang, J.; Ding, Y.; Sun, L.; Tu, T.; Jiang, X.; Du, S.; Hu, J.; et al. MiR-21: A Gene of Dual Regulation in Breast Cancer. Int. J. Oncol. 2015, 48, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Yan-Hui, L.; Liu, Y.-H.; Xiang, J.-W.; Wu, Q.-N.; Xu, L.-B.; Luo, X.-L.; Zhu, X.-L.; Liu, C.; Xu, F.-P.; Luo, D.-L.; et al. PIK3R1 Targeting by MiR-21 Suppresses Tumor Cell Migration and Invasion by Reducing PI3K/AKT Signaling and Reversing EMT, and Predicts Clinical Outcome of Breast Cancer. Int. J. Oncol. 2016, 48, 471–484. [Google Scholar] [CrossRef]
- Zhang, J.-G.; Wang, J.-J.; Zhao, F.; Liu, Q.; Jiang, K.; Yang, G.-H. MicroRNA-21 (miR-21) Represses Tumor Suppressor PTEN and Promotes Growth and Invasion in Non-Small Cell Lung Cancer (NSCLC). Clin. Chim. Acta 2010, 411, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Zang, C.; Sun, J.; Liu, W.; Chu, C.; Jiang, L.; Ge, R. MiRNA-21 Promotes Cell Proliferation and Invasion via VHL/PI3K/AKT in Papillary Thyroid Carcinoma. Hum. Cell 2019, 32, 428–436. [Google Scholar] [CrossRef]
- Li, L.; Zhou, L.; Li, Y.; Lin, S.; Tomuleasa, C. MicroRNA-21 Stimulates Gastric Cancer Growth and Invasion by Inhibiting the Tumor Suppressor Effects of Programmed Cell Death Protein 4 and Phosphatase and Tensin Homolog. J. BUON. 2014, 19, 228–236. [Google Scholar]
- Del Campo, S.E.M.; Latchana, N.; Levine, K.M.; Grignol, V.P.; Fairchild, E.T.; Jaime-Ramirez, A.C.; Dao, T.-V.; Karpa, V.I.; Carson, M.; Ganju, A.; et al. MiR-21 Enhances Melanoma Invasiveness via Inhibition of Tissue Inhibitor of Metalloproteinases 3 Expression: In Vivo Effects of MiR-21 Inhibitor. PLoS ONE 2015, 10, e0115919. [Google Scholar] [CrossRef]
- Luo, G.; Luo, W.; Sun, X.; Lin, J.; Wang, M.; Zhang, Y.; Luo, W.; Zhang, Y. MicroRNA-21 Promotes Migration and Invasion of Glioma Cells via Activation of Sox2 and β-Catenin Signaling. Mol. Med. Rep. 2016, 15, 187–193. [Google Scholar] [CrossRef]
- Han, L.; Yue, X.; Zhou, X.; Lan, F.; You, G.; Zhang, W.; Zhang, K.; Zhang, C.; Cheng, J.; Yu, S.; et al. MicroRNA-21 Expression Is Regulated by β-catenin/STAT3 Pathway and Promotes Glioma Cell Invasion by Direct Targeting RECK. CNS Neurosci. Ther. 2012, 18, 573–583. [Google Scholar] [CrossRef]
- Zheng, Y.; Xie, J.; Jiang, F.; Li, Y.; Chang, G.; Ma, H. Inhibition of miR-21 Promotes Cell Apoptosis in Oral Squamous Cell Carcinoma by Upregulating PTEN. Oncol. Rep. 2018, 40. [Google Scholar] [CrossRef]
- Kawakita, A.; Yanamoto, S.; Yamada, S.-I.; Naruse, T.; Takahashi, H.; Kawasaki, G.; Umeda, M. MicroRNA-21 Promotes Oral Cancer Invasion via the Wnt/β-Catenin Pathway by Targeting DKK2. Pathol. Oncol. Res. 2013, 20, 253–261. [Google Scholar] [CrossRef]
- Li, Y.; Yan, L.; Zhang, W.; Wang, H.; Chen, W.; Hu, N.; Ou, H. MiR-21 Inhibitor Suppresses Proliferation and Migration of Nasopharyngeal Carcinoma Cells through down-Regulation of BCL2 Expression. Int. J. Clin. Exp. Pathol. 2014, 7, 3478–3487. [Google Scholar]
- Xu, J.; Zhang, W.; Lv, Q.; Zhu, D. Overexpression of MiR-21 Promotes the Proliferation and Migration of Cervical Cancer Cells via the Inhibition of PTEN. Oncol. Rep. 2015, 33, 3108–3116. [Google Scholar] [CrossRef]
- Ahrend, H.; Kaul, A.; Ziegler, S.; Brandenburg, L.-O.; Zimmermann, U.; Mustea, A.; Burchardt, M.; Ziegler, P.; Stope, B.M. MicroRNA-1 and MicroRNA-21 Individually Regulate Cellular Growth of Non-Malignant and Malignant Renal Cells. Vivo 2017, 31, 625–630. [Google Scholar] [CrossRef][Green Version]
- Song, L.; Yang, J.; Duan, P.; Xu, J.; Luo, X.; Luo, F.; Zhang, Z.; Hou, T.; Liu, B.; Zhou, Q. MicroRNA-24 Inhibits Osteosarcoma Cell Proliferation Both in Vitro and in Vivo by Targeting LPAATβ. Arch. Biochem. Biophys. 2013, 535, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Du, L.; Li, J.; Qu, A.; Zhang, X.; Wang, L.; Wang, C. Down-Regulation of MiR-24-3p in Colorectal Cancer Is Associated With Malignant Behavior. Med. Oncol. 2014, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.J.; Song, B.; Mishra, P.J.; Wang, Y.; Humeniuk, R.; Banerjee, D.; Merlino, G.; Ju, J.; Bertino, J.R. MiR-24 Tumor Suppressor Activity Is Regulated Independent of p53 and through a Target Site Polymorphism. PLoS ONE 2009, 4, e8445. [Google Scholar] [CrossRef]
- Lu, K.; Wang, J.; Song, Y.; Zhao, S.; Liu, H.; Tang, D.; Pan, B.; Zhao, H.; Zhang, Q. MiRNA-24-3p Promotes Cell Proliferation and Inhibits Apoptosis in Human Breast Cancer by Targeting p27Kip1. Oncol. Rep. 2015, 34, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, Q.; Liu, C.; Wang, C.; Li, Y. Overexpression miR-24-3p Repressed Bim Expression to Confer Tamoxifen Resistance in Breast Cancer. J. Cell. Biochem. 2019, 120, 12966–12976. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, K.; Xu, Y.; Gao, Y.; Li, C.; Wang, R.; Chen, L. The Role of MicroRNA-26a in Human Cancer Progression and Clinical Application. Tumor Biol. 2016, 37, 7095–7108. [Google Scholar] [CrossRef]
- Gao, J.; Li, L.; Wu, M.; Liu, M.; Xie, X.; Guo, J.; Tang, H.; Xie, X. MiR-26a Inhibits Proliferation and Migration of Breast Cancer through Repression of MCL-1. PLoS ONE 2013, 8, e65138. [Google Scholar] [CrossRef]
- M’Hamed, I.F.; Privat, M.; Ponelle, F.; Penault-Llorca, F.; Kenani, A.; Bignon, Y.-J. Identification of MiR-10b, MiR-26a, MiR-146a and MiR-153 As Potential Triple-Negative Breast Cancer Biomarkers. Cell. Oncol. 2015, 38, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, Z.; Zheng, C.; Li, D.; Yang, K.; Cai, W. MiR-26b Inhibits Proliferation, Invasion, and Migration of Glioma by Targeting Cyclooxygenase-2. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2017, 42. [Google Scholar] [CrossRef]
- Xia, M.; Duan, M.; Tong, J.-H.; Xu, J.-G. MiR-26b Suppresses Tumor Cell Proliferation, Migration and Invasion by Directly Targeting COX-2 in Lung Cancer. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 4728–4737. [Google Scholar]
- Li, H.; Sun, Q.; Han, B.; Yu, X.; Hu, B.; Hu, S. MiR-26b Inhibits Hepatocellular Carcinoma Cell Proliferation, Migration, and Invasion by Targeting EphA2. Int. J. Clin. Exp. Pathol. 2015, 8, 4782–4790. [Google Scholar]
- Zhang, Z.; Liu, S.; Shi, R.; Zhao, G. MiR-27 Promotes Human Gastric Cancer Cell Metastasis by Inducing Epithelial-to-Mesenchymal Transition. Cancer Genet. 2011, 204, 486–491. [Google Scholar] [CrossRef]
- Kang, T.; Lu, W.; Xu, W.; Anderson, L.; Bacanamwo, M.; Thompson, W.; Chen, Y.E.; Liu, D. MicroRNA-27 (miR-27) Targets Prohibitin and Impairs Adipocyte Differentiation and Mitochondrial Function in Human Adipose-Derived Stem Cells. J. Biol. Chem. 2013, 288, 34394–34402. [Google Scholar] [CrossRef]
- Li, X.; Xu, M.; Ding, L.; Tang, J. MiR-27a: A Novel Biomarker and Potential Therapeutic Target in Tumors. J. Cancer 2019, 10, 2836–2848. [Google Scholar] [CrossRef]
- Yao, J.; Deng, B.; Zheng, L.; Dou, L.; Guo, Y.; Guo, K. MiR-27b Is Upregulated in Cervical Carcinogenesis and Promotes Cell Growth and Invasion by Regulating CDH11 and Epithelial-Mesenchymal Transition. Oncol. Rep. 2015, 35, 1645–1651. [Google Scholar] [CrossRef]
- Jiang, J.; Lv, X.; Fan, L.; Huang, G.; Zhan, Y.; Wang, M.; Lu, H. MicroRNA-27b Suppresses Growth and Invasion of NSCLC Cells by Targeting Sp1. Tumor Biol. 2014, 35, 10019–10023. [Google Scholar] [CrossRef]
- Bao, S.; Wang, X.; Wang, Z.; Yang, J.; Liu, F.; Yin, C. MicroRNA-30 Mediates Cell Invasion and Metastasis in Breast Cancer. Biochem. Cell Biol. 2018, 96, 825–831. [Google Scholar] [CrossRef]
- Yu, G.; Herazo-Maya, J.D.; Nukui, T.; Romkes, M.; Parwani, A.; Juan-Guardela, B.M.; Robertson, J.; Gauldie, J.; Siegfried, J.M.; Kaminski, N.; et al. Matrix Metalloproteinase-19 Promotes Metastatic BehaviorIn Vitroand Is Associated With Increased Mortality in Non–Small Cell Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 780–790. [Google Scholar] [CrossRef]
- Tsukasa, K.; Ding, Q.; Miyazaki, Y.; Matsubara, S.; Natsugoe, S.; Takao, S. MiR-30 Family Promotes Migratory and Invasive Abilities in CD133+ Pancreatic Cancer Stem-Like Cells. Hum. Cell 2016, 29, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Qiang, H.; Zhan, X.; Wang, W.; Cheng, Z.; Ma, S.; Jiang, C. A Study on the Correlations of the MiR-31 Expression With the Pathogenesis and Prognosis of Head and Neck Squamous Cell Carcinoma. Cancer Biother. Radiopharm 2019, 34, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.-F.; Wang, X.-D.; Sun, P.-D. Expression of MiR-31 in Rectal Cancer Patients and Its Effect on Proliferation Ability of Rectal Cancer Cells SW837. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8675–8681. [Google Scholar] [PubMed]
- Chen, T.; Yao, L.-Q.; Shi, Q.; Ren, Z.; Ye, L.-C.; Xu, J.-M.; Zhou, P.-H.; Zhong, Y.-S. MicroRNA-31 Contributes to Colorectal Cancer Development by Targeting Factor Inhibiting HIF-1α (FIH-1). Cancer Biol. Ther. 2014, 15, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. RETRACTED: A Pleiotropically Acting MicroRNA, MiR-31, Inhibits Breast Cancer Metastasis. Cell 2009, 137, 1032–1046. [Google Scholar] [CrossRef]
- Ivanov, S.V.; Goparaju, C.M.V.; Lopez, P.; Zavadil, J.; Toren-Haritan, G.; Rosenwald, S.; Hoshen, M.; Chajut, A.; Cohen, D.; Pass, H.I. Pro-Tumorigenic Effects of MiR-31 Loss in Mesothelioma. J. Biol. Chem. 2010, 285, 22809–22817. [Google Scholar] [CrossRef]
- Zhang, B.; Li, H.; Yin, C.; Sun, X.; Zheng, S.; Zhang, C.; Shi, L.; Liu, Y.; Lu, S. Dock1 Promotes the Mesenchymal Transition of Glioma and Is Modulated by MiR-31. Neuropathol. Appl. Neurobiol. 2016, 43, 419–432. [Google Scholar] [CrossRef]
- Creighton, C.J.; Fountain, M.D.; Yu, Z.; Nagaraja, A.K.; Zhu, H.; Khan, M.; Olokpa, E.; Zariff, A.; Gunaratne, P.H.; Matzuk, M.M.; et al. Molecular Profiling Uncovers a p53-Associated Role for MicroRNA-31 in Inhibiting the Proliferation of Serous Ovarian Carcinomas and Other Cancers. Cancer Res. 2010, 70, 1906–1915. [Google Scholar] [CrossRef]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. MiR-34a Repression of SIRT1 Regulates Apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef] [PubMed]
- Rui, X.; Zhao, H.; Xiao, X.; Wang, L.; Mo, L.; Yao, Y. MicroRNA-34a Suppresses Breast Cancer Cell Proliferation and Invasion by Targeting Notch1. Exp. Ther. Med. 2018, 16, 4387–4392. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Li, Y.; Shao, H.; Hu, R.; Wang, W.; Zhang, K.; Yang, Q. MiR-34a Inhibits Breast Cancer Proliferation and Progression by Targeting Wnt1 in Wnt/β-Catenin Signaling Pathway. Am. J. Med Sci. 2016, 352, 191–199. [Google Scholar] [CrossRef]
- Yang, S.; Li, Y.; Gao, J.; Zhang, T.; Li, S.; Luo, A.; Chen, H.; Ding, F.; Wang, X.; Liu, Z. MicroRNA-34 Suppresses Breast Cancer Invasion and Metastasis by Directly Targeting Fra-1. Oncogene 2012, 32, 4294–4303. [Google Scholar] [CrossRef]
- Christoffersen, N.R.; Shalgi, R.; Frankel, L.B.; Leucci, E.; Lees, M.; Klausen, M.; Pilpel, Y.; Nielsen, F.C.; Oren, M.; Lund, A. p53-Independent Upregulation of MiR-34a During Oncogene-Induced Senescence Represses MYC. Cell Death Differ. 2009, 17, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Javeri, A.; Ghaffarpour, M.; Taha, M.F.; Houshmand, M. Downregulation of MiR-34a in Breast Tumors Is Not Associated With Either p53 Mutations or Promoter Hypermethylation While It Correlates With Metastasis. Med. Oncol. 2013, 30, 1–10. [Google Scholar] [CrossRef]
- Welch, C.; Chen, Y.; Stallings, R.L. MicroRNA-34a Functions As a Potential Tumor Suppressor by Inducing Apoptosis in Neuroblastoma Cells. Oncogene 2007, 26, 5017–5022. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Neumann, J.; Jackstadt, R.; Mansmann, U.; Horst, D.; Kirchner, T.; Hermeking, H. Detection of MiR-34a Promoter Methylation in Combination With Elevated Expression of C-Met and β-Catenin Predicts Distant Metastasis of Colon Cancer. Clin. Cancer Res. 2013, 19, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Novello, C.; Pazzaglia, L.; Conti, A.; Quattrini, I.; Pollino, S.; Perego, P.; Picci, P.; Benassi, M.S. p53-Dependent Activation of MicroRNA-34a in Response to Etoposide-Induced DNA Damage in Osteosarcoma Cell Lines Not Impaired by Dominant Negative p53 Expression. PLoS ONE 2014, 9, e114757. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.-I.; Choi, J.; Zhou, Z.; Flesken-Nikitin, A.; Tarakhovsky, A.; Nikitin, A.Y. MET-Dependent Cancer Invasion May Be Preprogrammed by Early Alterations of p53-Regulated Feedforward Loop and Triggered by Stromal Cell-Derived HGF. Cell Cycle 2011, 10, 3834–3840. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Hwang, C.-I.; Matoso, A.; Vogt, M.; Flesken-Nikitin, A.; Godwin, A.K.; Kamat, A.A.; Sood, A.K.; Ellenson, L.H.; Hermeking, H.; et al. Frequent Downregulation of MiR-34 Family in Human Ovarian Cancers. Clin. Cancer Res. 2010, 16, 1119–1128. [Google Scholar] [CrossRef]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.R.; Honorio, S.; et al. The MicroRNA MiR-34a Inhibits Prostate Cancer Stem Cells and Metastasis by Directly Repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef]
- Bommer, G.T.; Gerin, I.; Feng, Y.; Kaczorowski, A.J.; Kuick, R.; Love, R.E.; Zhai, Y.; Giordano, T.J.; Qin, Z.S.; Moore, B.; et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr. Biol. 2007, 17, 1298–1307. [Google Scholar] [CrossRef]
- Mudduluru, G.; Ceppi, P.; Kumarswamy, R.; Scagliotti, G.V.; Papotti, M.; Allgayer, H. Regulation of Axl Receptor Tyrosine Kinase Expression by MiR-34a and MiR-199a/B in Solid Cancer. Oncogene 2011, 30, 2888–2899. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y. MiR-96 Targets SOX6 and Promotes Proliferation, Migration, and Invasion of Hepatocellular Carcinoma. Biochem. Cell Biol. 2018, 96, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-H.; Yeh, C.-T.; Ho, J.-Y.; Ng, K.-F.; Chen, T.-C. OncomiR MiR-96 and MiR-182 Promote Cell Proliferation and Invasion through Targeting EphrinA5 in Hepatocellular Carcinoma. Mol. Carcinog. 2016, 55, 366–375. [Google Scholar] [CrossRef]
- Hong, Y.; Liang, H.; Rehman, U.-U.; Wang, Y.; Zhang, W.; Zhou, Y.; Chen, S.; Yu, M.; Cui, S.; Liu, M.; et al. MiR-96 Promotes Cell Proliferation, Migration and Invasion by Targeting PTPN9 in Breast Cancer. Sci. Rep. 2016, 6, 37421. [Google Scholar] [CrossRef]
- Guttilla, I.K.; White, B.A. Coordinate Regulation of FOXO1 by MiR-27a, MiR-96, and MiR-182 in Breast Cancer Cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef]
- Lin, H.; Dai, T.; Xiong, H.; Zhao, X.; Chen, X.; Yu, C.; Li, J.; Wang, X.; Song, L. Unregulated MiR-96 Induces Cell Proliferation in Human Breast Cancer by Downregulating Transcriptional Factor FOXO3a. PLoS ONE 2010, 5, e15797. [Google Scholar] [CrossRef]
- Li, J.; Li, P.; Chen, T.; Gao, G.; Chen, X.; Du, Y.; Zhang, R.; Yang, R.; Zhao, W.; Dun, S.; et al. Expression of MicroRNA-96 and Its Potential Functions by Targeting FOXO3 in Non-Small Cell Lung Cancer. Tumor Biol. 2015, 36, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, S.; Chen, K.; Huang, H.; Ma, H. MiR-96 Promotes Proliferation and Chemo- or Radioresistance by down-Regulating RECK in Esophageal Cancer. Biomed. Pharmacother. 2014, 68, 951–958. [Google Scholar] [CrossRef]
- Feng, S.; Yao, J.; Zhang, Z.; Zhang, Y.; Zhang, Z.; Liu, J.; Tan, W.; Sun, C.; Chen, L.; Zhiyuan, Z. miR-96 Inhibits EMT by Targeting AEG-1 in Glioblastoma Cancer Cells. Mol. Med. Rep. 2017, 17, 2964–2972. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Fu, S.; Liu, Y.; Xu, Z.; Liu, Y.; Hao, J.; Wang, B.; Zhang, A. MiR-96 Suppresses Renal Cell Carcinoma Invasion via Downregulation of Ezrin Expression. J. Exp. Clin. Cancer Res. 2015, 34, 107. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lu, Z.; Liu, C.; Meng, Y.; Ma, Y.; Zhao, W.; Liu, J.; Yu, J.; Chen, J. MiRNA-96 Suppresses KRAS and Functions As a Tumor Suppressor Gene in Pancreatic Cancer. Cancer Res. 2010, 70, 6015–6025. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xue, S.; Dai, Y.; Yang, J.; Chen, Z.; Fang, X.; Zhou, W.; Wu, W.; Li, Q. Reduced Expression of MicroRNA-100 Confers Unfavorable Prognosis in Patients With Bladder Cancer. Diagn. Pathol. 2012, 7, 159. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, B.; Wang, C.; Chen, Y.; Du, C.; Zhao, G.; Zhou, Y.; Shi, Y. Prognostic Role of MicroRNA-100 in Various Carcinomas: Evidence from Six Studies. Tumor Biol. 2013, 35, 3067–3071. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Deregulation of MiR-100, MiR-99a and MiR-199b in Tissues and Plasma Coexists With Increased Expression of MTOR Kinase in Endometrioid Endometrial Carcinoma. BMC Cancer 2012, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Xi, Q.; Wang, Q.; Wei, P. Downregulation of MicroRNA-100 Correlates With Tumor Progression and Poor Prognosis in Colorectal Cancer. Med. Oncol. 2014, 31, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-W.; Ramasamy, K.; Bouamar, H.; Lin, A.-P.; Jiang, D.; Aguiar, R.C.T. MicroRNAs MiR-125a and MiR-125b Constitutively Activate the NF- B Pathway by Targeting the Tumor Necrosis Factor Alpha-Induced Protein 3 (TNFAIP3, A20). Proc. Natl. Acad. Sci. USA 2012, 109, 7865–7870. [Google Scholar] [CrossRef] [PubMed]
- So, A.Y.-L.; Sookram, R.; Chaudhuri, A.A.; Minisandram, A.; Cheng, D.; Xie, C.; Lim, E.L.; Flores, Y.G.; Jiang, S.; Kim, J.T.; et al. Dual Mechanisms by Which MiR-125b Represses IRF4 to Induce Myeloid and B-Cell Leukemias. Blood 2014, 124, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Theng, P.Y.; Le, M.T.N. Essential Functions of miR-125b in Cancer. Cell Prolif. 2021, 54, e12913. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Gopalan, V.; Smith, R.A.; Lam, A.K.-Y. MiR-126 in Human Cancers: Clinical Roles and Current Perspectives. Exp. Mol. Pathol. 2014, 96, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Lee, H.; Cho, Y.M.; Kwon, O.-J.; Kim, W.; Lee, E.K. TNFα-Induced MiR-130 Resulted in Adipocyte Dysfunction During Obesity-Related Inflammation. FEBS Lett. 2013, 587, 3853–3858. [Google Scholar] [CrossRef]
- Teoh, S.L.; Das, S. Tumour Biology of Obesity-Related Cancers: Understanding the Molecular Concept for Better Diagnosis and Treatment. Tumor Biol. 2016, 37, 14363–14380. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, X.; Liang, H.; Deng, T.; Chen, W.; Zhang, S.; Liu, M.; Gao, X.; Liu, Y.; Zhao, C.; et al. MiR-143 and MiR-145 Synergistically Regulate ERBB3 to Suppress Cell Proliferation and Invasion in Breast Cancer. Mol. Cancer 2014, 13, 1–14. [Google Scholar] [CrossRef]
- Tavanafar, F.; Safaralizadeh, R.; Hosseinpour-Feizi, M.A.; Mansoori, B.; Shanehbandi, D.; Mohammadi, A.; Baradaran, B. Restoration of MiR-143 Expression Could Inhibit Migration and Growth of MDA-MB-468 Cells through down-Regulating the Expression of Invasion-Related Factors. Biomed. Pharmacother. 2017, 91, 920–924. [Google Scholar] [CrossRef]
- Mao, Y.; Liu, J.; Zhang, D.; Li, B. MiR-143 Inhibits Tumor Progression by Targeting FAM83F in Esophageal Squamous Cell Carcinoma. Tumor Biol. 2016, 37, 9009–9022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Feng, Y.; Liu, P.; Yang, J. MiR-143 Inhibits Cell Proliferation and Invasion by Targeting DNMT3A in Gastric Cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Liu, X.; Gong, J.; Xu, B. MiR-143 down-Regulates TLR2 Expression in Hepatoma Cells and Inhibits Hepatoma Cell Proliferation and Invasion. Int. J. Clin. Exp. Pathol. 2015, 8, 12738–12747. [Google Scholar]
- Hirahata, M.; Osaki, M.; Kanda, Y.; Sugimoto, Y.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Kawai, A.; Ito, H.; et al. PAI -1, a Target Gene of miR-143, Regulates Invasion and Metastasis by Upregulating MMP -13 Expression of Human Osteosarcoma. Cancer Med. 2016, 5, 892–902. [Google Scholar] [CrossRef]
- Cheng, T.; Hu, C.; Yang, H.; Cao, L.; An, J. Transforming Growth Factor-β-Induced MiR-143 Expression in Regulation of Non-Small Cell Lung Cancer Cell Viability and Invasion Capacity in Vitro and in Vivo. Int. J. Oncol. 2014, 45, 1977–1988. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Tang, J.; Wang, P.; Li, L.; Yan, X.; Zheng, X.; Ren, S.; Zhang, M.; Xu, M. The Prognostic Value and Regulatory Mechanisms of MicroRNA-145 in Various Tumors: A Systematic Review and Meta-Analysis of 50 Studies. Cancer Epidemiol. Biomark. Prev. 2019, 28, 867–881. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Guo, J.; Wang, L. MiR-148a-3p Suppresses the Proliferation and Invasion of Esophageal Cancer by Targeting DNMT1. Genet. Test. Mol. Biomarkers 2019, 23, 98–104. [Google Scholar] [CrossRef]
- Tuysuz, E.C.; Gulluoglu, S.; Yaltirik, C.K.; Ozbey, U.; Kuskucu, A.; Çoban, E.A.; Sahin, F.; Türe, U.; Bayrak, O.F. Distinctive Role of Dysregulated MiRNAs in Chordoma Cancer Stem-Like Cell Maintenance. Exp. Cell Res. 2019, 380, 9–19. [Google Scholar] [CrossRef]
- Yao, J.; Lin, J.; He, L.; Huang, J.; Liu, Q. TNF-α/MiR-155 Axis Induces the Transformation of Osteosarcoma Cancer Stem Cells Independent of TP53INP1. Gene 2020, 726, 144224. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Liao, Q.S.; Tang, L. MiR-155 Affects Osteosarcoma Cell Proliferation and Invasion through Regulating NF-κB Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22. [Google Scholar] [CrossRef]
- Petrović, N.; Kolaković, A.; Stanković, A.; Lukic, S.; Řami, A.; Ivković, M.; Mandušić, V. MiR-155 Expression Level Changes Might Be Associated With Initial Phases of Breast Cancer Pathogenesis and Lymph-Node Metastasis. Cancer Biomark. 2016, 16, 385–394. [Google Scholar] [CrossRef]
- Soon, P.S.; Provan, P.J.; Kim, E.; Pathmanathan, N.; Graham, D.; Clarke, C.; Balleine, R.L. Profiling Differential MicroRNA Expression between in Situ, Infiltrative and Lympho-Vascular Space Invasive Breast Cancer: A Pilot Study. Clin. Exp. Metastasis 2018, 35, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Kono, H.; Nakamura, M.; Ohtsuka, T.; Nagayoshi, Y.; Mori, Y.; Takahata, S.; Aishima, S.; Tanaka, M. High Expression of MicroRNA-155 Is Associated With the Aggressive Malignant Behavior of Gallbladder Carcinoma. Oncol. Rep. 2013, 30, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Liu, H.; Liu, C. MiR-155 Promotes Uveal Melanoma Cell Proliferation and Invasion by Regulating NDFIP1 Expression. Technol. Cancer Res. Treat. 2017, 16, 1160–1167. [Google Scholar] [CrossRef]
- Wang, P.; Xu, L.-J.; Qin, J.-J.; Zhang, L.; Zhuang, G.-H. MicroRNA-155 Inversely Correlates With Esophageal Cancer Progression through Regulating Tumor-Associated Macrophage FGF2 Expression. Biochem. Biophys. Res. Commun. 2018, 503, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, Y.; Sun, Y.; Zheng, J.; Zhu, D. MiR-155 up-Regulation by LMP1 DNA Contributes to Increased Nasopharyngeal Carcinoma Cell Proliferation and Migration. Eur. Arch. Oto Rhino Laryngol. 2013, 271, 1939–1945. [Google Scholar] [CrossRef]
- Olivo-Marston, S.E.; Hursting, S.D.; Perkins, S.N.; Schetter, A.; Khan, M.; Croce, C.; Harris, C.C.; Lavigne, J. Effects of Calorie Restriction and Diet-Induced Obesity on Murine Colon Carcinogenesis, Growth and Inflammatory Factors, and MicroRNA Expression. PLoS ONE 2014, 9, e94765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-L.; Zhang, J.-H. MiR-181c Promotes Proliferation via Suppressing PTEN Expression in Inflammatory Breast Cancer. Int. J. Oncol. 2015, 46, 2011–2020. [Google Scholar] [CrossRef]
- Yamazaki, N.; Koga, Y.; Taniguchi, H.; Kojima, M.; Kanemitsu, Y.; Saito, N.; Matsumura, Y. High Expression of MiR-181c As a Predictive Marker of Recurrence in Stage II Colorectal Cancer. Oncotarget 2016, 8, 6970–6983. [Google Scholar] [CrossRef]
- Chen, M.; Wang, M.; Xu, S.; Guo, X.; Jiang, J. Upregulation of MiR-181c Contributes to Chemoresistance in Pancreatic Cancer by Inactivating the Hippo Signaling Pathway. Oncotarget 2015, 6, 44466–44479. [Google Scholar] [CrossRef]
- Zabaglia, L.M.; Bartolomeu, N.C.; Santos, M.; Peruquetti, R.L.; Chen, E.; Smith, M.; Payão, S.L.M.; Rasmussen, L.T. Decreased MicroRNA MiR-181c Expression Associated With Gastric Cancer. J. Gastrointest. Cancer 2017, 49, 97–101. [Google Scholar] [CrossRef]
- Lakomy, R.; Sana, J.; Hankeova, S.; Fadrus, P.; Kren, L.; Lzicarova, E.; Svoboda, M.; Dolezelova, H.; Smrcka, M.; Vyzula, R.; et al. MiR-195, MiR-196b, MiR-181c, MiR-21 Expression Levels and O-6-Methylguanine-DNA Methyltransferase Methylation Status Are Associated With Clinical Outcome in Glioblastoma Patients. Cancer Sci. 2011, 102, 2186–2190. [Google Scholar] [CrossRef]
- Wang, J.; Hao, F.; Fei, X.; Chen, Y. SPP1 Functions As an Enhancer of Cell Growth in Hepatocellular Carcinoma Targeted by MiR-181c. Am. J. Transl. Res. 2019, 11, 6924–6937. [Google Scholar]
- Fu, Y.; Tang, Y.; Wang, J.; Guo, Z. MicroRNA-181c Suppresses the Biological Progression of Osteosarcoma via Targeting SMAD7 and Regulating Transforming Growth Factor-β (TGF-β) Signaling Pathway. Med. Sci. Monit. 2019, 25, 4801–4810. [Google Scholar] [CrossRef]
- Cao, Y.-P.; Pan, M.; Song, Y.-L.; Zhang, H.-L.; Sui, H.-T.; Shan, B.-C.; Piao, H.-X. MiR-302 a/b/C Suppresses Tumor Angiogenesis in Hepatocellular Carcinoma by Targeting MACC1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7863–7873. [Google Scholar] [PubMed]
- Hulin, J.-A.; Tommasi, S.; Elliot, D.; Hu, D.G.; Lewis, B.C.; Mangoni, A.A. MiR-193b Regulates Breast Cancer Cell Migration and Vasculogenic Mimicry by Targeting Dimethylarginine Dimethylaminohydrolase 1. Sci. Rep. 2017, 7, 13996. [Google Scholar] [CrossRef] [PubMed]
- Khordadmehr, M.; Shahbazi, R.; Sadreddini, S.; Baradaran, B. miR-193: A New Weapon Against Cancer. J. Cell. Physiol. 2019, 234, 16861–16872. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Liu, G.; Huang, J.; Hu, H.; Jiang, W. miR-210 Promotes Lung Adenocarcinoma Proliferation, Migration, and Invasion by Targeting Lysyl oxidase-like 4. J. Cell. Physiol. 2019, 234, 14050–14057. [Google Scholar] [CrossRef] [PubMed]
- Barbano, R.; Palumbo, O.; Pasculli, B.; Galasso, M.; Volinia, S.; D’Angelo, V.; Icolaro, N.; Coco, M.; Dimitri, L.; Graziano, P.; et al. A MiRNA Signature for Defining Aggressive Phenotype and Prognosis in Gliomas. PLoS ONE 2014, 9, e108950. [Google Scholar] [CrossRef]
- Liu, C.; Tang, X. Downregulation of MicroRNA-210 Inhibits Osteosarcoma Growth in Vitro and in Vivo. Mol. Med. Rep. 2015, 12, 3674–3680. [Google Scholar] [CrossRef]
- Ni, J.; Zhou, S.; Yuan, W.; Cen, F.; Yan, Q. Mechanism of MiR-210 Involved in epithelial–mesenchymal Transition of Pancreatic Cancer Cells under Hypoxia. J. Recept. Signal. Transduct. 2019, 39, 399–406. [Google Scholar] [CrossRef]
- Wang, H.; Xu, C.; Kong, X.; Li, X.; Kong, X.; Wang, Y.; Ding, X.; Yang, Q. Trail Resistance Induces Epithelial-Mesenchymal Transition and Enhances Invasiveness by Suppressing PTEN via MiR-221 in Breast Cancer. PLoS ONE 2014, 9, e99067. [Google Scholar] [CrossRef]
- Ye, X.; Bai, W.; Zhu, H.; Zhang, X.; Chen, Y.; Wang, L.; Yang, A.; Zhao, J.; Jia, L. MiR-221 Promotes Trastuzumab-Resistance and Metastasis in HER2-Positive Breast Cancers by Targeting PTEN. BMB Rep. 2014, 47, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Penolazzi, L.; Bonaccorsi, G.; Gafà, R.; Ravaioli, N.; Gabriele, D.; Bosi, C.; Lanza, G.; Greco, P.; Piva, R. SLUG/HIF1-α/MiR-221 Regulatory Circuit in Endometrial Cancer. Gene 2019, 711, 143938. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Lai, R.; Chen, D.; Yan, W.; Zhang, Z.; Liu, Z.; Ding, X.; Chen, Y. Downregulation of MiR-221 Inhibits Cell Migration and Invasion through Targeting Methyl-CpG Binding Domain Protein 2 in Human Oral Squamous Cell Carcinoma Cells. BioMed Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Quintavalle, C.; Di Leva, G.; Zanca, C.; Romano, G.; Taccioli, C.; Liu, C.G.; Croce, C.M.; Condorelli, G. MicroRNA Signatures of TRAIL Resistance in Human Non-Small Cell Lung Cancer. Oncogene 2008, 27, 3845–3855. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, F.; Wu, Q.; Liu, X. MiR-221 Increases Osteosarcoma Cell Proliferation, Invasion and Migration Partly through the Downregulation of PTEN. Int. J. Mol. Med. 2015, 36, 1377–1383. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-C.; Chen, H.-H.; Qu, Y.-Y.; Xu, C.-W.; Yang, C.; Liu, Y. MicroRNA-221 Promotes Cisplatin Resistance in Osteosarcoma Cells by Targeting PPP2R2A. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Luo, F.; Zhou, J.; Wang, S.; Sun, Z.; Han, Q.; Bai, C. microRNA-222 Promotes Colorectal Cancer Cell Migration and Invasion by Targeting MST3. FEBS Open Bio 2019, 9, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yu, S.; Cao, S.; Yin, Y.; Hong, S.; Guan, H.; Li, Y.; Xiao, H. MicroRNA-222 Promotes Invasion and Metastasis of Papillary Thyroid Cancer Through Targeting Protein Phosphatase 2 Regulatory Subunit B Alpha Expression. Thyroid 2018, 28, 1162–1173. [Google Scholar] [CrossRef]
- Dentelli, P.; Traversa, M.; Rosso, A.; Togliatto, G.; Olgasi, C.; Marchiò, C.; Provero, P.; Lembo, A.; Bon, G.; Annaratone, L.; et al. MiR-221/222 Control Luminal Breast Cancer Tumor Progression by Regulating Different Targets. Cell Cycle 2014, 13, 1811–1826. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Filkowski, J.N.; Tryndyak, V.P.; Golubov, A.; Shpyleva, S.I.; Kovalchuk, O. Alterations of MicroRNAs and Their Targets Are Associated With Acquired Resistance of MCF-7 Breast Cancer Cells to Cisplatin. Int. J. Cancer 2010, 127, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, Z.; Liu, Z.; Shi, S.; Zhang, Z.; Zhang, J.; Lin, H. MiR-221/222 Activate the Wnt/β-Catenin Signaling to Promote Triple-Negative Breast Cancer. J. Mol. Cell Biol. 2018, 10, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, W.; Zhou, C.; Xi, W.; Yuan, L.; Chen, X.; Li, Y.; Yang, A.; Zhang, J.; Wang, T. MiR-221/222 Promote Human Glioma Cell Invasion and Angiogenesis by Targeting TIMP2. Tumor Biol. 2015, 36, 3763–3773. [Google Scholar] [CrossRef]
- Liu, H.; Cao, B.; Zhao, Y.; Liang, H.; Liu, X.; Hao, F. Upregulated MiR-221/222 Promotes Cell Proliferation and Invasion and Is Associated With Invasive Features in Retinoblastoma. Cancer Biomark. 2018, 22, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhou, Z. Downregulation of MiR-302b Is Associated With Poor Prognosis and Tumor Progression of Breast Cancer. Breast Cancer 2019, 27, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Binmadi, N.O.; Basile, J.R.; Perez, P.; Gallo, A.; Tandon, M.; Elias, W.; Jang, S.-I.; Alevizos, I. MiRNA Expression Profile of Mucoepidermoid Carcinoma. Oral Dis. 2017, 24, 537–543. [Google Scholar] [CrossRef]
- Wang, H.; Wang, G.; Gao, Y.; Zhao, C.; Li, X.; Zhang, F.; Jiang, C.; Wu, B. Lnc-SNHG1 Activates the TGFBR2/SMAD3 and RAB11A/Wnt/β-Catenin Pathway by Sponging MiR-302/372/373/520 in Invasive Pituitary Tumors. Cell. Physiol. Biochem. 2018, 48, 1291–1303. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique MicroRNA Molecular Profiles in Lung Cancer Diagnosis and Prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Hosseini, M.; Ghasemi, F.; Shahidsales, S.; Maftouh, M.; Akbarzade, H.; Parizadeh, S.A.R.; Hassanian, S.M.; Avan, A. Circulating MicroRNAs As Potential Diagnostic, Prognostic and Therapeutic Targets in Pancreatic Cancer. Curr. Pharm. Des. 2017, 22, 6444–6450. [Google Scholar] [CrossRef]
- Zhan, B.; Lu, D.; Luo, P.; Wang, B. Prognostic Value of Expression of MicroRNAs in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. Clin. Lab. 2016, 62, 2203–2211. [Google Scholar]
- Liu, F.; Zhuang, L.; Wu, R.; Li, D. MiR-365 Inhibits Cell Invasion and Migration of Triple Negative Breast Cancer through ADAM10. J. BUON. 2019, 24, 1905–1912. [Google Scholar] [PubMed]
- Yan, J.-W.; Lin, J.-S.; He, X.-X. The Emerging Role of MiR-375 in Cancer. Int. J. Cancer 2013, 135, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Emdad, L.; Lee, S.-G.; Su, Z.Z.; Jeon, H.Y.; Boukerche, H.; Sarkar, D.; Fisher, P.B. Astrocyte Elevated Gene-1 (AEG-1) Functions As an Oncogene and Regulates Angiogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 21300–21305. [Google Scholar] [CrossRef]
- Hui, A.B.; Bruce, J.P.; Alajez, N.M.; Shi, W.; Yue, S.; Perez-Ordonez, B.; Xu, W.; O’Sullivan, B.; Waldron, J.; Cummings, B.; et al. Significance of Dysregulated Metadherin and MicroRNA-375 in Head and Neck Cancer. Clin. Cancer Res. 2011, 17, 7539–7550. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.L.; Kwong, D.L.W.; Chan, T.H.-M.; Law, S.Y.-K.; Chen, L.; Li, Y.; Qin, Y.-R.; Guan, X.-Y. MicroRNA-375 Inhibits Tumour Growth and Metastasis in Oesophageal Squamous Cell Carcinoma through Repressing Insulin-Like Growth Factor 1 Receptor. Gut 2011, 61, 33–42. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced Expression of the Let-7 MicroRNAs in Human Lung Cancers in Association With Shortened Postoperative Survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Togashi, Y.; Nomura, K.; Ninomiya, H.; Hiramatsu, M.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Ishikawa, Y. Let-7 MicroRNA Expression Is Reduced in Bronchioloalveolar Carcinoma, a Non-Invasive Carcinoma, and Is Not Correlated With Prognosis. Lung Cancer 2007, 58, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Feng, Y.; Zeng, X.; He, M.; Gong, Y.; Liu, Y. Extracellular vesicles-encapsulated let-7i Shed from Bone Mesenchymal Stem Cells Suppress Lung Cancer via KDM3A/DCLK1/FXYD3 Axis. J. Cell. Mol. Med. 2021, 25, 1911–1926. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The Tumor Suppressor MicroRNA Let-7 Represses the HMGA2 Oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- Akao, Y.; Nakagawa, Y.; Naoe, T. Let-7 MicroRNA Functions As a Potential Growth Suppressor in Human Colon Cancer Cells. Biol. Pharm. Bull. 2006, 29, 903–906. [Google Scholar] [CrossRef]
- Tu, H.-C.; Schwitalla, S.; Qian, Z.; LaPier, G.S.; Yermalovich, A.; Ku, Y.-C.; Chen, S.-C.; Viswanathan, S.R.; Zhu, H.; Nishihara, R.; et al. LIN28 Cooperates With WNT Signaling to Drive Invasive Intestinal and Colorectal Adenocarcinoma in Mice and Humans. Genes Dev. 2015, 29, 1074–1086. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Song, Z.; Chen, H.; Chen, Z.; Yang, P.; Li, W.; Yang, Z.; Zhang, T.; Wang, F.; Wei, J.; et al. Long Noncoding RNA PVT1-214 Promotes Proliferation and Invasion of Colorectal Cancer by Stabilizing Lin28 and Interacting With MiR-128. Oncogene 2019, 38, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Paz, E.A.; LaFleur, B.J.; Gerner, E.W. Polyamines Are Oncometabolites That Regulate the LIN28/Let-7 Pathway in Colorectal Cancer Cells. Mol. Carcinog. 2013, 53, E96–E106. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Meng, P.; Wang, T.; Qin, W.; Qin, W.; Wang, F.; Yuan, J.; Chen, Z.; Yang, A.; Wang, H. MicroRNA Let-7a Inhibits Proliferation of Human Prostate Cancer Cells In Vitro and In Vivo by Targeting E2F2 and CCND2. PLoS ONE 2010, 5, e10147. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.J.; Yoon, H.; Kim, S.W.; Kim, H.; Kim, Y.T.; Kim, J.H.; Kim, S. MicroRNA Expression Profiles in Serous Ovarian Carcinoma. Clin. Cancer Res. 2008, 14, 2690–2695. [Google Scholar] [CrossRef]
- Buechner, J.; Tømte, E.; Haug, B.H.; Henriksen, J.R.; Løkke, C.; Flægstad, T.; Einvik, C. Tumour-Suppressor MicroRNAs Let-7 and Mir-101 Target the Proto-Oncogene MYCN and Inhibit Cell Proliferation in MYCN-Amplified Neuroblastoma. Br. J. Cancer 2011, 105, 296–303. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; Van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B Induces Neuroblastoma and Enhances MYCN Levels via Let-7 Suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Liu, J.M.; Long, X.H.; Zhang, G.M.; Zhou, Y.; Chen, X.Y.; Huang, S.H.; Liu, Z.L.; Zhang, Z.H. Let-7g Reverses Malignant Phenotype of Osteosarcoma Cells by Targeting Aurora-B. Int. J. Clin. Exp. Pathol. 2014, 7, 4596–4606. [Google Scholar]
- Wang, J.; Lin, Y.; Jiang, T.; Gao, C.; Wang, D.; Wang, X.; Wei, Y.; Liu, T.; Zhu, L.; Wang, P.; et al. Up-Regulation of TIMP-3 and RECK Decrease the Invasion and Metastasis Ability of Colon Cancer. Arab. J. Gastroenterol. 2019, 20, 127–134. [Google Scholar] [CrossRef]
- Wang, P.; Guan, Q.; Zhou, D.; Yu, Z.; Song, Y.; Qiu, W. MiR-21 Inhibitors Modulate Biological Functions of Gastric Cancer Cells via PTEN/PI3K/MTOR Pathway. DNA Cell Biol. 2018, 37, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Scorilas, A.; Fragoulis, E.G.; Christodoulou, M.-I. Blood-Based Analysis of 84 MicroRNAs Identifies Molecules Deregulated in Individuals With Type-2 Diabetes, Risk Factors for the Disease or Metabolic Syndrome. Diabetes Res. Clin. Pract. 2020, 164, 108187. [Google Scholar] [CrossRef]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.-I. Decreased Expression of MicroRNAs Targeting Type-2 Diabetes Susceptibility Genes in Peripheral Blood of Patients and Predisposed Individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef]
- Wang, M.; Li, L.; Liu, R.; Song, Y.; Zhang, X.; Niu, W.; Kumar, A.K.; Guo, Z.; Hu, Z. Obesity-Induced Overexpression of MiRNA-24 Regulates Cholesterol Uptake and Lipid Metabolism by Targeting SR-B1. Gene 2018, 668, 196–203. [Google Scholar] [CrossRef]
- Fu, X.; Dong, B.; Tian, Y.; Lefebvre, P.; Meng, Z.; Wang, X.; Pattou, F.; Han, W.; Wang, X.; Lou, F.; et al. MicroRNA-26a Regulates Insulin Sensitivity and Metabolism of Glucose and Lipids. J. Clin. Investig. 2015, 125, 2497–2509. [Google Scholar] [CrossRef]
- Acharya, A.; Berry, D.C.; Zhang, H.; Jiang, Y.; Jones, B.; Hammer, R.E.; Graff, J.M.; Mendell, J.T. MiR-26 Suppresses Adipocyte Progenitor Differentiation and Fat Production by Targeting Fbxl19. Genes Dev. 2019, 33, 1367–1380. [Google Scholar] [CrossRef]
- Batchu, R.B.; Gruzdyn, O.V.; Qazi, A.; Kaur, J.; Mahmud, E.M.; Weaver, D.W.; Gruber, S.A. Enhanced Phosphorylation of p53 by MicroRNA-26a Leading to Growth Inhibition of Pancreatic Cancer. Surgery 2015, 158, 981–987. [Google Scholar] [CrossRef]
- Xu, G.; Ji, C.; Song, G.; Zhao, C.; Shi, C.; Song, L.; Chen, L.; Yang, L.; Huang, F.; Pang, L.; et al. MiR-26b Modulates Insulin Sensitivity in Adipocytes by Interrupting the PTEN/PI3K/AKT Pathway. Int. J. Obes. 2015, 39, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, Y.; Liu, Y.; Zhu, D.; Yu, J.; Li, G.; Sun, Z.; Wang, W.; Jiang, H.; Hong, Z. MiR-27a Promotes Insulin Resistance and Mediates Glucose Metabolism by Targeting PPAR-γ-Mediated PI3K/AKT Signaling. Aging 2019, 11, 7510–7524. [Google Scholar] [CrossRef]
- Roldan, M.; Macias-Gonzalez, M.; Garcia, R.; Tinahones, F.J.; Martin, M. Obesity short-circuits Stemness Gene Network in Human Adipose Multipotent Stem Cells. FASEB J. 2011, 25, 4111–4126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rathinam, R.; Walch, A.; Alahari, S.K. ST14 (Suppression of Tumorigenicity 14) Gene Is a Target for MiR-27b, and the Inhibitory Effect of ST14 on Cell Growth Is Independent of MiR-27b Regulation. J. Biol. Chem. 2009, 284, 23094–23106. [Google Scholar] [CrossRef]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. MiR-27b-3p Inhibits Proliferation and Potentially Reverses Multi-Chemoresistance by Targeting CBLB/GRB2 in Breast Cancer Cells. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Srivastava, A.; Shankar, K.; Beg, M.; Rajan, S.; Gupta, A.; Varshney, S.; Kumar, D.; Gupta, S.; Mishra, R.K.; Gaikwad, A.N. Chronic Hyperinsulinemia Induced MiR-27b Is Linked to Adipocyte Insulin Resistance by Targeting Insulin Receptor. J. Mol. Med. 2018, 96, 315–331. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, L.; Fan, K.; Wang, J. MiR-27b Targets LIMK1 to Inhibit Growth and Invasion of NSCLC Cells. Mol. Cell. Biochem. 2014, 390, 85–91. [Google Scholar] [CrossRef]
- Miranda, K.; Yang, X.; Bam, M.; Murphy, E.A.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA-30 Modulates Metabolic Inflammation by Regulating Notch Signaling in Adipose Tissue Macrophages. Int. J. Obes. 2018, 42, 1140–1150. [Google Scholar] [CrossRef]
- Gottmann, P.; Ouni, M.; Saussenthaler, S.; Roos, J.; Stirm, L.; Jähnert, M.; Kamitz, A.; Hallahan, N.; Jonas, W.; Fritsche, A.; et al. A Computational Biology Approach of a Genome-Wide Screen Connected MiRNAs to Obesity and Type 2 Diabetes. Mol. Metab. 2018, 11, 145–159. [Google Scholar] [CrossRef]
- Kang, T.; Jones, T.M.; Naddell, C.; Bacanamwo, M.; Calvert, J.W.; Thompson, W.E.; Bond, V.C.; Chen, Y.E.; Liu, D. Adipose-Derived Stem Cells Induce Angiogenesis via Microvesicle Transport of MiRNA-31. STEM CELLS Transl. Med. 2016, 5, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, S.A.K.; Teo, C.R.; Beillard, E.J.; Voorhoeve, P.M.; Zhou, W.; Ghosh, S.; Casey, P.J. MicroRNA-31 Controls G Protein Alpha-13 (GNA13) Expression and Cell Invasion in Breast Cancer Cells. Mol. Cancer 2015, 14, 1–10. [Google Scholar] [CrossRef]
- Sun, K.-K.; Shen, X.-J.; Yang, D.; Gan, M.-Q.; Liu, G.; Zhang, Y.-F.; Hua, P.; Wang, H.-D.; Wu, X.-Y. MicroRNA-31 Triggers G2/M Cell Cycle Arrest, Enhances the Chemosensitivity and Inhibits Migration and Invasion of Human Gastric Cancer Cells by Downregulating the Expression of Zeste Homolog 2 (ZH2). Arch. Biochem. Biophys. 2019, 663, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-Secreted Exosomal MicroRNA-34a Inhibits M2 Macrophage Polarization to Promote Obesity-Induced Adipose Inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating MicroRNAs in Patients With Chronic Hepatitis C and Non-Alcoholic Fatty Liver Disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef]
- Fu, T.; Choi, S.-E.; Kim, D.-H.; Seok, S.; Suino-Powell, K.M.; Xu, H.E.; Kemper, J.K. Aberrantly Elevated MicroRNA-34a in Obesity Attenuates Hepatic Responses to FGF19 by Targeting a Membrane Coreceptor -Klotho. Proc. Natl. Acad. Sci. USA 2012, 109, 16137–16142. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a Inhibits Beige and Brown Fat Formation in Obesity in Part by Suppressing Adipocyte Fibroblast Growth Factor 21 Signaling and SIRT1 Function. Mol. Cell. Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of MiR-34a on Cell Growth and Chemoresistance in Prostate Cancer PC3 Cells. Biochem. Biophys. Res. Commun. 2008, 377, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-M.; Min, K.-H.; Lee, W. Induction of MiR-96 by Dietary Saturated Fatty Acids Exacerbates Hepatic Insulin Resistance through the Suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jia, M.; Xu, L.; Fang, Z.; Wu, W.; Zhang, Q.; Chung, P.; Lin, Y.; Wang, S.; Zhang, Y. miR-96 and Autophagy Are Involved in the Beneficial Effect of Grape Seed Proanthocyanidins Against high-fat-diet-induced Dyslipidemia in Mice. Phytother. Res. 2019, 33, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Du, Y.; Liu, P.; Zhang, J.; Li, Y.; Shen, H.; Xing, L.; Xue, X.; Chen, J.; et al. Long Noncoding RNA TP53TG1 Promotes Pancreatic Ductal Adenocarcinoma Development by Acting As a Molecular Sponge of microRNA-96. Cancer Sci. 2019, 110, 2760–2772. [Google Scholar] [CrossRef]
- Pek, S.L.T.; Sum, C.F.; Lin, M.X.; Cheng, A.K.S.; Wong, M.T.K.; Lim, S.C.; Tavintharan, S. Circulating and Visceral Adipose MiR-100 Is down-Regulated in Patients With Obesity and Type 2 Diabetes. Mol. Cell. Endocrinol. 2016, 427, 112–123. [Google Scholar] [CrossRef]
- Li, C.; Gao, Y.; Zhang, K.; Chen, J.; Han, S.; Feng, B.; Wang, R.; Chen, L. Multiple Roles of MicroRNA-100 in Human Cancer and Its Therapeutic Potential. Cell. Physiol. Biochem. 2015, 37, 2143–2159. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, X.; Chen, L.; Zhang, M.; Lei, L.; Gao, W.; Shi, Z.; Dong, Y.; Wang, Z.; Li, X.; et al. Hepatic MiR-125b Inhibits Insulin Signaling Pathway by Targeting PIK3CD. J. Cell. Physiol. 2018, 233, 6052–6066. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-Y.; Yang, C.-Y.; Rui, Z.-L. MicroRNA-125b-5p Improves Pancreatic β-Cell Function through Inhibiting JNK Signaling Pathway by Targeting DACT1 in Mice With Type 2 Diabetes Mellitus. Life Sci. 2019, 224, 67–75. [Google Scholar] [CrossRef]
- Kurylowicz, A.; Owczarz, M.; Polosak, J.; I Jonas, M.; Lisik, W.; Chmura, A.; Puzianowska-Kuznicka, M. SIRT1 and SIRT7 Expression in Adipose Tissues of Obese and Normal-Weight Individuals Is Regulated by MicroRNAs But Not by Methylation Status. Int. J. Obes. 2016, 40, 1635–1642. [Google Scholar] [CrossRef]
- Wang, J.K.; Wang, Z.; Li, G. MicroRNA-125 in Immunity and Cancer. Cancer Lett. 2019, 454, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Budd, W.T.; Seashols-Williams, S.; Clark, G.C.; Weaver, D.; Calvert, V.; Petricoin, E.; Dragoescu, E.A.; O’Hanlon, K.; Zehner, Z.E. Dual Action of MiR-125b As a Tumor Suppressor and OncomiR-22 Promotes Prostate Cancer Tumorigenesis. PLoS ONE 2015, 10, e0142373. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Arner, E.; Mejhert, N.; Kulyté, A.; Balwierz, P.J.; Pachkov, M.; Cormont, M.; Lorente-Cebrián, S.; Ehrlund, A.; Laurencikiene, J.; Hedén, P.; et al. Adipose Tissue MicroRNAs As Regulators of CCL2 Production in Human Obesity. Diabetes 2012, 61, 1986–1993. [Google Scholar] [CrossRef] [PubMed]
- Yiew, N.K.H.; Chatterjee, T.K.; Tang, Y.L.; Pellenberg, R.; Stansfield, B.K.; Bagi, Z.; Fulton, D.J.; Stepp, D.W.; Chen, W.; Patel, V.; et al. A Novel Role for the Wnt Inhibitor APCDD1 in Adipocyte Differentiation: Implications for Diet-Induced Obesity. J. Biol. Chem. 2017, 292, 6312–6324. [Google Scholar] [CrossRef] [PubMed]
- Al-Rawaf, H.A. Circulating MicroRNAs and Adipokines As Markers of Metabolic Syndrome in Adolescents With Obesity. Clin. Nutr. 2019, 38, 2231–2238. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.J.; Zhao, N.; Shen, H.; Wang, H. Long Noncoding RNA MRPL39 Inhibits Gastric Cancer Proliferation and Progression by Directly Targeting MiR-130. Genet. Test. Mol. Biomarkers 2018, 22, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-Induced Overexpression of MiRNA-143 Inhibits Insulin-Stimulated AKT Activation and Impairs Glucose Metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef]
- Takanabe, R.; Ono, K.; Abe, Y.; Takaya, T.; Horie, T.; Wada, H.; Kita, T.; Satoh, N.; Shimatsu, A.; Hasegawa, K. Up-Regulated Expression of MicroRNA-143 in Association With Obesity in Adipose Tissue of Mice Fed High-Fat Diet. Biochem. Biophys. Res. Commun. 2008, 376, 728–732. [Google Scholar] [CrossRef]
- Kilic, I.D.; Dodurga, Y.; Uludag, B.; Alihanoglu, Y.I.; Yildiz, B.S.; Enli, Y.; Secme, M.; Bostancı, H.E. MicroRNA -143 and -223 in Obesity. Gene 2015, 560, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Collares, R.V.A.; Salgado, W., Jr.; da Cunha Tirapelli, D.P.; dos Santos, J.S. The Expression of LEP, LEPR, IGF1 and IL10 in Obesity and the Relationship With MicroRNAs. PLoS ONE 2014, 9, e93512. [Google Scholar] [CrossRef]
- Wang, C.-J.; Zhou, Z.-G.; Wang, L.; Yang, L.; Bin Zhou, B.; Gu, J.; Chen, H.-Y.; Sun, X.-F. Clinicopathological Significance of MicroRNA-31, -143 and -145 Expression in Colorectal Cancer. Dis. Markers 2009, 26, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Jančík, S.; Drábek, J.; Radzioch, D.; Hajdúch, M. Clinical Relevance of KRAS in Human Cancers. J. Biomed. Biotechnol. 2010, 2010, 1–13. [Google Scholar] [CrossRef]
- Kirby, T.J.; Walton, R.G.; Finlin, B.S.; Zhu, B.; Unal, R.; Rasouli, N.; Peterson, C.A.; Kern, P.A. Integrative MRNA-MicroRNA Analyses Reveal Novel Interactions Related to Insulin Sensitivity in Human Adipose Tissue. Physiol. Genom. 2016, 48, 145–153. [Google Scholar] [CrossRef]
- Liu, H.-T.; Xu, Y.-T.; Li, H.-Y.; Zhao, J.; Zhai, H.-Y.; Chen, Y. Loss of MicroRNA Expression Is Involved in the Development and Prognosis of Breast Cancer Complicated by Type 2 Diabetes Mellitus. Int. J. Biol. Markers 2016, 31, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Qin, Y.; Jia, J.; Lin, T.; Lin, X.; Chen, L.; Zeng, H.; Han, Y.; Wu, L.; Huang, S.; et al. MiR-155 Enhances Insulin Sensitivity by Coordinated Regulation of Multiple Genes in Mice. PLoS Genet. 2016, 12, e1006308. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. MiR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef]
- Karkeni, E.; Astier, J.; Tourniaire, F.; El Abed, M.; Romier, B.; Gouranton, E.; Wan, L.; Borel, P.; Salles, J.; Walrand, S.; et al. Obesity-Associated Inflammation Induces MicroRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function. J. Clin. Endocrinol. Metab. 2016, 101, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Tryggestad, J.B.; Teague, A.M.; Sparling, D.P.; Jiang, S.; Chernausek, S.D. Macrophage-Derived microRNA-155 Increases in Obesity and Influences Adipocyte Metabolism by Targeting Peroxisome Proliferator-Activated Receptor Gamma. Obesity 2019, 27, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Virtue, A.; Johnson, C.; Lopez-Pastraña, J.; Shao, Y.; Fu, H.; Li, X.; Li, Y.-F.; Yin, Y.; Mai, J.; Rizzo, V.; et al. MicroRNA-155 Deficiency Leads to Decreased Atherosclerosis, Increased White Adipose Tissue Obesity, and Non-Alcoholic Fatty Liver Disease. J. Biol. Chem. 2017, 292, 1267–1287. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, S.; Li, Z.; Yang, Q.; Li, B.; Zhu, S.; Wang, L.; Wu, J.; Yuan, J.; Yang, C.; et al. Tumour-Originated Exosomal MiR-155 Triggers Cancer-Associated Cachexia to Promote Tumour Progression. Mol. Cancer 2018, 17, 1–7. [Google Scholar] [CrossRef]
- Flowers, E.; Aouizerat, B.E.; Abbasi, F.; Lamendola, C.; Grove, K.M.; Fukuoka, Y.; Reaven, G.M. Circulating MicroRNA-320a and MicroRNA-486 Predict Thiazolidinedione Response: Moving towards Precision Health for Diabetes Prevention. Metabolism 2015, 64, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Parrizas, M.; Brugnara, L.; Esteban, Y.; González-Franquesa, A.; Canivell, S.; Murillo, S.; Gordillo-Bastidas, E.; Cussó, R.; Cadefau, J.A.; Garcia-Roves, P.M.; et al. Circulating MiR-192 and MiR-193b Are Markers of Prediabetes and Are Modulated by an Exercise Intervention. J. Clin. Endocrinol. Metab. 2015, 100, E407–E415. [Google Scholar] [CrossRef]
- Liu, J.; Xing, Y.; Rong, L. MiR-181 Regulates Cisplatin-Resistant Non-Small Cell Lung Cancer via Downregulation of Autophagy through the PTEN/PI3K/AKT Pathway. Oncol. Rep. 2018, 39, 1631–1639. [Google Scholar] [CrossRef]
- Tian, F.; Tang, P.; Sun, Z.; Zhang, R.; Zhu, D.; He, J.; Liao, J.; Wan, Q.; Shen, J. MiR-210 in Exosomes Derived from Macrophages under High Glucose Promotes Mouse Diabetic Obesity Pathogenesis by Suppressing NDUFA4 Expression. J. Diabetes Res. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Qin, Q.; Furong, W.; Baosheng, L. Multiple Functions of Hypoxia-Regulated MiR-210 in Cancer. J. Exp. Clin. Cancer Res. 2014, 33, 50. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Mercader, J.M.; Catalán, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gómez-Ambrosi, J.; Anglada, R.; Fernández-Formoso, J.A.; Ricart, W.; et al. Targeting the Circulating MicroRNA Signature of Obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; López-Bermejo, A.; Fernández-Real, J.M. Changes in Circulating MicroRNAs Are Associated With Childhood Obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Badosa, G.; Bonmatí, A.; Ortega, F.-J.; Mercader, J.-M.; Guindo-Martínez, M.; Torrents, D.; Prats-Puig, A.; Martinez-Calcerrada, J.-M.; Platero-Gutierrez, E.; De Zegher, F.; et al. Altered Circulating MiRNA Expression Profile in Pregestational and Gestational Obesity. J. Clin. Endocrinol. Metab. 2015, 100, E1446–E1456. [Google Scholar] [CrossRef]
- Lopez, Y.N.; Coen, P.; Goodpaster, B.H.; A Seyhan, A. Gastric Bypass Surgery With Exercise Alters Plasma MicroRNAs That Predict Improvements in Cardiometabolic Risk. Int. J. Obes. 2017, 41, 1121–1130. [Google Scholar] [CrossRef]
- Peng, J.; Zhou, Y.; Deng, Z.; Zhang, H.; Wu, Y.; Song, T.; Yang, Y.; Wei, H.; Peng, J. miR-221 Negatively Regulates Inflammation and Insulin Sensitivity in White Adipose Tissue by Repression of sirtuin-1 (SIRT1). J. Cell. Biochem. 2018, 119, 6418–6428. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, G.; Fan, C.; Diao, Y.; Wu, B.; Li, J. Increased Expression of MicroRNA-221 in Gastric Cancer and Its Clinical Significance. J. Int. Med. Res. 2012, 40, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Segal, C.V.; Koufaris, C.; Powell, C.; Gooderham, N.J. Effects of Treatment With Androgen Receptor Ligands on MicroRNA Expression of Prostate Cancer Cells. Toxicology 2015, 333, 45–52. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Gan, R.; Zhao, L.; Li, W.; Zhou, H.; Wang, X.; Lü, J.; Meng, Q.H. Down-Regulation of Mir-221 and Mir-222 Restrain Prostate Cancer Cell Proliferation and Migration That Is Partly Mediated by Activation of SIRT1. PLoS ONE 2014, 9, e98833. [Google Scholar] [CrossRef]
- Oses, M.; Sanchez, J.M.; Portillo, M.P.; Aguilera, C.M.; Labayen, I. Circulating MiRNAs As Biomarkers of Obesity and Obesity-Associated Comorbidities in Children and Adolescents: A Systematic Review. Nutrients 2019, 11, 2890. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Rovira, O.; Guerra, E.; Esteve, E.; Xifra, G.; Martínez, C.; Ricart, W.; Rieusset, J.; et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change With Insulin Sensitization. Diabetes Care 2014, 37, 1375–1383. [Google Scholar] [CrossRef]
- De Mendonça, M.; De Sousa, É.; Da Paixão, A.O.; Dos Santos, B.A.; Spagnol, A.R.; Murata, G.M.; Araújo, H.N.; De Lima, T.I.; Guimarães, D.S.P.S.F.; Silveira, L.R.; et al. MicroRNA MiR-222 Mediates Pioglitazone Beneficial Effects on Skeletal Muscle of Diet-Induced Obese Mice. Mol. Cell. Endocrinol. 2020, 501, 110661. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential Expression of MicroRNAs in Omental Adipose Tissue From Gestational Diabetes Mellitus Subjects Reveals MiR-222 As a Regulator of ERα Expression in Estrogen-Induced Insulin Resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef]
- Gan, M.; Shen, L.; Wang, S.; Guo, Z.; Zheng, T.; Tan, Y.; Fan, Y.; Liu, L.; Chen, L.; Jiang, A.; et al. Genistein Inhibits High Fat Diet-Induced Obesity through MiR-222 by Targeting BTG2 and Adipor1. Food Funct. 2020, 11, 2418–2426. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Shi, B.; Ma, S.; Xu, Z.; Ge, Y.; Liu, Y.; Zheng, D.; Shi, J. Functions of MiR-146a and MiR-222 in Tumor-Associated Macrophages in Breast Cancer. Sci. Rep. 2015, 5, 18648. [Google Scholar] [CrossRef]
- Vivacqua, A.; De Marco, P.; Belfiore, A.; Maggiolini, M. Recent Advances on the Role of MicroRNAs in Both Insulin Resistance and Cancer. Curr. Pharm. Des. 2017, 23, 3658–3666. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, F.; Zhang, H.; Tseng, Y.-H. MicroRNA Regulation of Brown Adipogenesis and Thermogenic Energy Expenditure. Front. Endocrinol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of Circulating MiR-101, MiR-375 and MiR-802 As Biomarkers for Type 2 Diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef]
- Ling, H.-Y.; Wen, G.-B.; Feng, S.-D.; Tuo, Q.-H.; Ou, H.-S.; Yao, C.H.; Zhu, B.-Y.; Gao, Z.-P.; Zhang, L.; Liao, D.-F. MicroRNA-375 Promotes 3T3-L1 Adipocyte Differentiation through Modulation of Extracellular Signal-Regulated Kinase Signalling. Clin. Exp. Pharmacol. Physiol. 2011, 38, 239–246. [Google Scholar] [CrossRef]
- Kasiappan, R.; Rajarajan, D. Role of MicroRNA Regulation in Obesity-Associated Breast Cancer: Nutritional Perspectives. Adv. Nutr. 2017, 8, 868–888. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Fu, M.; Bookout, A.L.; Kliewer, S.A.; Mangelsdorf, D.J. MicroRNA Let-7 Regulates 3T3-L1 Adipogenesis. Mol. Endocrinol. 2009, 23, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.H.; Leroith, D. Obesity, Type 2 Diabetes, and Cancer: The Insulin and IGF Connection. Endocr. Relat. Cancer 2012, 19, F27–F45. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the Pairing between Let-7 and Hmga2 Enhances Oncogenic Transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the Let-7 MicroRNA Family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- O’Day, E.; Lal, A. MicroRNAs and Their Target Gene Networks in Breast Cancer. Breast Cancer Res. 2010, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Divella, R.; Daniele, A.; Savino, E.; Paradiso, A. Anticancer Effects of Nutraceuticals in the Mediterranean Diet: An Epigenetic Diet Model. Cancer Genom. Proteom. 2020, 17, 335–350. [Google Scholar] [CrossRef]
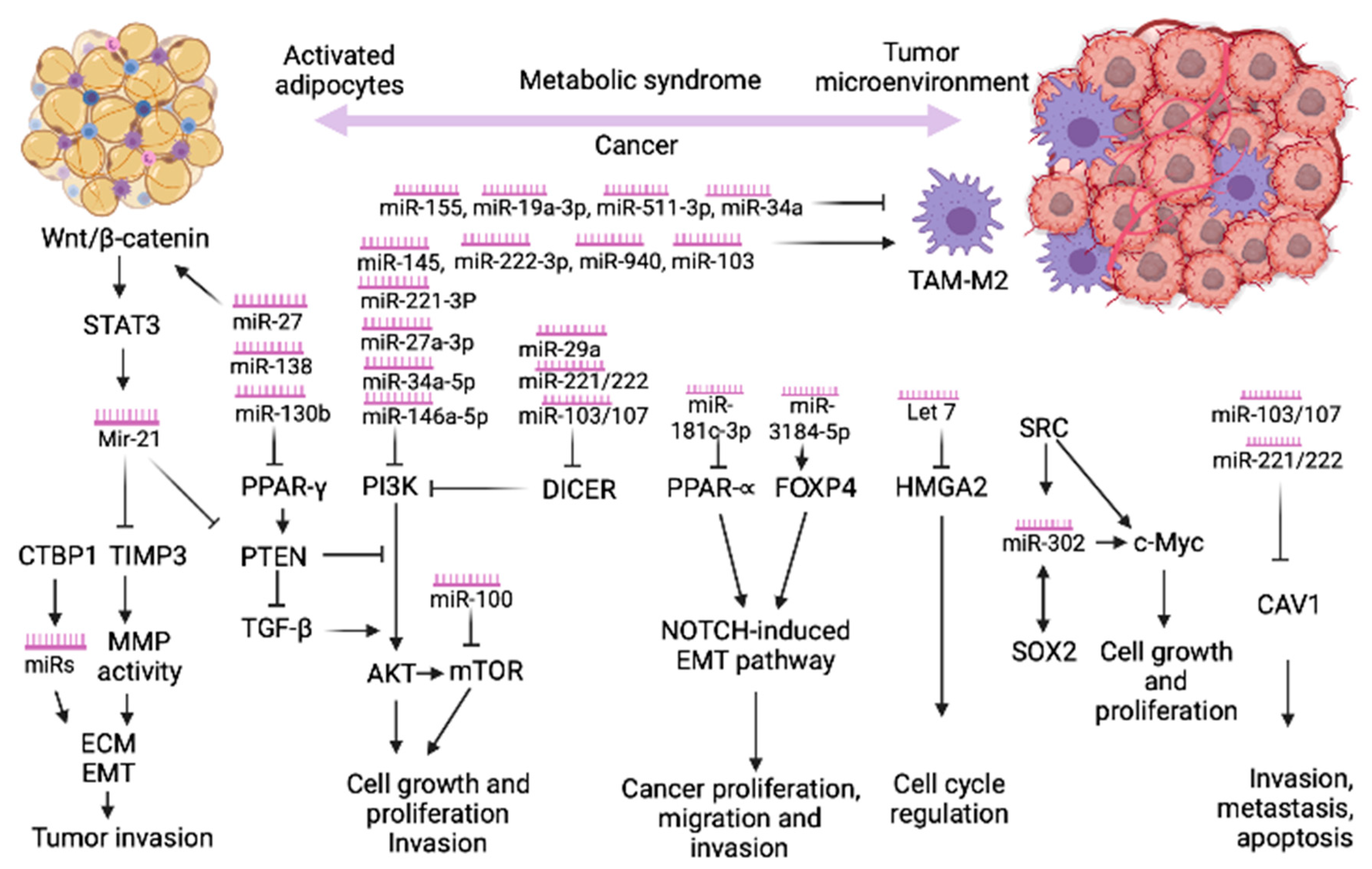
| miRNAs | Target Genes and Functions in Obesity/IR/MeS | Target Genes or Pathway in Cancer | Type of Cancer (Oncomir/Tumor Suppressor) | References for Cancer Genes or Pathway |
|---|---|---|---|---|
| miR-21 | TIMP3 PPAR-c, GLUT4 PTEN/PI3K/Akt pathway; STAT3, TGFRB2, PTEN, Sprouty1 and Sprouty 2 | RhoB | HCC, Breast cancer ↑ | [96] |
| PDCD4 and Maspin | Prostate cancer ↑ | [97,98] | ||
| Prostate cancer ↓ | [99] | |||
| AKT/ERK pathways | HCC ↑ | [100] | ||
| RECK | Osteosarcoma ↑ | [101] | ||
| CASC7 and IGN3 | Colorectal cancer ↑ | [102] | ||
| PTEN | Colorectal cancer ↑ | [103] | ||
| RhoB | Colorectal cancer ↑ | [104] | ||
| Sec23A | Colorectal cancer ↑ | [105] | ||
| TIMP-3 and RECK | Colorectal cancer ↑ | [105] | ||
| STAT3, PIK3R1 | Breast cancer ↑ | [106,107] | ||
| PTEN | NSCLC ↑ | [108] | ||
| VHL/PI3K/AKT | Papillary thyroid cancer ↑ | [109] | ||
| PTEN and PDCD4 | Gastric cancer ↑ | [110] | ||
| TIMP3 | Melanoma ↑ | [111] | ||
| Sox2/β-catenin, RECK | Glioma ↑ | [112,113] | ||
| PTEN, DKK, BCL2 | OSCC ↑ | [114,115,116] | ||
| PTEN | Cervical cancer ↑ | [117] | ||
| RCC ↑ | [118] | |||
| miR-24-3p | SR-B1, HMGCR, DHCR24, SREBP2 KCNQ1 | LPAATβ | Osteosarcoma ↓ | [119] |
| CCK8 | Colorectal cancer ↓ | [120,121] | ||
| p27Kip1, Bim | Breast cancer ↓ | [122,123] | ||
| miR-26a | GSK3β, PKCδ, PKCθ, ACSL3, ACSL4, PCK1, TCF7L2, FXl19 | p53, SMAD1, EZH2, IL-6-Stat3, CTDSP1/2/L, SODD, CKS2, FGF9 | Lung, breast, HCC, rabdomyosarcoma, prostate, melanoma, papillary thyroid, gastric, pancreatic cancer ↓ | [124] |
| MCL-1 BRCA1 | Breast cancer ↓ Triple-negative BC↓ | [125] [126] | ||
| PTEN and PHB, ERα, GSK3 | Glioma, ovarian cancer, colon cancer, cholangiocarcinoma ↑ | [124] | ||
| miR-26b | Glut 4, PTEN/PI3K/AK, Fbxl19 | Cox-2 | Glioma, NSCLC ↓ | [127,128] |
| EphA2 | HCC ↓ | [129] | ||
| miR-27 | PPAR-γ, Wnt1, GLUT-4 PI3K, PRDM16, PPARα, CREB, PGC1β | ZEB1, ZEB2, Slug, Vimentin, E-cadherin | Gastric cancer ↓ | [130] |
| MDR1/P-glycoprotein | Cancer cells ↑ | [130] | ||
| SPRY1, BAK, FOXO1, CBLB/GRB2 | Breast cancer ↓ | [131,132] | ||
| miR-27b | PHB, INSR, PPARγ | PPARγ | CRC ↑ | [37] |
| CDH11, EMT, PPARγ- NHE1 pathway | Cervical cancer ↑ | [39,133] | ||
| LIMK1, Sp1 | NSCLC ↓ | [48,134] | ||
| miR-30 | DDL40-Notch-1 | Breast cancer ↑ | [135] | |
| MMP19 | NSCLC ↑ | [136] | ||
| Fibronectin, Vimentin, N-cadherin | Pancreatic cancer ↑ | [137] | ||
| miR-31 | PPARg, PRKAA1, ACACA, GLUT4, IRS1, HIF-1a | ARIDIA | HNSCC ↑ | [138] |
| Rectal cancer ↑ | [139] | |||
| FIH-1 | CRC ↑ | [140] | ||
| RhoA, GNA13 | Breast cancer ↑ | [140,141] | ||
| PPP6C | Mezothelioma ↓ | [142] | ||
| Dock1, NF-jB/Snail | Glioma ↓ | [143] | ||
| ZH2, p53 pathway | Gastric, ovarian, osteosarcoma, prostate cancer ↓ | [63,144] | ||
| miR-34a | Inhibit macrophage M2 induced adipose inflammation | BCL-2 and SIRT1 | Breast cancer↓ | [145] |
| Notch1 | Breast cancer↓ | [146] | ||
| Wnt/β-catenin signaling pathway | Breast cancer↓ | [147] | ||
| Fra-1 | Breast cancer↓ | [148] | ||
| MYC, P53 | Breast cancer↓ | [149,150] | ||
| E2F3 | Neuroblastoma ↓ | [151] | ||
| c-Met and β-catenin | Colon cancer ↓ | [152] | ||
| P53 | Osteosarcoma ↓ | [153] | ||
| MET, P53 | Ovarian cancer ↓ | [154,155] | ||
| CD44 | Prostatic cancer ↓ | [156] | ||
| AXL | Solid cancer ↓ | [157,158] | ||
| miR96 | INSR, IRS | SOX6, EphrinA5 | HCC ↑ | [159,160] |
| PTPN9, FOXO1, FOXO3a | Breast cancer ↑ | [161,162,163] | ||
| FOXO3 | NSCLC ↑ | [164] | ||
| RECK | ESCC ↑ | [165] | ||
| AEG-1 | Glioblastoma ↓ | [166] | ||
| Ezrin | RCC ↓ | [167] | ||
| KRAS | Pancreatic cancer ↓ | [168] | ||
| miR-100 | mTOR, IGFR, VLDLR | Bladder cancer | [169] | |
| HOXA1, Rac1, ICMT, EphB6, AGO2, Plk1, Wnt, β-catenin or RBSP3 | HCC, RCC, bladder cancer, NSCLC, and epithelial ovarian cancer | [170] [171] | ||
| mTOR kinase | Endometrioid endometrial carcinoma | [171] | ||
| CRC | [172] | |||
| miR-125b | PI3K/AKT JNK signaling pathway, SIRTs | NF-κB, | DLBCL ↑ | [173] |
| MAPK11, IRF4, TET2-VEGFA | Acute leukemia ↑ | [174] | ||
| Wnt, PI3K/Akt, STAT-3, MAPK, NF-κB, p53 | HCC, CRC, RCC, thyroid larynx, osteosarcoma, prostate melanoma, Ewing sarcoma, glioblastoma, gallbladder, ovarian cancer | [175] | ||
| miR-126 | IRS-1, CCL2 | PI3K, KRAS, EGFL7, CRK, ADAM9, HOXA9, IRS-1, SOX-2, SLC7A5 and VEGF | Gastrointestinal tract, genital tracts, breast, thyroid, lung cancers | [176] |
| NSCLC | [177] | |||
| CRC | [176] | |||
| RCC | [176] | |||
| miR-143 | ORP8/insulin-AKT pathway, PPARg and aP2 Leptin | Bcl2, ERK5, KRAS | Cervical, prostate, CRC ↓ | [178] |
| ERBB3 | Breast cancer ↓ | [179] | ||
| KRAS, Vimentin, CXCR4, MMP-9 | Breast cancer ↓ | [180] | ||
| FAM83F | Esophageal squamous cell carcinoma ↓ | [181] | ||
| DNMT3A | Gastric cancer ↓ | [182] | ||
| TLR2, NF-κB, MMP-2, MMP-9, CD44, MMP14, integrin β1, integrin β4 | HCC ↓ | [183] | ||
| PAI-1/MMP-13 | Osteosarcoma ↓ | [184] | ||
| Smad3, CD44, and K-Ras | NSCLC ↓ | [185] | ||
| miR-145 | AKT/PI3K/GLUT4 ADAM22, MYO5A, LOX, and GM2A | PI3K/AKT, MRP1, SMAD, KLF4, c-myc, Ets1, E-cadherin, FSCN1, BCL2 | BC, gastric, CRC, NSCLC, glioma, HCC, osteosarcoma, ovarian, cervical, prostate, bladder, nasopharyngeal cancer ↓ | [186] |
| miR-148a-3p | inhibit DNMT1 | DNMT1 | Esophageal Cancer↓ | [187] |
| WNT5A, TGF-α, BTG2 and MYCBP | Chordoma ↑ | [188] | ||
| miR-155 | C/EBPβ, HDAC4, PPARγ, GLUT4, IRS1, PPAR-c, Creb1, Cebpb, Pparg, Pnpla2, Fabp4, Fasn, AdipoQ, | TNF∝, NF-kB pathway, ERK pathway, Caspase 3 | Osteosarcoma ↑ | [189] [190] |
| RhoA, PEG10 and MYB | Breast cancer ↑ | [191,192] | ||
| Gallbladder cancer ↑ | [193] | |||
| NDFIP1 | Melanoma ↑ | [194] | ||
| FGF2 | ESCC ↑ | [195] | ||
| Nasopharyngeal carcinoma ↑ | [196] | |||
| IGF-1 | Colon cancer ↑ | [197] | ||
| 181c-3p | PPARα; reduced inhibition of PPARα, BC proliferation | PPARα | Breast cancer ↓ | [77] |
| PTEN | Breast cancer ↑ | [198] | ||
| PTEN/PI3K/pAkt | CRC ↓ | [199] | ||
| PTEN/PI3K/AKT | NSCLC ↓ | [200] | ||
| XIAP, caspase 9, caspase 3 | Gastric cancer ↓ | [201] | ||
| CTGF, BIRC5, BLC2L1 | Pancreatic cancer ↑ | [202] | ||
| MGMT | Glioblastoma ↓ | [203] | ||
| SPP1 | HCC ↓ | [204] | ||
| SMAD7, TGF-β | Osteosarcoma ↓ | [205] | ||
| MiR-193b | CCL2, NTFYα si NRIP1 | DDAH1 | Triple-negative breast cancer ↓ | [206] |
| TGF-𝜷, SMAD3, NF1 | CRC, Glioma, Head and neck SCC↑ | [207] | ||
| K-Ras, ERBB4 | Lung cancer↓ | [207] | ||
| MAX, KRAS, TGF-𝜷, CCND1, ETS1, MAPK | ESCC, Gastric cancer, HCC, Pancreatic cancer ↓ | [207] | ||
| Mcl-1, c-kit, MYB | Melanoma, Leukemia ↓ | [207] | ||
| caspase 3 and 7, uPA | Ovarian, Prostate cancer ↓ | [207] | ||
| miR-210 | NDUFA4 GPD1L | LOXL4 | Lung adenocarcinoma ↑ | [208] |
| Glioma ↑ | [209] | |||
| Osteosarcoma ↑ | [210] | |||
| HOXA 9 | Pancreatic cancer ↑ | [211] | ||
| miR-221 | SIRT1 IRS/PI3K/AKT | PTEN/TRAIL | Breast cancer↑ | [212,213] |
| ERα, PR, HIF1-α, SLUG | Endometrial cancer↑ | [214] | ||
| Prostate cancer ↑ | [98], | |||
| MBD2 | OSCC ↑ | [215] | ||
| Kit | NSCLC ↑ | [216] | ||
| PTEN, PPP2R2A | Osteosarcoma↑ | [217,218] | ||
| AKT/ERK pathway | HCC ↑ | [100] | ||
| miR-222 | CXCR4 GLUT4 ERs, BTG2, adipor1 | p27 (kip1) | NSLCC | [216] |
| MST3 | CRC ↑ | [219] | ||
| PPP2R2A | Papillary thyroid cancer ↑ | [220] | ||
| miR-221/222 | CAV1 | CAV1 | Breast cancer↑ | [51] |
| β4 integrin, STAT5A, and ADAM-17 | Breast cancer↑ | [221] | ||
| p27, p57, ER∝ | Breast cancer↑ | [222] | ||
| Wnt/β-catenin, WIF1, SFRP2, DKK2, AXIN2 | Breast cancer↑ | [223] | ||
| TIMP2 | Gliomas ↑ | [224] | ||
| Retinoblastomas ↑ | [225] | |||
| miR-302 | Maintain SOX2 and c-Myc by targeting repressor of c-Myc | MACC1 | HCC ↓ | [83] |
| Sox2, c-Myc, Nanog | Breast cancer ↑ | [226] | ||
| RUNX2 | Breast cancer ↓ | [227] | ||
| TGF-β | Mucoepidermoid carcinoma of salivary glands ↑ | [228] | ||
| TGFBR2/SMAD3 RAB11A/Wnt/β-Catenin | Pituitary Tumors ↑ | [229] | ||
| miR-365 | Cebpα, Fabp4, and Pparγ | BTG2 | Pancreas ↑ | [230] |
| ETS1 | NSCLC ↓ | [231] | ||
| ADAM1 | Triple negative breast cancer ↓ | [232] | ||
| miR-375 | ERK ½ Myotrophin | PSAT1 | ESCC ↓ | [233] |
| AEG-1 | HCC, Head and neck cancers ↓ | [234,235] | ||
| PDK1, YWHAZ | Gastric ↓ | [236] | ||
| miR 3184-3p | FOXP4–NOTCH induced EMT | N-cadherin, vimentin, E-cadherin | Breast cancer ↓ | [77] |
| Let 7 | Inhibit HMGA2, inhibit preadipocyte proliferation, insulin-PI3K-mTOR IGF1R, INSR, IRS2 | HMGA2 | Breast cancer ↓ | [302] |
| lin-41, hbl-1/lin-57 RAS | Lung cancer ↓ | [229,237,238] | ||
| KDM3A/DCLK1/FXYD3 | Lung cancer ↓ | [239] | ||
| HGMA2 | Lung cancer ↓ | [240] | ||
| RAS, c-MYC | CRC ↑ | [241] | ||
| HGMA2 LIN 28 | CRC ↓ | [242,243,244] | ||
| E2F2, CCND2 | Prostate cancer↓ | [245] | ||
| RAS | Ovarian cancer ↓ | [246] | ||
| MYCN | Neuroblastoma ↓ | [247,248] | ||
| Aurora-B | Osteosarcoma ↓ | [249] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fodor, A.; Lazar, A.L.; Buchman, C.; Tiperciuc, B.; Orasan, O.H.; Cozma, A. MicroRNAs: The Link between the Metabolic Syndrome and Oncogenesis. Int. J. Mol. Sci. 2021, 22, 6337. https://doi.org/10.3390/ijms22126337
Fodor A, Lazar AL, Buchman C, Tiperciuc B, Orasan OH, Cozma A. MicroRNAs: The Link between the Metabolic Syndrome and Oncogenesis. International Journal of Molecular Sciences. 2021; 22(12):6337. https://doi.org/10.3390/ijms22126337
Chicago/Turabian StyleFodor, Adriana, Andrada Luciana Lazar, Cristina Buchman, Brandusa Tiperciuc, Olga Hilda Orasan, and Angela Cozma. 2021. "MicroRNAs: The Link between the Metabolic Syndrome and Oncogenesis" International Journal of Molecular Sciences 22, no. 12: 6337. https://doi.org/10.3390/ijms22126337
APA StyleFodor, A., Lazar, A. L., Buchman, C., Tiperciuc, B., Orasan, O. H., & Cozma, A. (2021). MicroRNAs: The Link between the Metabolic Syndrome and Oncogenesis. International Journal of Molecular Sciences, 22(12), 6337. https://doi.org/10.3390/ijms22126337







