RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center
Abstract
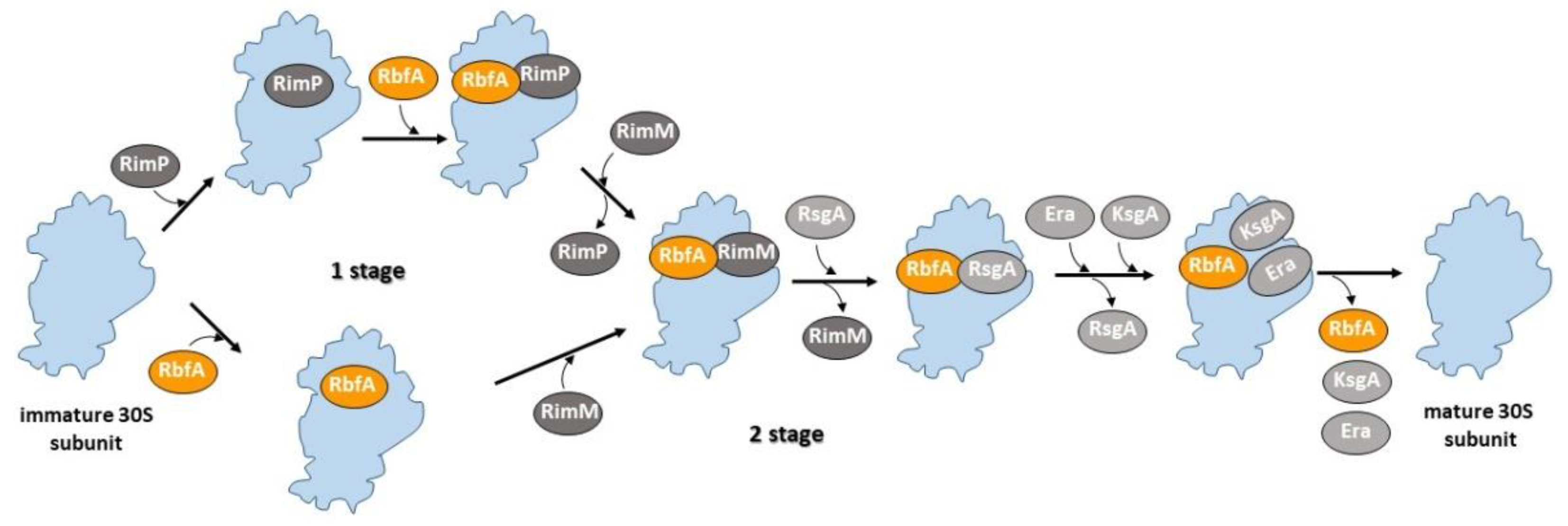
1. Introduction
2. Results
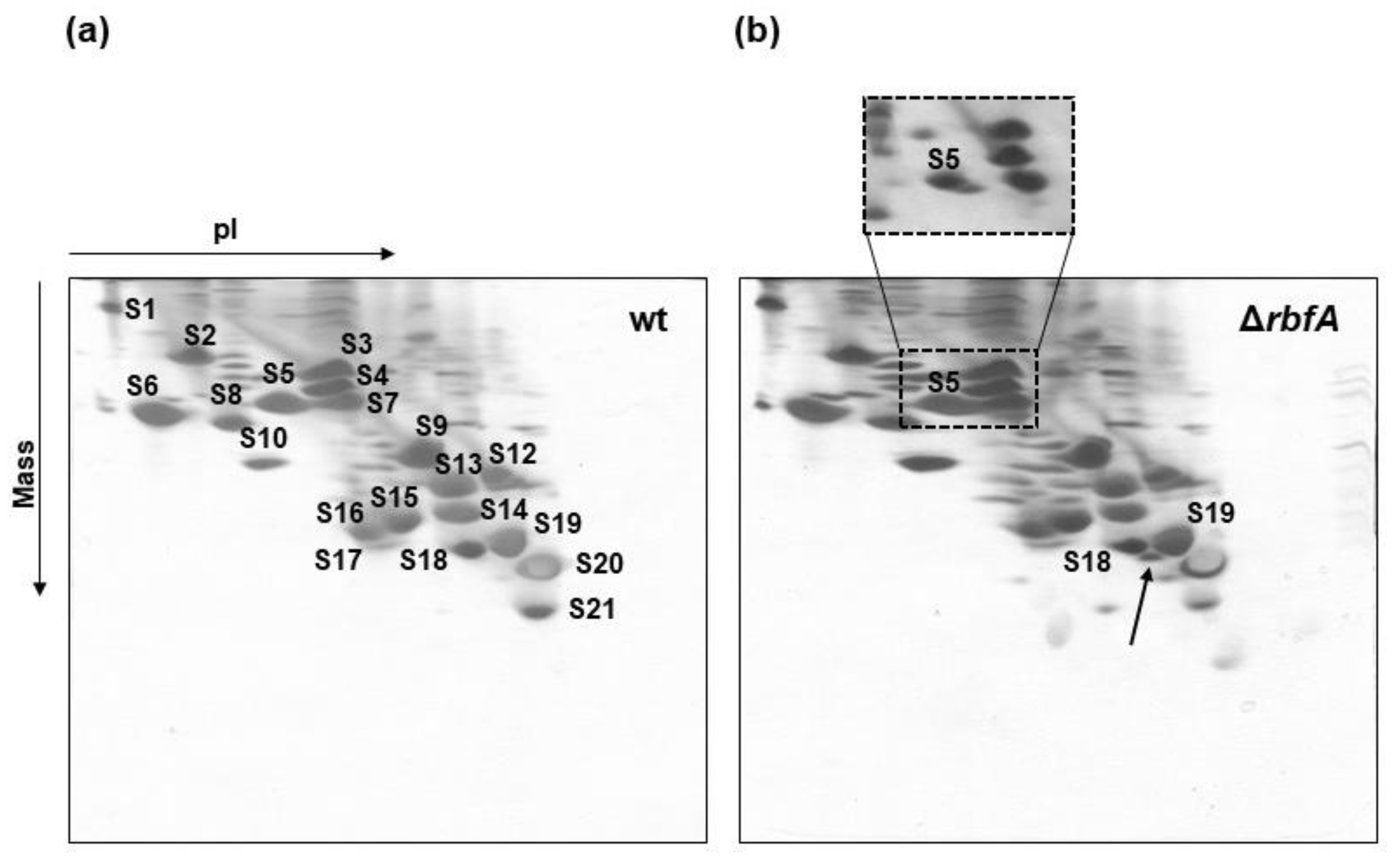
2.1. Characterization of the rbfA Null Strain of E. coli and Protein Complement of Pre-30S Particles from ∆rbfA Cells
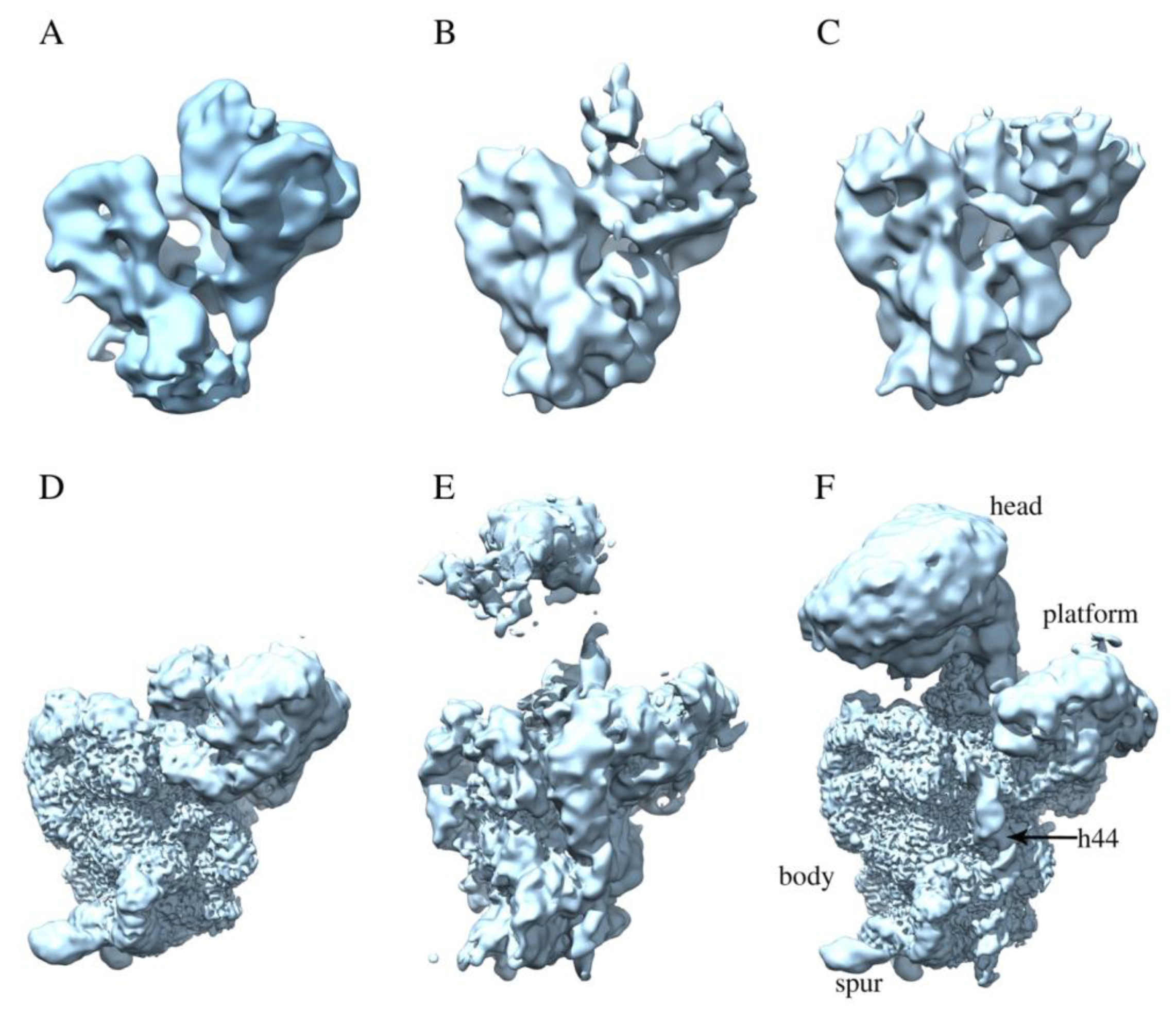
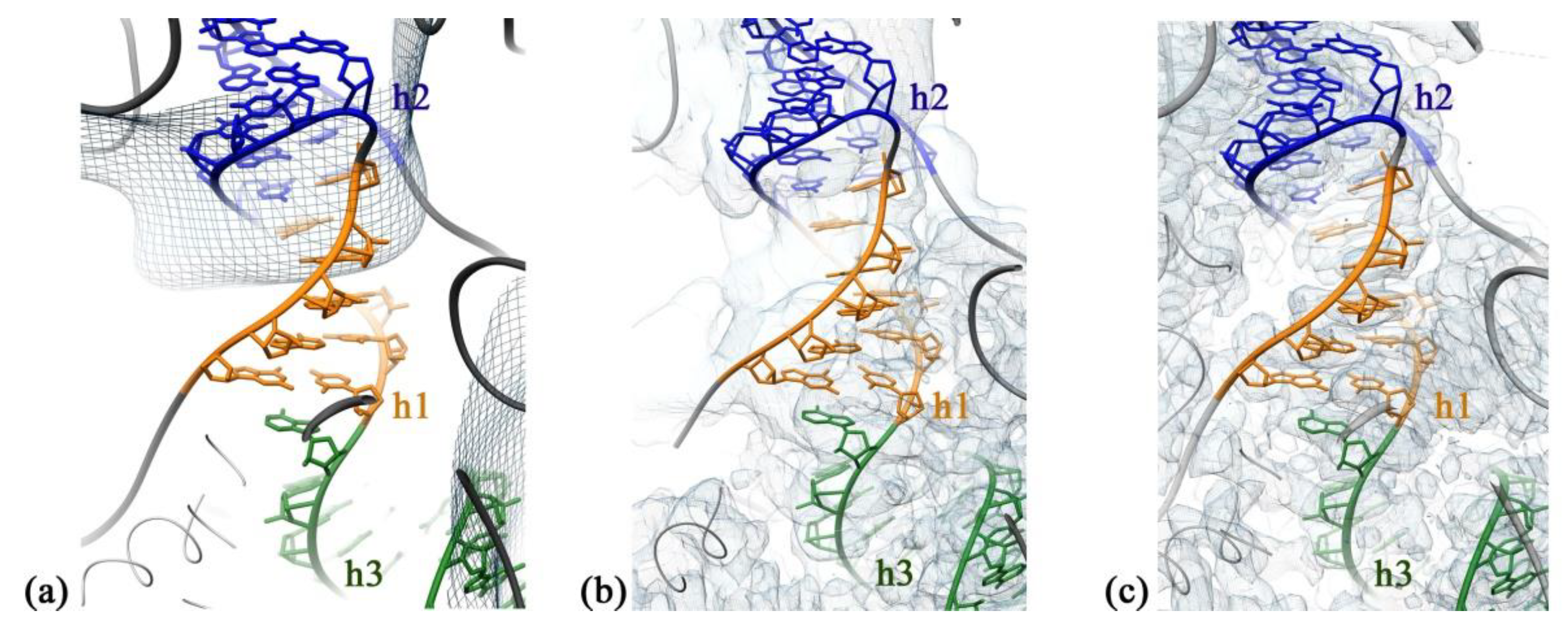
2.2. RbfA Participates in the Formation of the Central Pseudoknot
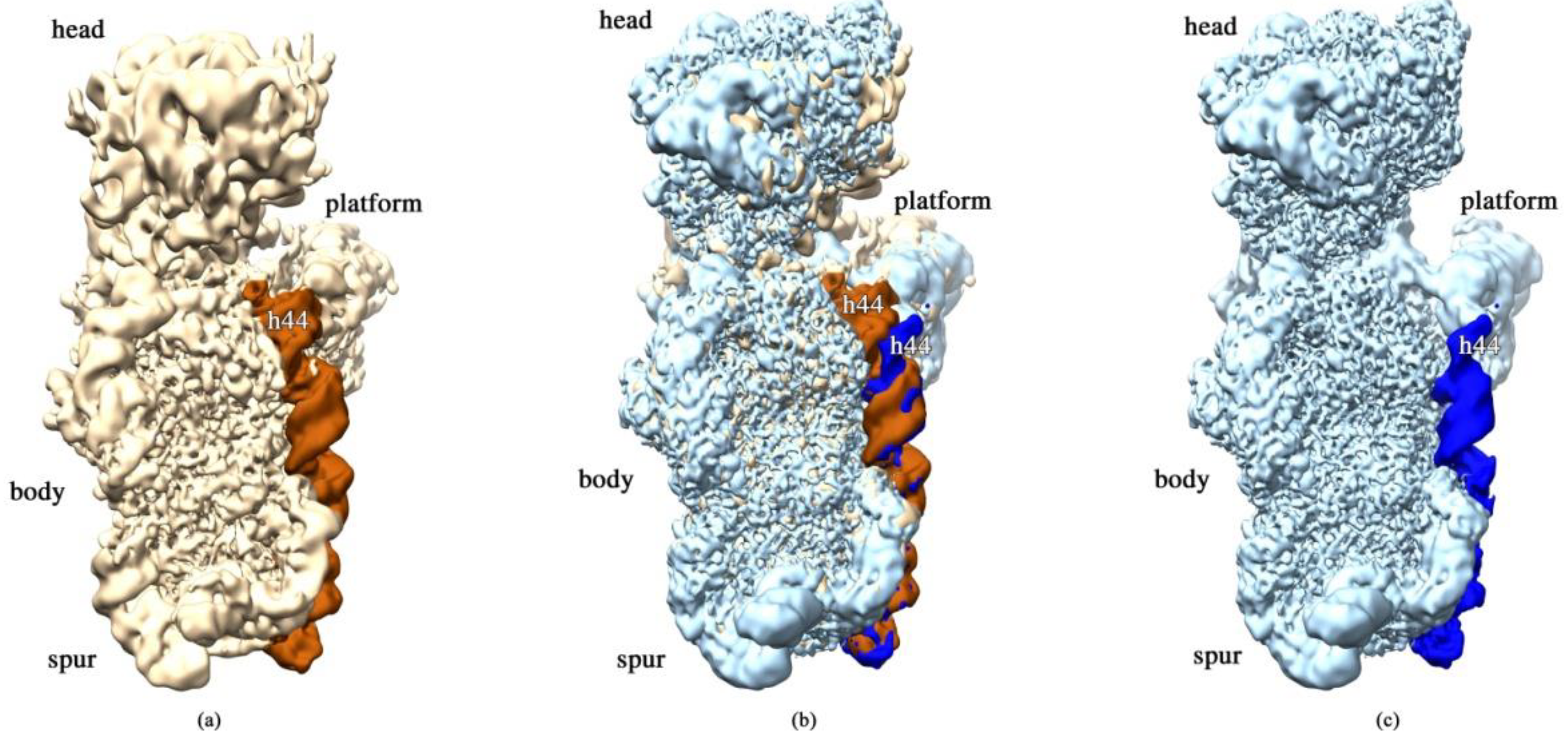
2.3. RbfA Is Necessary for Correct Docking of Helix 44
3. Discussion
4. Materials and Methods
4.1. E. coli Strains
4.2. Isolation of Wild-Type 30S Ribosomal Subunits and Immature ΔrbfA 30S Particles
4.3. Two-Dimensional Gel Electrophoresis
4.4. Cryo-Sample Preparation
4.5. Cryo-EM Data Collecting
4.6. Cryo-EM Processing
4.7. Model Building
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woodson, S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 2000, 57, 796–808. [Google Scholar] [CrossRef]
- Gulati, M.; Jain, N.; Davis, J.H.; Williamson, J.R.; Britton, R.A. Functional interaction between ribosomal protein L6 and RbgA during ribosome assembly. PLoS Genet. 2014, 10, e1004694. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, Y.J.; Swapna, G.V.T.; Rajan, P.K.; Ke, H.; Xia, B.; Shukla, K.; Inouye, M.; Montelione, G.T. Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J. Mol. Biol 2003, 327, 521–536. [Google Scholar] [CrossRef]
- Rubin, S.M.; Pelton, J.G.; Yokota, H.; Kim, R.; Wemmer, D.E. Solution structure of a putative ribosome binding protein from Mycoplasma pneumoniae and comparison to a distant homolog. J. Struct. Funct. Genom. 2003, 4, 235–243. [Google Scholar] [CrossRef]
- Dammel, C.S.; Noller, H.F. Suppression of a cold-sensitive mutation in 16s rRNA by overexpression of a novel ribosome-binding factor, RbfA. Genes Dev. 1995, 9, 626–637. [Google Scholar] [CrossRef]
- Xia, B.; Ke, H.; Shinde, U.; Inouye, M. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J. Mol. Biol. 2003, 332, 575–584. [Google Scholar] [CrossRef]
- Dammel, C.S.; Noller, H.F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993, 7, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.G.; Inouye, M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 1996, 21, 1207–1218. [Google Scholar] [CrossRef]
- Datta, P.P.; Wilson, D.N.; Kawazoe, M.; Swami, N.K.; Kaminishi, T.; Sharma, M.R.; Booth, T.M.; Takemoto, C.; Fucini, P.; Yokoyama, S.; et al. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol. Cell. 2007, 28, 434–445. [Google Scholar] [CrossRef]
- Bylund, G.O.; Wipemo, L.C.; Lundberg, L.A.; Wikstrom, P.M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli. J. Bacteriol. 1998, 180, 73–82. [Google Scholar] [CrossRef]
- Valverde, R.; Edwards, L.; Regan, L. Structure and function of KH domains. FEBS J. 2008, 275, 2712–2726. [Google Scholar] [CrossRef]
- Soper, S.F.C.; Dator, R.P.; Limbach, P.A.; Woodson, S.A. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell 2013, 52, 506–516. [Google Scholar] [CrossRef]
- Yang, Z.; Guo, Q.; Goto, S.; Chen, Y.; Li, N.; Yan, K.; Zhang, Y.; Muto, A.; Deng, H.; Himeno, H.; et al. Structural insights into the assembly of the 30S ribosomal subunit in vivo: Functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 2014, 5, 394–407. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Milon, P.; Konevega, A.L.; Peske, F.; Fabbretti, A.; Gualerzi, C.O.; Rodnina, M.V. Transient kinetics, fluorescence, and FRET in studies of initiation of translation in bacteria. Methods Enzymol. 2007, 430, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Wittmann-Liebold, B.; Greuer, B. The primary structure of protein S5 from the small subunit of the Escherichia coli ribosome. FEBS Lett. 1978, 95, 91–98. [Google Scholar] [CrossRef]
- Yaguchi, M. Primary structure of protein S18 from the small Escherichia coli ribosomal subunit. FEBS Lett. 1975, 59, 217–220. [Google Scholar] [CrossRef]
- Isono, K.; Krauss, J.; Hirota, Y. Isolation and characterization of temperature-sensitive mutants of Escherichta coli with altered ribosomal proteins. Mol. Gen. Genet. 1976, 149, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Isono, K.; Isono, S. Ribosomal Protein Modification in Escherichia coli. II. Studies of a Mutant Lacking the N-terminal Acetylation of Protein S18. Mol. Gen. Genet. 1980, 177, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Poot, R.A.; Jeeninga, R.E.; Pleij, C.W.; van Duin, J. Acetylation of ribosomal protein S5 affected by defects in the central pseudoknot in 16S ribosomal RNA? FEBS Lett. 1997, 401, 175–179. [Google Scholar] [CrossRef]
- Schluenzen, F.; Tocilj, A.; Zarivach, R.; Harms, J.; Gluehmann, M.; Janell, D.; Bashan, A.; Bartels, H.; Agmon, I.; Franceschi, F.; et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 2000, 102, 615–623. [Google Scholar] [CrossRef]
- Jahagirdar, D.; Jha, V.; Basu, K.; Gomez-Blanco, J.; Vargas, J.; Ortega, J. Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy. RNA 2020, 26, 2017–2030. [Google Scholar] [CrossRef] [PubMed]
- Jomaa, A.; Stewart, G.; Mears, J.A.; Kireeva, I.; Brown, E.D.; Ortega, J. Cryo-electron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA 2011, 17, 2026–2038. [Google Scholar] [CrossRef][Green Version]
- Guo, Q.; Goto, S.; Chen, Y.; Feng, B.; Xu, Y.; Muto, A.; Himeno, H.; Deng, H.; Lei, J.; Gao, N. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 2013, 41, 2609–2620. [Google Scholar] [CrossRef]
- Leong, V.; Kent, M.; Jomaa, A.; Ortega, J. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 2013, 19, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Razi, A.; Davis, J.H.; Hao, Y.; Jahagirdar, D.; Thurlow, B.; Basu, K.; Jain, N.; Gomez-Blanco, J.; Britton, R.A.; Vargas, J.; et al. Role of Era in assembly and homeostasis of the ribosomal small subunit. Nucl. Acids Res. 2019, 47, 8301–8317. [Google Scholar] [CrossRef]
- Mulder, A.M.; Yoshioka, C.; Beck, A.H.; Bunner, A.E.; Milligan, R.A.; Potter, C.S.; Bridget Carragher, B.; Williamson, J.R. Visualizing ribosome biogenesis: Parallel assembly pathways for 30S subunit. Science 2010, 330, 673–677. [Google Scholar] [CrossRef]
- Sashital, D.G.; Greeman, C.A.; Lyumkis, D.; Potter, C.S.; Carragher, B.; Williamson, J.R. A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. Elife 2014, 3, e04491. [Google Scholar] [CrossRef] [PubMed]
- Yusupov, M.M.; Yusupova, G.Z.; Baucom, A.; Lieberman, K.; Earnest, T.N.; Cate, J.H.; Noller, H.F. Crystal structure of the ribosome at 5.5 A resolution. Science 2001, 292, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Talkington, M.W.T.; Siuzdak, G.; Williamson, J.R. An assembly landscape for the 30S ribosomal subunit. Nature 2005, 438, 628–632. [Google Scholar] [CrossRef]
- Williamson, J.R. Assembly of the 30S ribosomal subunit. Q. Rev. Biophys. 2005, 38, 397–403. [Google Scholar] [CrossRef]
- Adilakshmi, T.; Bellur, D.L.; Woodson, S.A. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 2008, 455, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Kaczanowska, M.; Rydén-Aulin, M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007, 71, 477–494. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G., 3rd; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Madjar, J.J.; Michel, S.; Cozzone, A.J.; Reboud, J.P. A method to identify individual proteins in four different two-dimensional gel electrophoresis systems: Application to Escherichia coli ribosomal proteins. Anal. Biochem. 1979, 92, 174–182. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Tegunov, D.; Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 2019, 16, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Punjani, A.; Fleet, D.J. 3D Variability Analysis: Resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021, 213, 107702. [Google Scholar] [CrossRef]
- Punjani, A.; Zhang, H.; Fleet, D.J. Non-uniform refinement: Adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 2020, 17, 1214–1221. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Wachi, M.; Umitsuki, G.; Shimizu, M.; Takada, A.; Nagai, K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 1999, 259, 483–488. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maksimova, E.M.; Korepanov, A.P.; Kravchenko, O.V.; Baymukhametov, T.N.; Myasnikov, A.G.; Vassilenko, K.S.; Afonina, Z.A.; Stolboushkina, E.A. RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center. Int. J. Mol. Sci. 2021, 22, 6140. https://doi.org/10.3390/ijms22116140
Maksimova EM, Korepanov AP, Kravchenko OV, Baymukhametov TN, Myasnikov AG, Vassilenko KS, Afonina ZA, Stolboushkina EA. RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center. International Journal of Molecular Sciences. 2021; 22(11):6140. https://doi.org/10.3390/ijms22116140
Chicago/Turabian StyleMaksimova, Elena M., Alexey P. Korepanov, Olesya V. Kravchenko, Timur N. Baymukhametov, Alexander G. Myasnikov, Konstantin S. Vassilenko, Zhanna A. Afonina, and Elena A. Stolboushkina. 2021. "RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center" International Journal of Molecular Sciences 22, no. 11: 6140. https://doi.org/10.3390/ijms22116140
APA StyleMaksimova, E. M., Korepanov, A. P., Kravchenko, O. V., Baymukhametov, T. N., Myasnikov, A. G., Vassilenko, K. S., Afonina, Z. A., & Stolboushkina, E. A. (2021). RbfA Is Involved in Two Important Stages of 30S Subunit Assembly: Formation of the Central Pseudoknot and Docking of Helix 44 to the Decoding Center. International Journal of Molecular Sciences, 22(11), 6140. https://doi.org/10.3390/ijms22116140






