Magnesium Isoglycyrrhizinate Reduces Hepatic Lipotoxicity through Regulating Metabolic Abnormalities
Abstract
1. Introduction
2. Results
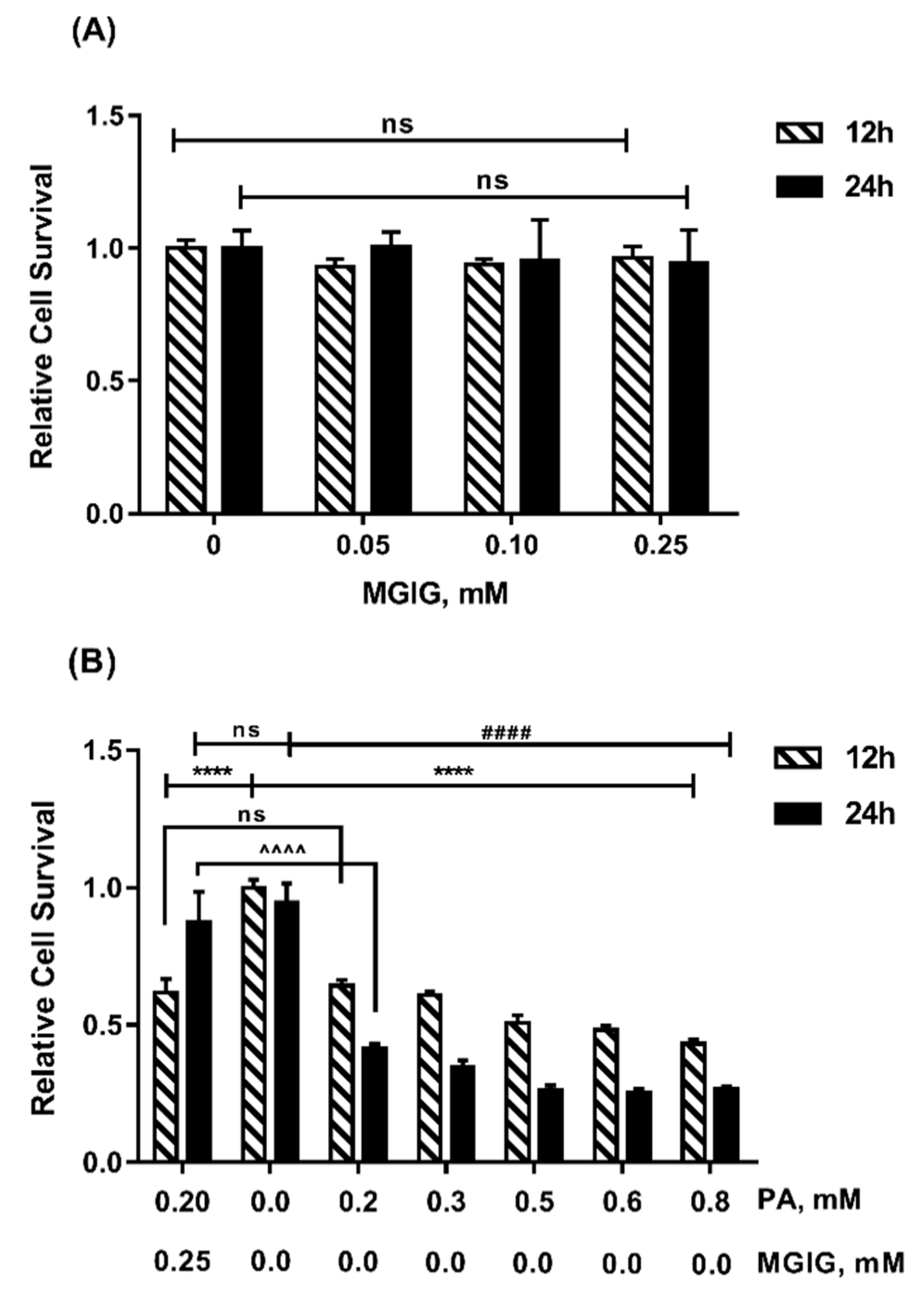
2.1. The Protective Effect of MGIG on PA-INDUCED Cytotoxicity
2.2. The Protective Effect of MGIG against PA-Induced Organelle Damage
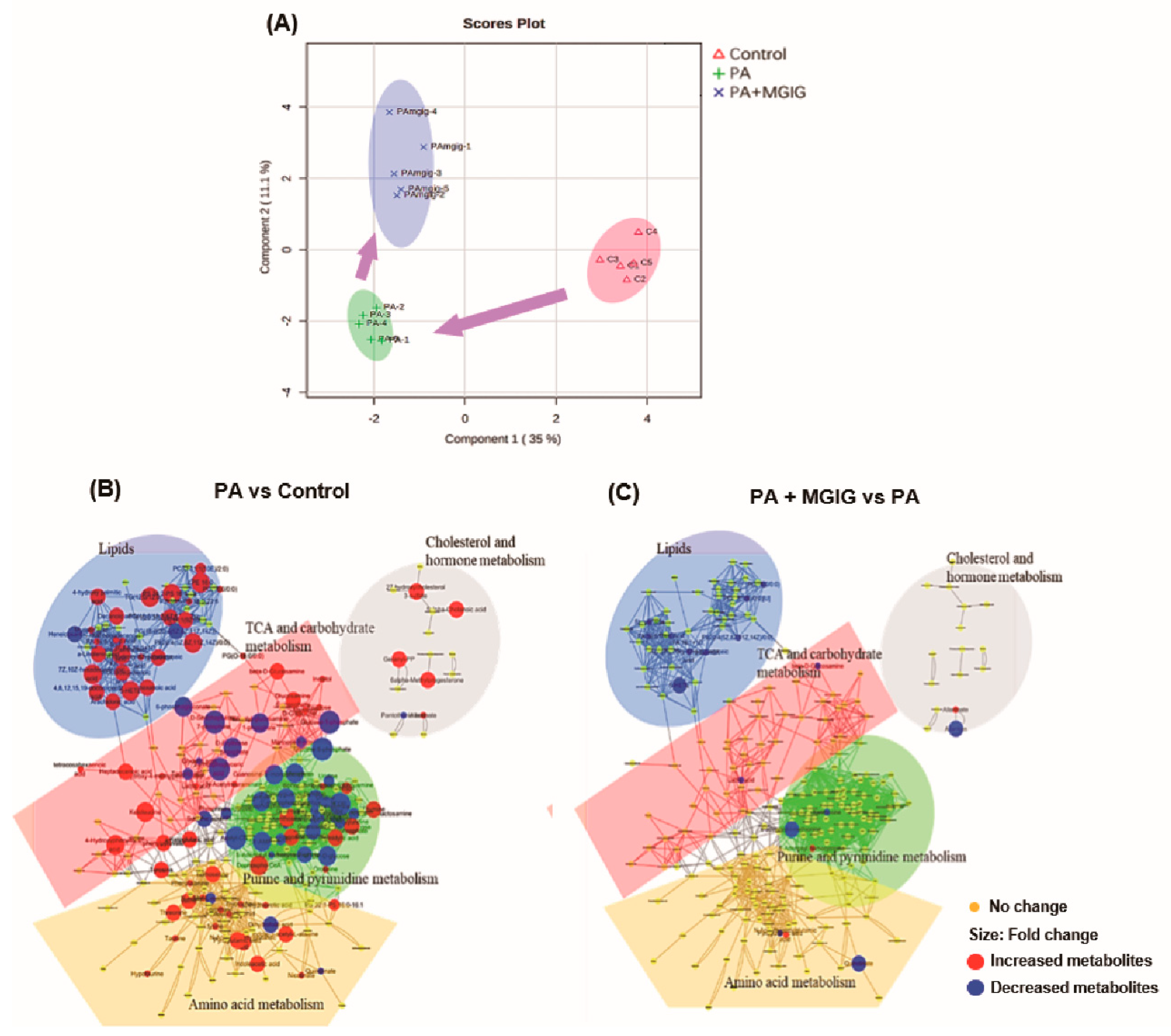
2.3. The Regulatory Effect of MGIG on PA-Induced Metabolic Reprogramming
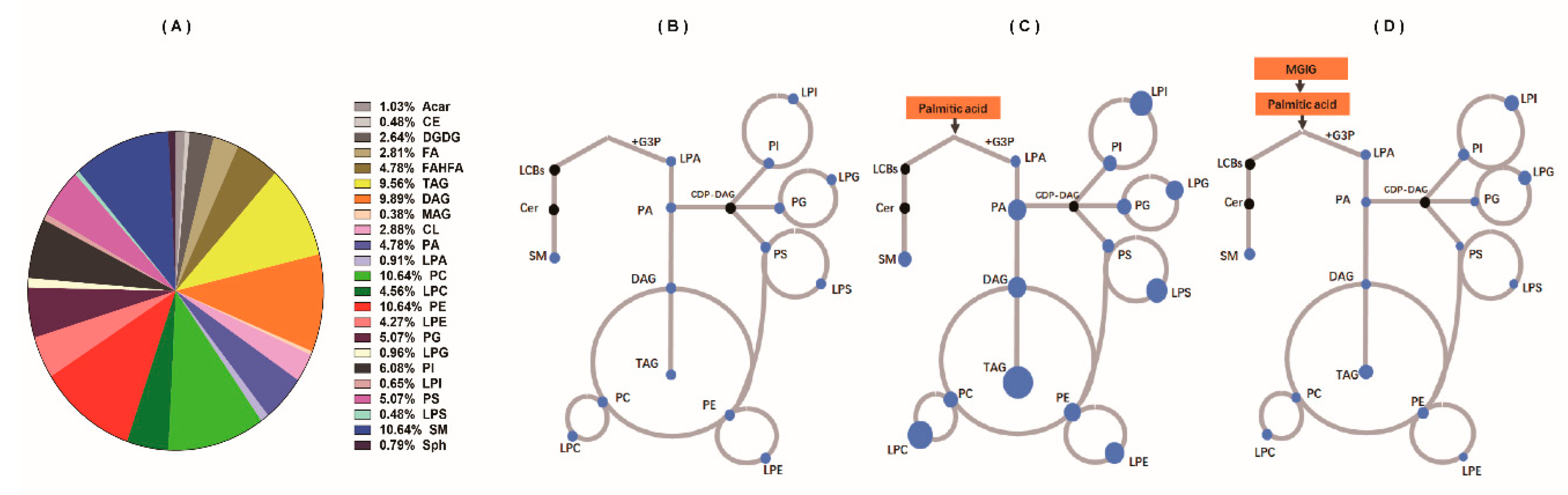
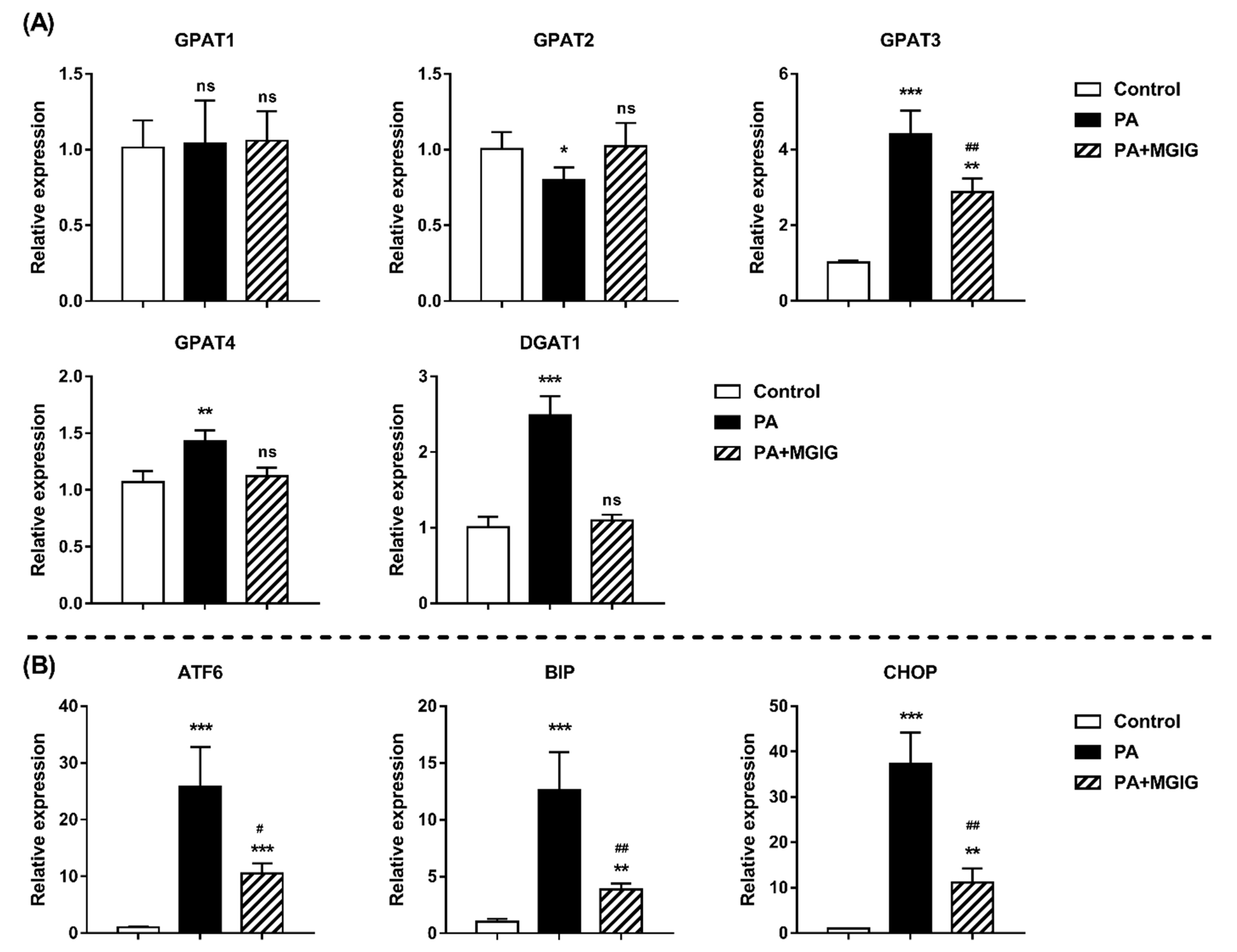
2.4. The Regulatory Effect of MGIG on PA-Induced Abnormal Lipid Metabolism
3. Discussion and Conclusions
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. Cell Proliferation Assay
4.5. Cell Damage Assay
4.6. Gene Expression Profiling and qPCR
4.7. Metabolomics Analysis
4.7.1. Metabolomics Sample Preparation
4.7.2. Metabolomics Sample Detection
4.7.3. Metabolomics Data Analysis
4.8. Lipidomics Analysis
4.8.1. Lipidomics Sample Preparation
4.8.2. Lipidomics Sample Detection
4.8.3. Lipidomics Data Analysis
4.9. Cytoscape Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Henne, W.M.; Reese, M.L.; Goodman, J.M. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Brookheart, R.T.; Michel, C.I.; Schaffer, J.E. As a Matter of Fat. Cell Metab. 2009, 10, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Van Herpen, N.A.; Schrauwen-Hinderling, V.B. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol. Behav. 2008, 94, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, H.; Goto, M.; Tamai, N. Membrane States of Saturated Glycerophospholipids: A Thermodynamic Study of Bilayer Phase Transitions. Chem. Pharm. Bull. 2019, 67, 300–307. [Google Scholar] [CrossRef]
- Lara-Castro, C.; Garvey, W.T. Intracellular Lipid Accumulation in Liver and Muscle and the Insulin Resistance Syndrome. Endocrinol. Metab. Clin. N. Am. 2008, 37, 841–856. [Google Scholar] [CrossRef]
- Yazıcı, D.; Sezer, H. Insulin Resistance, Obesity and Lipotoxicity. Adv. Exp. Med. Biol. 2017, 960, 277–304. [Google Scholar] [CrossRef]
- Rada, P.; Gonzalez-Rodriguez, A.; Garcia-Monzon, C.; Valverde, A.M. Understanding lipotoxicity in NAFLD pathogenesis: Is CD36 a key driver? Cell Death Dis. 2020, 11, 802. [Google Scholar] [CrossRef]
- Mota, M.; Banini, B.A.; Cazanave, S.C.; Sanyal, A.J. Molecular mechanisms of lipotoxicity and glucotoxicity in nonalcoholic fatty liver disease. Metabolism 2016, 65, 1049–1061. [Google Scholar] [CrossRef]
- Neuschwander-Tetri, B.A. Non-alcoholic fatty liver disease. BMC Med. 2017, 15, 45. [Google Scholar] [CrossRef]
- Basaranoglu, M.; Ormeci, N. Nonalcoholic fatty liver disease: Diagnosis, pathogenesis, and management. Turk. J. Gastroenterol. 2014, 25, 127–132. [Google Scholar] [CrossRef]
- Sui, M.; Jiang, X.; Chen, J.; Yang, H.; Zhu, Y. Magnesium isoglycyrrhizinate ameliorates liver fibrosis and hepatic stellate cell activation by regulating ferroptosis signaling pathway. Biomed. Pharmacother. 2018, 106, 125–133. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Liu, Z.H.; Wang, S.C.; Zhang, X.Q.; Xu, H.J.; Yang, L.; Kong, L.D. Magnesium isoglycyrrhizinate alleviates fructose-induced liver oxidative stress and inflammatory injury through suppressing NOXs. Eur. J. Pharmacol. 2020, 883, 173314. [Google Scholar] [CrossRef]
- Zou, X.; Wang, Y.; Peng, C.; Wang, B.; Niu, Z.; Li, Z.; Niu, J. Magnesium isoglycyrrhizinate has hepatoprotective effects in an oxaliplatininduced model of liver injury. Int. J. Mol. Med. 2018, 42, 2020–2030. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Gao, M.; Zhong, H.; Chen, C.; Yao, Y.; Zhang, Z.; Zhang, X.; Li, F.; Zhang, J.; et al. Efficacy and safety of magnesium isoglycyrrhizinate injection in patients with acute drug-induced liver injury: A phase II trial. Liver Int. 2019, 39, 2102–2111. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, J.; Shang, J.; Zhang, L. Prevention of Free Fatty Acid-Induced Hepatic Lipotoxicity in HepG2 Cells by Magnesium Isoglycyrrhizinate in vitro. Pharmacology 2009, 84, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, J.; Chen, F.; Lin, K.; Zhu, M.; Chen, L.; Zhou, X.; Li, C.; Zhu, H. Protective role of magnesium isoglycyrrhizinate in non-alcoholic fatty liver disease and the associated molecular mechanisms. Int. J. Mol. Med. 2016, 38, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; de Sousa, M.C.; Foti, M. miRNAs and NAFLD: From pathophysiology to therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef]
- Li, B.; He, X.; Jia, W.; Li, H. Novel Applications of Metabolomics in Personalized Medicine: A Mini-Review. Molecules 2017, 22, 1173. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Hammock, B.D.; Watkins, S.M. Metabolomics: Building on a century of biochemistry to guide human health. Metabolomics 2005, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Rattray, N.J.W.; Deziel, N.C.; Wallach, J.D.; Khan, S.A.; Vasiliou, V.; Ioannidis, J.P.A.; Johnson, C.H. Beyond genomics: Understanding exposotypes through metabolomics. Hum. Genom. 2018, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.R. Lipidomics: New Tools and Applications. Cell 2010, 143, 888–895. [Google Scholar] [CrossRef]
- Brügger, B. Lipidomics: Analysis of the Lipid Composition of Cells and Subcellular Organelles by Electrospray Ionization Mass Spectrometry. Annu. Rev. Biochem. 2014, 83, 79–98. [Google Scholar] [CrossRef]
- Engin, A.B. What Is Lipotoxicity? Adv. Exp. Med. Biol. 2017, 960, 197–220. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.D.; Serra-Caetano, A.; Cabrita, M.; Bekman, E.; Braga, J.; Rino, J.; Santus, R.; Filipe, P.L.; Sousa, A.E.; Ferreira, J.A. Quantification of cell cycle kinetics by EdU (5-ethynyl-2′-deoxyuridine)-coupled-fluorescence-intensity analysis. Oncotarget 2017, 8, 40514–40532. [Google Scholar] [CrossRef]
- Shen, Y.; Zhao, Z.; Zhang, L.; Shi, L.; Shahriar, S.; Chan, R.B.; Di Paolo, G.; Min, W. Metabolic activity induces membrane phase separation in endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2017, 114, 13394–13399. [Google Scholar] [CrossRef]
- Nagle, C.A.; Vergnes, L.; Dejong, H.; Wang, S.; Lewin, T.M.; Reue, K.; Coleman, R.A. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 2008, 49, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Beigneux, A.P.; Vergnes, L.; Qiao, X.; Quatela, S.; Davis, R.; Watkins, S.M.; Coleman, R.A.; Walzem, R.L.; Philips, M.; Reue, K.; et al. Agpat6--a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J. Lipid Res. 2006, 47, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Kuo, M.S.; Li, S.; Bui, H.H.; Peake, D.A.; Sanders, P.E.; Thibodeaux, S.J.; Chu, S.; Qian, Y.W.; Zhao, Y.; et al. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J. Biol. Chem. 2008, 283, 10048–10057. [Google Scholar] [CrossRef]
- Unger, R.H.; Scherer, P.E. Gluttony, sloth and the metabolic syndrome: A roadmap to lipotoxicity. Trends Endocrinol. Metab. 2010, 21, 345–352. [Google Scholar] [CrossRef]
- Wang, S.; Kaufman, R.J. How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr. Opin. Lipidol. 2014, 25, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Fagone, P.; Jackowski, S. Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res. 2009, 50, S311–S316. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, E.; Artemenko, K.; Manukyan, L.; Bergquist, J.; Bergsten, P. Oleate protects beta-cells from the toxic effect of palmitate by activating pro-survival pathways of the ER stress response. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2016, 1861, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kaufman, R.J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Hao, K.; Hong, Y.; Liu, J.; Zhu, J.; Jiang, W.; Zhu, Z.; Wang, G.; Peng, Y. Magnesium Isoglycyrrhizinate Reduces Hepatic Lipotoxicity through Regulating Metabolic Abnormalities. Int. J. Mol. Sci. 2021, 22, 5884. https://doi.org/10.3390/ijms22115884
Lu L, Hao K, Hong Y, Liu J, Zhu J, Jiang W, Zhu Z, Wang G, Peng Y. Magnesium Isoglycyrrhizinate Reduces Hepatic Lipotoxicity through Regulating Metabolic Abnormalities. International Journal of Molecular Sciences. 2021; 22(11):5884. https://doi.org/10.3390/ijms22115884
Chicago/Turabian StyleLu, Li, Kun Hao, Yu Hong, Jie Liu, Jinwei Zhu, Wenjiao Jiang, Zheying Zhu, Guangji Wang, and Ying Peng. 2021. "Magnesium Isoglycyrrhizinate Reduces Hepatic Lipotoxicity through Regulating Metabolic Abnormalities" International Journal of Molecular Sciences 22, no. 11: 5884. https://doi.org/10.3390/ijms22115884
APA StyleLu, L., Hao, K., Hong, Y., Liu, J., Zhu, J., Jiang, W., Zhu, Z., Wang, G., & Peng, Y. (2021). Magnesium Isoglycyrrhizinate Reduces Hepatic Lipotoxicity through Regulating Metabolic Abnormalities. International Journal of Molecular Sciences, 22(11), 5884. https://doi.org/10.3390/ijms22115884





