Assessment of a Small Molecule Synthetic Lignan in Enhancing Oxidative Balance and Decreasing Lipid Accumulation in Human Retinal Pigment Epithelia
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Generation of RPE-LC3B
2.3. Maintenance of RPE Cell Culture
2.4. Cell Stress Studies
2.5. OS Phagocytic Challenge and LGM2605 Treatment
2.5.1. Challenge with Photoreceptor Outer Segments (OS)
2.5.2. Treatment with LGM2605
2.6. RNA Isolation and Gene Expression Analysis
2.7. Immunoblotting
2.8. Staining for Neutral Lipids
2.9. Staining for Mitochondria
2.10. Staining for Actin
2.11. Transepithelial Resistance (TER)
2.12. TUNEL Staining
2.13. Ketone Body Measurements, β-Hydroxybutyrate (β-HB) Assay
2.14. Lactate Dehydrogenase (LDH) Assay
2.15. Lipid Peroxidation
2.16. Cytokine Analysis
2.17. Statistical Analyses
3. Results
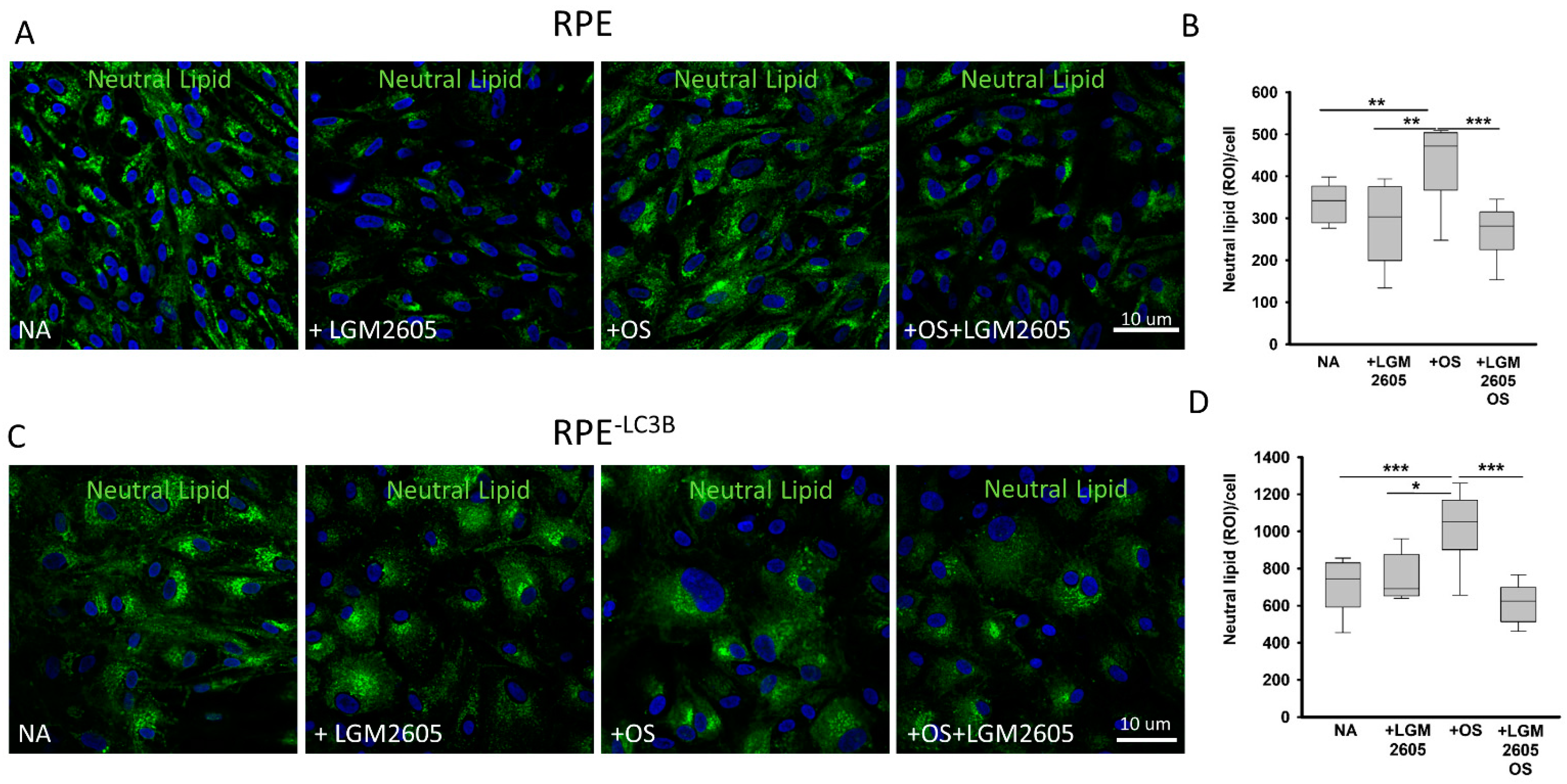
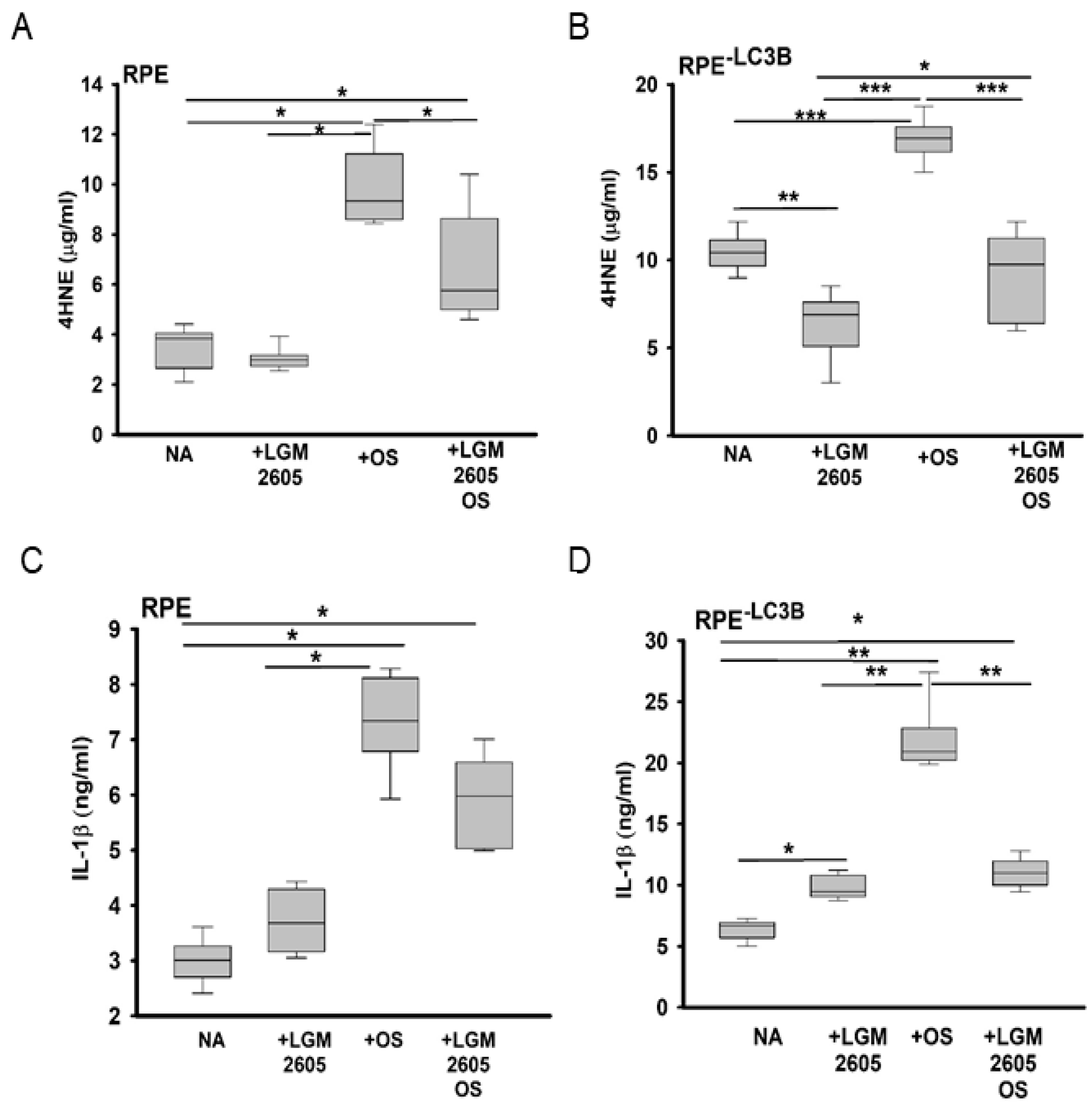
3.1. Lipid Overload and Cytokine Release in RPE
3.2. LGM2605 Restores Oxidative Capacity and Decrease Cytokine Release in RPE
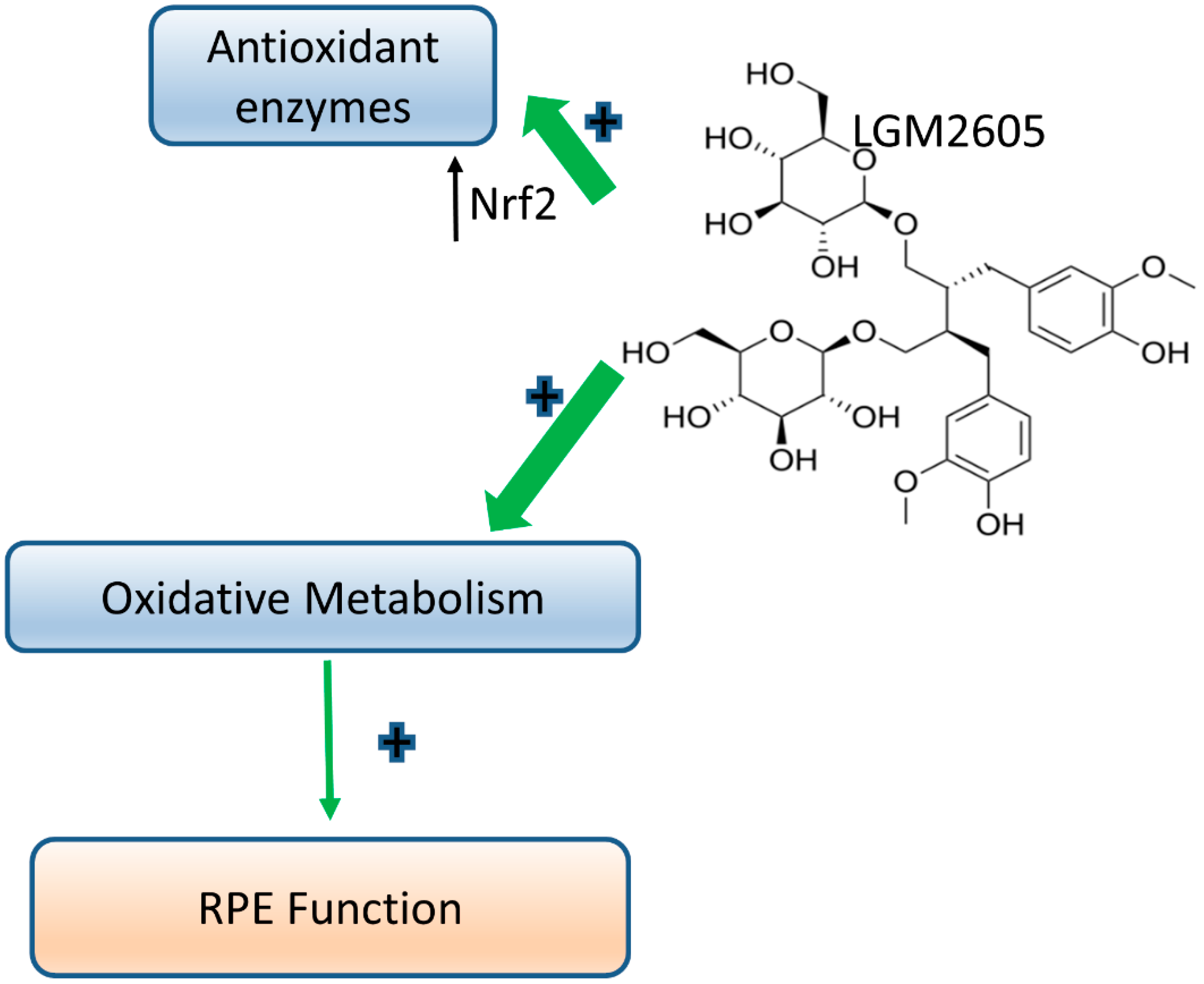
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Léveillard, T.; Philp, N.J.; Sennlaub, F. Is Retinal Metabolic Dysfunction at the Center of the Pathogenesis of Age-related Macular Degeneration? Int. J. Mol. Sci. 2019, 20, 762. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, E.M.; Emri, E.; Merle, B.M.J.; Colijn, J.M.; Kersten, E.; Cougnard-Gregoire, A.; Dammeier, S.; Meester-Smoor, M.; Pool, F.M.; de Jong, E.K.; et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye Res. 2018, 67, 56–86. [Google Scholar] [CrossRef]
- Curcio, C.A. Soft Drusen in Age-Related Macular Degeneration: Biology and Targeting Via the Oil Spill Strategies. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD160–AMD181. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Batliwala, S.; Xavier, C.; Liu, Y.; Wu, H.; Pang, I.H. Involvement of Nrf2 in Ocular Diseases. Oxid. Med. Cell Longev. 2017, 2017, 1703810. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, M.T.; Maldonado, R.S.; Toth, C.A.; O’Connell, R.V.; Chen, B.B.; Chiu, S.J.; Farsiu, S.; Wallace, D.K.; Stinnett, S.S.; Panayotti, G.M.; et al. Subfoveal fluid in healthy full-term newborns observed by handheld spectral-domain optical coherence tomography. Am. J. Ophthalmol. 2012, 153, 167–175.e163. [Google Scholar] [CrossRef]
- Cabrera, M.T.; Freedman, S.F.; Kiely, A.E.; Chiang, M.F.; Wallace, D.K. Combining ROPtool measurements of vascular tortuosity and width to quantify plus disease in retinopathy of prematurity. J. Aapos 2011, 15, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Asatryan, A.; Bazan, N.G. Molecular mechanisms of signaling via the docosanoid neuroprotectin D1 for cellular homeostasis and neuroprotection. J. Biol. Chem. 2017, 292, 12390–12397. [Google Scholar] [CrossRef]
- Datta, S.; Cano, M.; Ebrahimi, K.; Wang, L.; Handa, J.T. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 2017, 60, 201–218. [Google Scholar] [CrossRef]
- Framme, C.; Schuele, G.; Birngruber, R.; Roider, J.; Schuett, F.; Kopitz, J.; Holz, F.; Brinkmann, R. Temperature dependent fluorescence of A2-E, the main fluorescent lipofuscin component in the RPE. Curr. Eye Res. 2004, 29, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.; Umapathy, A.; Tan, L.X.; Daniele, L.; Philp, N.J.; Boesze-Battaglia, K.; Williams, D.S. The cell biology of the retinal pigment epithelium. Prog. Retin. Eye Res. 2020, 100846. [Google Scholar] [CrossRef] [PubMed]
- Volland, S.; Esteve-Rudd, J.; Hoo, J.; Yee, C.; Williams, D.S. A comparison of some organizational characteristics of the mouse central retina and the human macula. PLoS ONE 2015, 10, e0125631. [Google Scholar] [CrossRef] [PubMed]
- Adijanto, J.; Du, J.; Moffat, C.; Seifert, E.L.; Hurle, J.B.; Philp, N.J. The retinal pigment epithelium utilizes fatty acids for ketogenesis. J. Biol. Chem. 2014, 289, 20570–20582. [Google Scholar] [CrossRef]
- Reyes-Reveles, J.; Dhingra, A.; Alexander, D.; Bragin, A.; Philp, N.J.; Boesze-Battaglia, K. Phagocytosis-dependent ketogenesis in retinal pigment epithelium. J. Biol. Chem. 2017, 292, 8038–8047. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Rudd, J.; Hazim, R.A.; Diemer, T.; Paniagua, A.E.; Volland, S.; Umapathy, A.; Williams, D.S. Defective phagosome motility and degradation in cell nonautonomous RPE pathogenesis of a dominant macular degeneration. Proc. Natl. Acad. Sci. USA 2018, 115, 5468–5473. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Zhao, H.; Martinez, J.; Doggett, T.A.; Kolesnikov, A.V.; Tang, P.H.; Ablonczy, Z.; Chan, C.C.; Zhou, Z.; Green, D.R.; et al. Noncanonical autophagy promotes the visual cycle. Cell 2013, 154, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Guziewicz, K.E.; Sinha, D.; Gómez, N.M.; Zorych, K.; Dutrow, E.V.; Dhingra, A.; Mullins, R.F.; Stone, E.M.; Gamm, D.M.; Boesze-Battaglia, K.; et al. Bestrophinopathy: An RPE-photoreceptor interface disease. Prog. Retin. Eye Res. 2017, 58, 70–88. [Google Scholar] [CrossRef]
- Zhang, Y.; Cross, S.D.; Stanton, J.B.; Marmorstein, A.D.; Le, Y.Z.; Marmorstein, L.Y. Early AMD-like defects in the RPE and retinal degeneration in aged mice with RPE-specific deletion of Atg5 or Atg7. Mol. Vis. 2017, 23, 228. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Mitchell, C.H.; Boesze-Battaglia, K. Autophagy in the eye: Implications for ocular cell health. Exp. Eye Res. 2014, 124, 56–66. [Google Scholar] [CrossRef]
- Ferrington, D.A.; Sinha, D.; Kaarniranta, K. Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration. Prog. Retin. Eye Res. 2016, 51, 69–89. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Tokarz, P.; Koskela, A.; Paterno, J.; Blasiak, J. Autophagy regulates death of retinal pigment epithelium cells in age-related macular degeneration. Cell Biol. Toxicol. 2017, 33, 113–128. [Google Scholar] [CrossRef]
- Song, C.; Mitter, S.K.; Qi, X.; Beli, E.; Rao, H.V.; Ding, J.; Ip, C.S.; Gu, H.; Akin, D.; Dunn, W.A., Jr.; et al. Oxidative stress-mediated NFκB phosphorylation upregulates p62/SQSTM1 and promotes retinal pigmented epithelial cell survival through increased autophagy. PLoS ONE 2017, 12, e0171940. [Google Scholar] [CrossRef] [PubMed]
- Ando, S.; Hashida, N.; Yamashita, D.; Kawabata, T.; Asao, K.; Kawasaki, S.; Sakurai, K.; Yoshimori, T.; Nishida, K. Rubicon regulates A2E-induced autophagy impairment in the retinal pigment epithelium implicated in the pathology of age-related macular degeneration. Biochem. Biophys. Res. Commun. 2021, 551, 148–154. [Google Scholar] [CrossRef]
- Szatmári-Tóth, M.; Kristóf, E.; Veréb, Z.; Akhtar, S.; Facskó, A.; Fésüs, L.; Kauppinen, A.; Kaarniranta, K.; Petrovski, G. Clearance of autophagy-associated dying retinal pigment epithelial cells—A possible source for inflammation in age-related macular degeneration. Cell Death Dis. 2016, 7, e2367. [Google Scholar] [CrossRef] [PubMed]
- Mitter, S.K.; Rao, H.V.; Qi, X.; Cai, J.; Sugrue, A.; Dunn, W.A., Jr.; Grant, M.B.; Boulton, M.E. Autophagy in the retina: A potential role in age-related macular degeneration. Adv. Exp. Med. Biol. 2012, 723, 83–90. [Google Scholar] [CrossRef]
- Rodríguez-Muela, N.; Koga, H.; García-Ledo, L.; de la Villa, P.; de la Rosa, E.J.; Cuervo, A.M.; Boya, P. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 2013, 12, 478–488. [Google Scholar] [CrossRef]
- Dhingra, A.; Bell, B.A.; Peachey, N.S.; Daniele, L.L.; Reyes-Reveles, J.; Sharp, R.C.; Jun, B.; Bazan, N.G.; Sparrow, J.R.; Kim, H.J.; et al. Microtubule-Associated Protein 1 Light Chain 3B, (LC3B) Is Necessary to Maintain Lipid-Mediated Homeostasis in the Retinal Pigment Epithelium. Front. Cell Neurosci. 2018, 12, 351. [Google Scholar] [CrossRef] [PubMed]
- Muniz-Feliciano, L.; Doggett, T.A.; Zhou, Z.; Ferguson, T.A. RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy 2017, 13, 2072–2085. [Google Scholar] [CrossRef]
- Martinez, J. LAP it up, fuzz ball: A short history of LC3-associated phagocytosis. Curr. Opin. Immunol. 2018, 55, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J. Detection of LC3-Associated Phagocytosis (LAP). Curr. Protoc. Cell Biol. 2020, 87, e104. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Cunha, L.D.; Park, S.; Yang, M.; Lu, Q.; Orchard, R.; Li, Q.Z.; Yan, M.; Janke, L.; Guy, C.; et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 2016, 533, 115–119. [Google Scholar] [CrossRef]
- Frost, L.S.; Lopes, V.S.; Bragin, A.; Reyes-Reveles, J.; Brancato, J.; Cohen, A.; Mitchell, C.H.; Williams, D.S.; Boesze-Battaglia, K. The Contribution of Melanoregulin to Microtubule-Associated Protein 1 Light Chain 3 (LC3) Associated Phagocytosis in Retinal Pigment Epithelium. Mol. Neurobiol. 2015, 52, 1135–1151. [Google Scholar] [CrossRef]
- Yao, J.; Jia, L.; Shelby, S.J.; Ganios, A.M.; Feathers, K.; Thompson, D.A.; Zacks, D.N. Circadian and noncircadian modulation of autophagy in photoreceptors and retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 2014, 55, 3237–3246. [Google Scholar] [CrossRef]
- Daniele, L.L.; Caughey, J.; Volland, S.; Sharp, R.C.; Dhingra, A.; Williams, D.S.; Philp, N.J.; Boesze-Battaglia, K. Peroxisome turnover and diurnal modulation of antioxidant activity in retinal pigment epithelia utilizes microtubule-associated protein 1 light chain 3B (LC3B). Am. J. Physiol. Cell Physiol. 2019, 317, C1194–C1204. [Google Scholar] [CrossRef]
- Kokkinaki, D.; Hoffman, M.; Kalliora, C.; Kyriazis, I.D.; Maning, J.; Lucchese, A.M.; Shanmughapriya, S.; Tomar, D.; Park, J.Y.; Wang, H.; et al. Chemically synthesized Secoisolariciresinol diglucoside (LGM2605) improves mitochondrial function in cardiac myocytes and alleviates septic cardiomyopathy. J. Mol. Cell Cardiol. 2019, 127, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Mishra, O.P.; Pietrofesa, R.; Christofidou-Solomidou, M. Novel synthetic (S,S) and (R,R)-secoisolariciresinol diglucosides (SDGs) protect naked plasmid and genomic DNA From gamma radiation damage. Radiat. Res. 2014, 182, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Mishra, O.P.; Popov, A.V.; Pietrofesa, R.A.; Christofidou-Solomidou, M. Gamma-irradiation produces active chlorine species (ACS) in physiological solutions: Secoisolariciresinol diglucoside (SDG) scavenges ACS—A novel mechanism of DNA radioprotection. Biochim. Biophys. Acta 2016, 1860, 1884–1897. [Google Scholar] [CrossRef]
- Velalopoulou, A.; Tyagi, S.; Pietrofesa, R.A.; Arguiri, E.; Christofidou-Solomidou, M. The Flaxseed-Derived Lignan Phenolic Secoisolariciresinol Diglucoside (SDG) Protects Non-Malignant Lung Cells from Radiation Damage. Int. J. Mol. Sci. 2015, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Rom, S.; Zuluaga-Ramirez, V.; Reichenbach, N.L.; Erickson, M.A.; Winfield, M.; Gajghate, S.; Christofidou-Solomidou, M.; Jordan-Sciutto, K.L.; Persidsky, Y. Secoisolariciresinol diglucoside is a blood-brain barrier protective and anti-inflammatory agent: Implications for neuroinflammation. J. Neuroinflamm. 2018, 15, 25. [Google Scholar] [CrossRef]
- Kartha, S.; Weisshaar, C.L.; Pietrofesa, R.A.; Christofidou-Solomidou, M.; Winkelstein, B.A. Synthetic Secoisolariciresinol Diglucoside Attenuates Established Pain, Oxidative Stress and Neuroinflammation in a Rodent Model of Painful Radiculopathy. Antioxidants 2020, 9, 1209. [Google Scholar] [CrossRef]
- Christofidou-Solomidou, M.; Pietrofesa, R.A.; Park, K.; Albelda, S.M.; Serve, K.M.; Keil, D.E.; Pfau, J.C. Synthetic secoisolariciresinol diglucoside (LGM2605) inhibits Libby amphibole fiber-induced acute inflammation in mice. Toxicol. Appl. Pharmacol. 2019, 375, 81–93. [Google Scholar] [CrossRef]
- Flayer, C.H.; Larson, E.D.; Joseph, A.; Kao, S.; Qu, W.; Van Haren, A.; Royer, C.M.; Miller, L.A.; Capitanio, J.P.; Sielecki, T.; et al. Ozone-induced enhancement of airway hyperreactivity in rhesus macaques: Effects of antioxidant treatment. J. Allergy Clin. Immunol. 2020, 145, 312–323. [Google Scholar] [CrossRef]
- Velalopoulou, A.; Chatterjee, S.; Pietrofesa, R.A.; Koziol-White, C.; Panettieri, R.A.; Lin, L.; Tuttle, S.; Berman, A.; Koumenis, C.; Christofidou-Solomidou, M. Synthetic Secoisolariciresinol Diglucoside (LGM2605) Protects Human Lung in an Ex Vivo Model of Proton Radiation Damage. Int. J. Mol. Sci. 2017, 18, 2525. [Google Scholar] [CrossRef] [PubMed]
- Mishra, O.P.; Simmons, N.; Tyagi, S.; Pietrofesa, R.; Shuvaev, V.V.; Valiulin, R.A.; Heretsch, P.; Nicolaou, K.C.; Christofidou-Solomidou, M. Synthesis and antioxidant evaluation of (S,S)- and (R,R)-secoisolariciresinol diglucosides (SDGs). Bioorg. Med. Chem. Lett. 2013, 23, 5325–5328. [Google Scholar] [CrossRef]
- Frost, L.S.; Lopes, V.S.; Stefano, F.P.; Bragin, A.; Williams, D.S.; Mitchell, C.H.; Boesze-Battaglia, K. Loss of melanoregulin (MREG) enhances cathepsin-D secretion by the retinal pigment epithelium. Vis. Neurosci. 2013, 30, 55–64. [Google Scholar] [CrossRef]
- Frost, L.S.; Dhingra, A.; Reyes-Reveles, J.; Boesze-Battaglia, K. The Use of DQ-BSA to Monitor the Turnover of Autophagy-Associated Cargo. Methods Enzymol. 2017, 587, 43–54. [Google Scholar] [CrossRef]
- Weng, S.; Mao, L.; Gong, Y.; Sun, T.; Gu, Q. Role of quercetin in protecting ARPE-19 cells against H2O2-induced injury via nuclear factor erythroid 2 like 2 pathway activation and endoplasmic reticulum stress inhibition. Mol. Med. Rep. 2017, 16, 3461–3468. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Marcheselli, V.L.; Serhan, C.N.; Bazan, N.G. Neuroprotectin D1: A docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc. Natl. Acad. Sci. USA 2004, 101, 8491–8496. [Google Scholar] [CrossRef] [PubMed]
- Guha, S.; Baltazar, G.C.; Coffey, E.E.; Tu, L.A.; Lim, J.C.; Beckel, J.M.; Patel, S.; Eysteinsson, T.; Lu, W.; O’Brien-Jenkins, A.; et al. Lysosomal alkalinization, lipid oxidation, and reduced phagosome clearance triggered by activation of the P2X7 receptor. FASEB J. 2013, 27, 4500–4509. [Google Scholar] [CrossRef] [PubMed]
- Pietrofesa, R.A.; Velalopoulou, A.; Albelda, S.M.; Christofidou-Solomidou, M. Asbestos Induces Oxidative Stress and Activation of Nrf2 Signaling in Murine Macrophages: Chemopreventive Role of the Synthetic Lignan Secoisolariciresinol Diglucoside (LGM2605). Int. J. Mol. Sci. 2016, 17, 322. [Google Scholar] [CrossRef]
- Pietrofesa, R.A.; Woodruff, P.; Hwang, W.T.; Patel, P.; Chatterjee, S.; Albelda, S.M.; Christofidou-Solomidou, M. The Synthetic Lignan Secoisolariciresinol Diglucoside Prevents Asbestos-Induced NLRP3 Inflammasome Activation in Murine Macrophages. Oxid. Med. Cell Longev. 2017, 2017, 7395238. [Google Scholar] [CrossRef] [PubMed]
- Christofidou-Solomidou, M.; Tyagi, S.; Pietrofesa, R.; Dukes, F.; Arguiri, E.; Turowski, J.; Grieshaber, P.A.; Solomides, C.C.; Cengel, K.A. Radioprotective role in lung of the flaxseed lignan complex enriched in the phenolic secoisolariciresinol diglucoside (SDG). Radiat. Res. 2012, 178, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.M.; Gombart, Z.J.; Chen, J.W. Chloroquine treatment of ARPE-19 cells leads to lysosome dilation and intracellular lipid accumulation: Possible implications of lysosomal dysfunction in macular degeneration. Cell Biosci. 2011, 1, 10. [Google Scholar] [CrossRef] [PubMed]
- Shenker, B.J.; Walker, L.M.; Zekavat, Z.; Ojcius, D.M.; Huang, P.R.; Boesze-Battaglia, K. Cytolethal distending toxin-induced release of interleukin-1β by human macrophages is dependent upon activation of glycogen synthase kinase 3β, spleen tyrosine kinase (Syk) and the noncanonical inflammasome. Cell Microbiol. 2020, 22, e13194. [Google Scholar] [CrossRef]
- Shenker, B.J.; Walker, L.P.; Zekavat, A.; Dlakić, M.; Boesze-Battaglia, K. Blockade of the PI-3K signalling pathway by the Aggregatibacter actinomycetemcomitans cytolethal distending toxin induces macrophages to synthesize and secrete pro-inflammatory cytokines. Cell Microbiol. 2014, 16, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Opoku, E.; Traughber, C.A.; Zhang, D.; Iacano, A.J.; Khan, M.; Han, J.; Smith, J.D.; Gulshan, K. Gasdermin D mediates inflammation-induced defects in reverse cholesterol transport and promotes atherosclerosis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Doyle, S.L.; Campbell, M.; Ozaki, E.; Salomon, R.G.; Mori, A.; Kenna, P.F.; Farrar, G.J.; Kiang, A.S.; Humphries, M.M.; Lavelle, E.C.; et al. NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components. Nat. Med. 2012, 18, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Doyle, S.L.; Ozaki, E.; Brennan, K.; Humphries, M.M.; Mulfaul, K.; Keaney, J.; Kenna, P.F.; Maminishkis, A.; Kiang, A.S.; Saunders, S.P.; et al. IL-18 attenuates experimental choroidal neovascularization as a potential therapy for wet age-related macular degeneration. Sci. Transl. Med. 2014, 6, 230–244. [Google Scholar] [CrossRef]
- Ozaki, E.; Campbell, M.; Kiang, A.S.; Humphries, M.; Doyle, S.L.; Humphries, P. Inflammation in age-related macular degeneration. Adv. Exp. Med. Biol. 2014, 801, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Tarallo, V.; Hirano, Y.; Gelfand, B.D.; Dridi, S.; Kerur, N.; Kim, Y.; Cho, W.G.; Kaneko, H.; Fowler, B.J.; Bogdanovich, S.; et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012, 149, 847–859. [Google Scholar] [CrossRef]
- Zhen, Y.; Zhang, H. NLRP3 Inflammasome and Inflammatory Bowel Disease. Front. Immunol. 2019, 10, 276. [Google Scholar] [CrossRef]
- Engler, D.B.; Leonardi, I.; Hartung, M.L.; Kyburz, A.; Spath, S.; Becher, B.; Rogler, G.; Müller, A. Helicobacter pylori-specific protection against inflammatory bowel disease requires the NLRP3 inflammasome and IL-18. Inflamm. Bowel. Dis. 2015, 21, 854–861. [Google Scholar] [CrossRef] [PubMed]
- Oficjalska, K.; Raverdeau, M.; Aviello, G.; Wade, S.C.; Hickey, A.; Sheehan, K.M.; Corr, S.C.; Kay, E.W.; O’Neill, L.A.; Mills, K.H.; et al. Protective role for caspase-11 during acute experimental murine colitis. J. Immunol. 2015, 194, 1252–1260. [Google Scholar] [CrossRef] [PubMed]
- Seregin, S.S.; Golovchenko, N.; Schaf, B.; Chen, J.; Eaton, K.A.; Chen, G.Y. NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol. 2017, 10, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.H.; Boyd, K.L.; Vogel, P.; Kastan, M.B.; Lamkanfi, M.; Kanneganti, T.D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010, 32, 379–391. [Google Scholar] [CrossRef]
- Ferguson, T.A.; Green, D.R. Autophagy and phagocytosis converge for better vision. Autophagy 2014, 10, 165–167. [Google Scholar] [CrossRef]
- Shenker, B.J.; Walker, L.M.; Zekavat, A.; Weiss, R.H.; Boesze-Battaglia, K. The Cell-Cycle Regulatory Protein p21(CIP1/WAF1) Is Required for Cytolethal Distending Toxin (Cdt)-Induced Apoptosis. Pathogens 2020, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Kerur, N.; Fukuda, S.; Banerjee, D.; Kim, Y.; Fu, D.; Apicella, I.; Varshney, A.; Yasuma, R.; Fowler, B.J.; Baghdasaryan, E.; et al. cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 2018, 24, 50–61. [Google Scholar] [CrossRef]
- Imran, M.; Ahmad, N.; Anjum, F.M.; Khan, M.K.; Mushtaq, Z.; Nadeem, M.; Hussain, S. Potential protective properties of flax lignan secoisolariciresinol diglucoside. Nutr. J. 2015, 14, 71. [Google Scholar] [CrossRef]
- Wang, Y.; Grenell, A.; Zhong, F.; Yam, M.; Hauer, A.; Gregor, E.; Zhu, S.; Lohner, D.; Zhu, J.; Du, J. Metabolic signature of the aging eye in mice. Neurobiol. Aging 2018, 71, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.R.; Ferrington, D.A. Perspective on AMD Pathobiology: A Bioenergetic Crisis in the RPE. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD41–AMD47. [Google Scholar] [CrossRef]
- Cotter, D.G.; Ercal, B.; Huang, X.; Leid, J.M.; d’Avignon, D.A.; Graham, M.J.; Dietzen, D.J.; Brunt, E.M.; Patti, G.J.; Crawford, P.A. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. J. Clin. Investig. 2014, 124, 5175–5190. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The emerging role of Nrf2 in mitochondrial function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Kazantsev, A.G. The role of Nrf2 signaling in counteracting neurodegenerative diseases. FEBS J. 2018, 285, 3576–3590. [Google Scholar] [CrossRef]
- d’Avignon, D.A.; Puchalska, P.; Ercal, B.; Chang, Y.; Martin, S.E.; Graham, M.J.; Patti, G.J.; Han, X.; Crawford, P.A. Hepatic ketogenic insufficiency reprograms hepatic glycogen metabolism and the lipidome. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, K.; Nzirorera, C.; Kienesberger, P.C. Lipid metabolism and signaling in cardiac lipotoxicity. Biochim. Biophys. Acta 2016, 1861, 1513–1524. [Google Scholar] [CrossRef]



| SAMPLE | IL-18 Released ng/mL | |||
|---|---|---|---|---|
| NA | +LGM2605 | +OS | +OS and LGM2605 | |
| RPE | ||||
| Day 2 | 0.11 ± 0.01 | 0.73 ± 0.87 | 1.47 ± 0.18 * | 2.21 ± 0.12 ** |
| Day 6 | 0.48 ± 0.04 | 0.98 ± 0.12 | 0.73 ± 0.84 | 6.04 ± 0.44 ** |
| RPE-LC3B | ||||
| Day 2 | 0.17 ± 0.02 | 1.47 ± 0.12 * | 2.09 ± 0.33 ** | 4.56 ± 0.89 ** |
| Day 6 | 0.11 ± 0.01 | 0.36 ± 0.44 | 1.10 ± 0.0.81 * | 0.73 ± 0.09 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhingra, A.; Sharp, R.C.; Kim, T.; Popov, A.V.; Ying, G.-S.; Pietrofesa, R.A.; Park, K.; Christofidou-Solomidou, M.; Boesze-Battaglia, K. Assessment of a Small Molecule Synthetic Lignan in Enhancing Oxidative Balance and Decreasing Lipid Accumulation in Human Retinal Pigment Epithelia. Int. J. Mol. Sci. 2021, 22, 5764. https://doi.org/10.3390/ijms22115764
Dhingra A, Sharp RC, Kim T, Popov AV, Ying G-S, Pietrofesa RA, Park K, Christofidou-Solomidou M, Boesze-Battaglia K. Assessment of a Small Molecule Synthetic Lignan in Enhancing Oxidative Balance and Decreasing Lipid Accumulation in Human Retinal Pigment Epithelia. International Journal of Molecular Sciences. 2021; 22(11):5764. https://doi.org/10.3390/ijms22115764
Chicago/Turabian StyleDhingra, Anuradha, Rachel C. Sharp, Taewan Kim, Anatoliy V. Popov, Gui-Shuang Ying, Ralph A. Pietrofesa, Kyewon Park, Melpo Christofidou-Solomidou, and Kathleen Boesze-Battaglia. 2021. "Assessment of a Small Molecule Synthetic Lignan in Enhancing Oxidative Balance and Decreasing Lipid Accumulation in Human Retinal Pigment Epithelia" International Journal of Molecular Sciences 22, no. 11: 5764. https://doi.org/10.3390/ijms22115764
APA StyleDhingra, A., Sharp, R. C., Kim, T., Popov, A. V., Ying, G.-S., Pietrofesa, R. A., Park, K., Christofidou-Solomidou, M., & Boesze-Battaglia, K. (2021). Assessment of a Small Molecule Synthetic Lignan in Enhancing Oxidative Balance and Decreasing Lipid Accumulation in Human Retinal Pigment Epithelia. International Journal of Molecular Sciences, 22(11), 5764. https://doi.org/10.3390/ijms22115764








