A Computer-Based Methodology to Design Non-Standard Peptides Potentially Able to Prevent HOX-PBX1-Associated Cancer Diseases
Abstract
1. Introduction
2. Results and Discussion
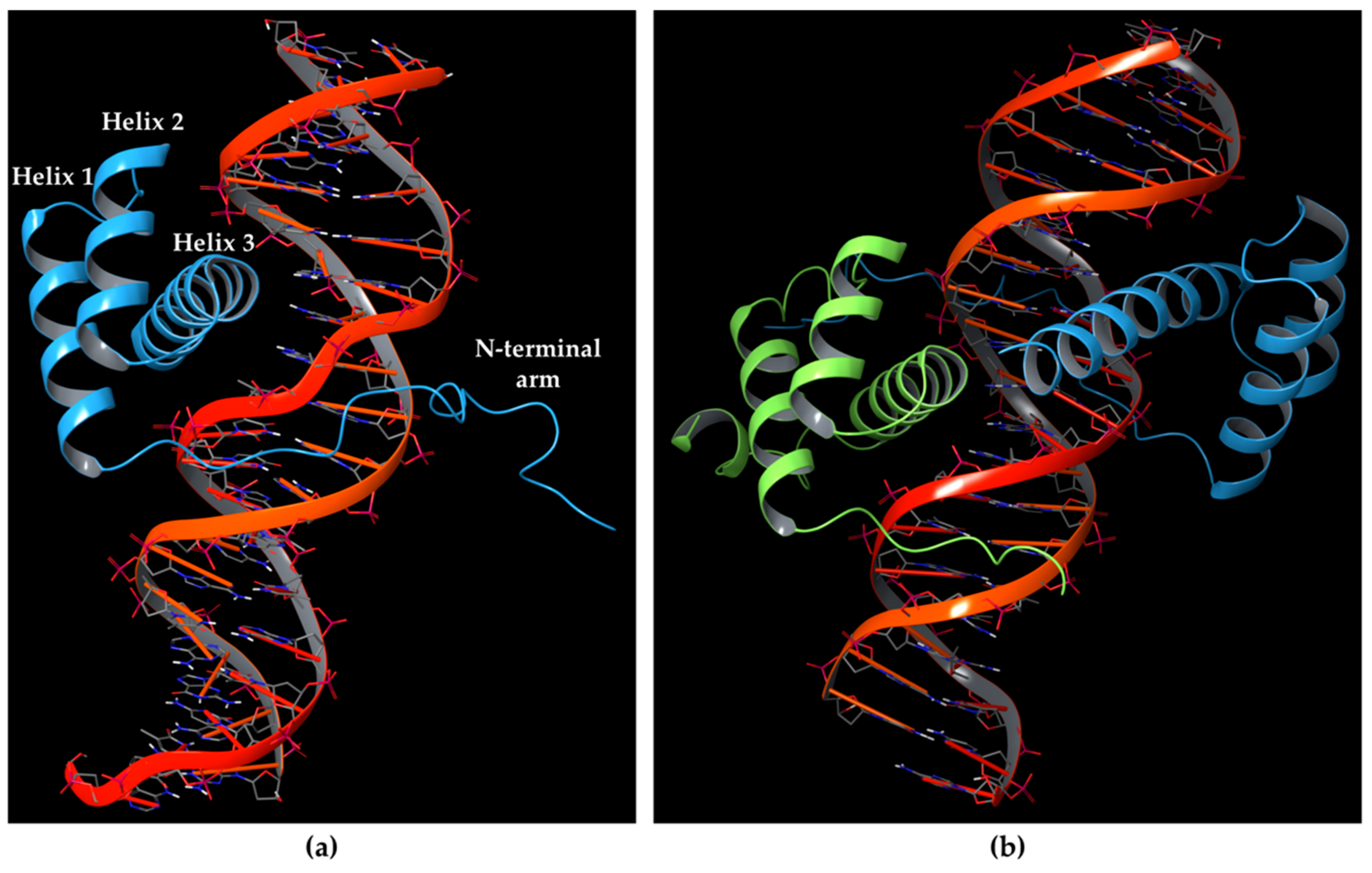
2.1. Molecular Dynamics Simulation of HOXA9-PBX1-DNA Complex
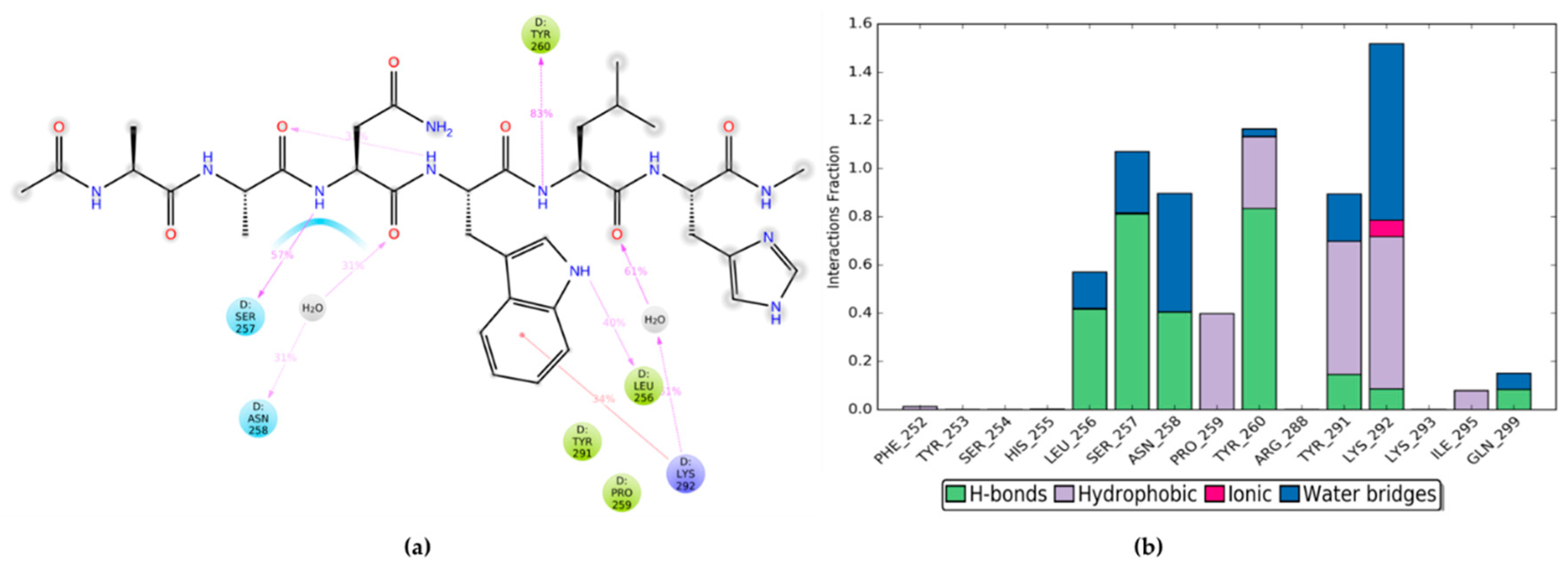
2.2. MD Simulations of HOXA9 Hexapeptide and HTL001 Core Peptide in Complex with PBX1 Protein
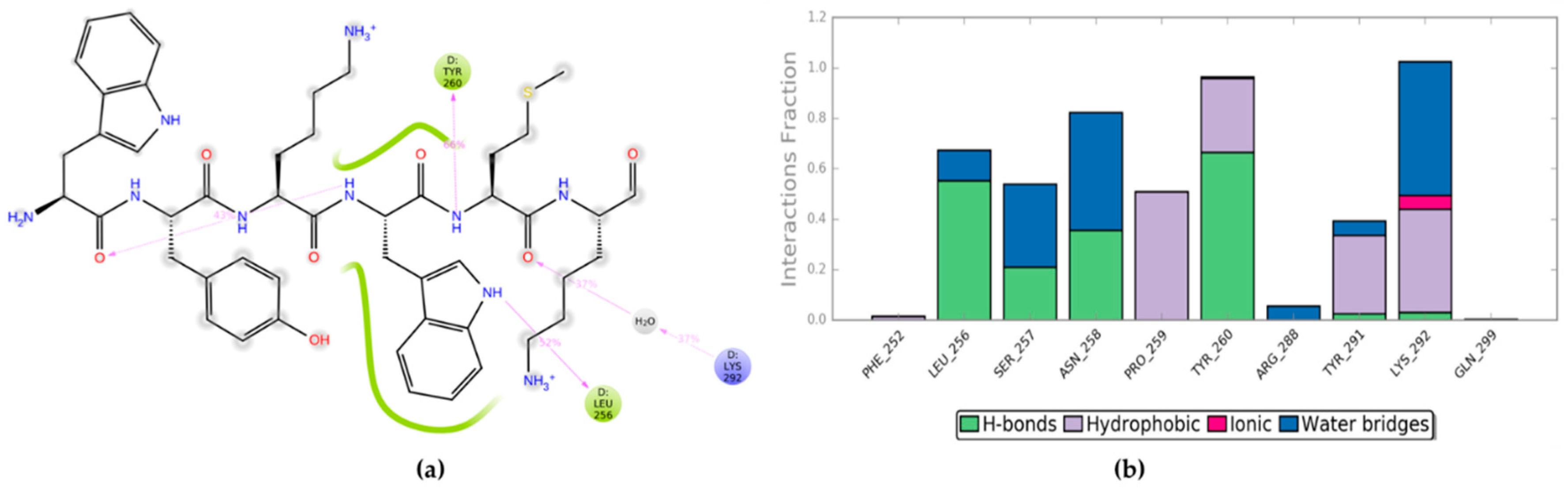
2.3. Residue Scanning of Point-mutated Peptides and Related MD Simulations and MM-GBSA Calculations
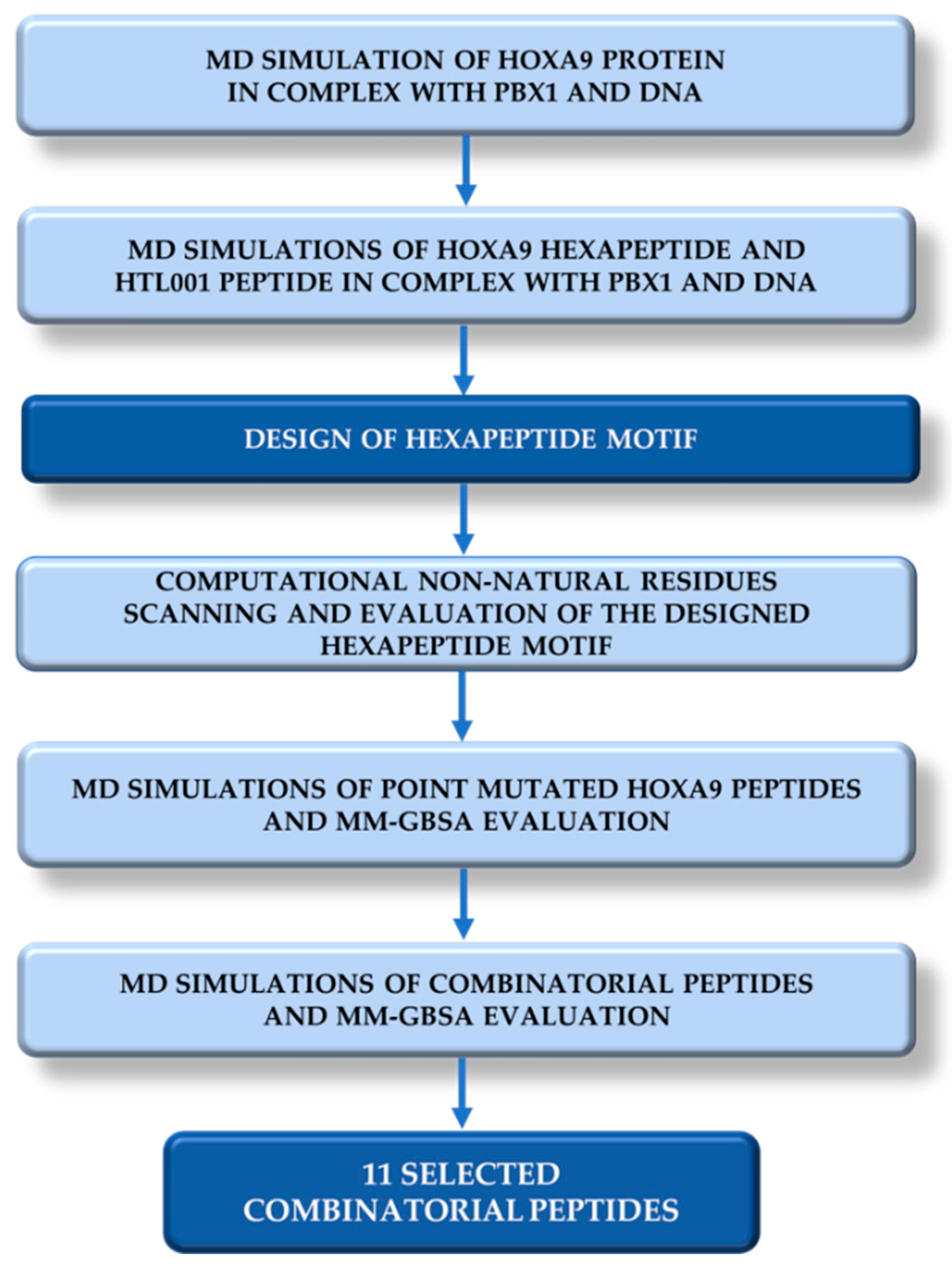
2.4. Combinatorial Peptides Generation and Related MD Simulations and MM-GBSA Calculations
- 1
- CIR–TBP–0BN–Trp–PBF–His
- 2
- CIR–TBP–0BN–Trp-CP3–His
- 3
- CIR–TBP–0BN–Trp-QU4–His
- 4
- CIR–TBP–0BN–Trp–ANT–His
- 5
- ALC–TBP–0BN–Trp–PBF–His
- 6
- ALC–TBP–0BN–Trp–CP3–His
- 7
- ALC–TBP–0BN–Trp–QU4–His
- 8
- ALC–TBP–0BN–Trp–ANT–His
- 9
- MTR–TBP–0BN–Trp–ANT–His
- 10
- MTR–TBP–0BN–Trp–CP3–His
- 11
- MTR–TBP–0BN–Trp–QU4–His
- 12
- MTR–TBP–0BN–Trp–ANT–His
3. Methods
3.1. Preparation of HOXA9-PBX1-DNA Complex
3.2. HOXA9 Hexapeptide Residues Scanning Using Non-standard “SwissSidechain” Amino Acids
3.3. MD Simulations of PBX1-DNA in Complex with HOXA9 Protein, HOXA9 Hexapeptide, HTL001 Core Peptide, Point-mutated Peptides and Combinatorial Peptides
- (1)
- Minimization with the solute restrained;
- (2)
- Minimization without restraints;
- (3)
- Simulation in the NVT ensemble using a Berendsen thermostat with a simulation time of 12 ps, a temperature of 10 K, a fast temperature relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained;
- (4)
- Simulation in the NPT ensemble using a Berendsen thermostat and a Berendsen barostat with a simulation time of 12 ps, a temperature of 10 K and a pressure of 1 atm, a fast temperature relaxation constant, a slow pressure relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained;
- (5)
- Simulation in the NPT ensemble using a Berendsen thermostat and a Berendsen barostat with a simulation time of 24 ps, a temperature of 300 K and a pressure of 1 atm, a fast temperature relaxation constant, a slow pressure relaxation constant, velocity resampling every 1 ps, and non-hydrogen solute atoms restrained;
- (6)
- Simulation in the NPT ensemble using a Berendsen thermostat and a Berendsen barostat with a simulation time of 24 ps, a temperature of 300 K and a pressure of 1 atm, a fast temperature relaxation constant and a normal pressure relaxation constant.
3.4. MM-GBSA Calculations Performed on the Protein-peptide Complexes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pearson, J.C.; Lemons, D.; McGinnis, W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005, 6, 893–904. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Gehring, W.J.; Qian, Y.Q.; Billeter, M.; Furukubo-Tokunaga, K.; Schier, A.F.; Resendez-Perez, D.; Affolter, M.; Otting, G.; Wüthrich, K. Homeodomain-DNA recognition. Cell 1994, 78, 211–223. [Google Scholar] [CrossRef]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, N.; Kuziora, M.A.; McGinnis, W. Human Hox-4.2 and Drosophila Deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell 1990, 63, 969–976. [Google Scholar] [CrossRef]
- Quinonez, S.C.; Innis, J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014, 111, 4–15. [Google Scholar] [CrossRef]
- Hombría, J.C.-G.; Lovegrove, B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation 2003, 71, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Herrero, E. Hox Targets and Cellular Functions. Scientifica 2013, 2013, 738257. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Chan, S.-K. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996, 12, 258–262. [Google Scholar] [CrossRef]
- Hayashi, S.; Scott, M.P. What determines the specificity of action of Drosophila homeodomain proteins? Cell 1990, 63, 883–894. [Google Scholar] [CrossRef]
- Berger, M.F.; Badis, G.; Gehrke, A.R.; Talukder, S.; Philippakis, A.A.; Peña-Castillo, L.; Alleyne, T.M.; Mnaimneh, S.; Botvinnik, O.B.; Chan, E.T.; et al. Variation in Homeodomain DNA Binding Revealed by High-Resolution Analysis of Sequence Preferences. Cell 2008, 133, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of Homeodomain Specificities Allows the Family-wide Prediction of Preferred Recognition Sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Knoepfler, P.S.; Kamps, M.P. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol. Cell. Biol. 1995, 15, 5811–5819. [Google Scholar] [CrossRef] [PubMed]
- Moens, C.B.; Selleri, L. Hox cofactors in vertebrate development. Dev. Biol. 2006, 291, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Longobardi, E.; Penkov, D.; Mateos, D.; De Florian, G.; Torres, M.; Blasi, F. Biochemistry of the tale transcription factors PREP, MEIS, and PBX in vertebrates. Dev. Dyn. 2014, 243, 59–75. [Google Scholar] [CrossRef]
- Peifer, M.; Wieschaus, E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990, 4, 1209–1223. [Google Scholar] [CrossRef]
- Piper, D.E.; Batchelor, A.H.; Chang, C.-P.; Cleary, M.L.; Wolberger, C. Structure of a HoxB1–Pbx1 Heterodimer Bound to DNA. Cell 1999, 96, 587–597. [Google Scholar] [CrossRef]
- Brendolan, A. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 2005, 132, 3113–3126. [Google Scholar] [CrossRef]
- Allen, T.D.; Zhlp, Y.-X.; Hawley, T.S.; Hawley, R.G. TALE Homeoproteins as HOX11-Interacting Partners in T-cell Leukemia. Leuk. Lymphoma 2000, 39, 241–256. [Google Scholar] [CrossRef]
- Chan, S.-K.; Jaffe, L.; Capovilla, M.; Botas, J.; Mann, R.S. The DNA binding specificity of ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 1994, 78, 603–615. [Google Scholar] [CrossRef]
- van Dijk, M.A.; Murre, C. extradenticle Raises the DNA binding specificity of homeotic selector gene products. Cell 1994, 78, 617–624. [Google Scholar] [CrossRef]
- Phelan, M.L.; Rambaldi, I.; Featherstone, M.S. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol. Cell. Biol. 1995, 15, 3989–3997. [Google Scholar] [CrossRef]
- Abu-Shaar, M.; Ryoo, H.D.; Mann, R.S. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 1999, 13, 935–945. [Google Scholar] [CrossRef]
- Stevens, K.E.; Mann, R.S. A Balance Between Two Nuclear Localization Sequences and a Nuclear Export Sequence Governs Extradenticle Subcellular Localization. Genetics 2007, 175, 1625–1636. [Google Scholar] [CrossRef][Green Version]
- Saleh, M.; Huang, H.; Green, N.C.; Featherstone, M.S. A Conformational Change in PBX1A Is Necessary for Its Nuclear Localization. Exp. Cell Res. 2000, 260, 105–115. [Google Scholar] [CrossRef]
- LaRonde-LeBlanc, N.A.; Wolberger, C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003, 17, 2060–2072. [Google Scholar] [CrossRef]
- Merabet, S.; Hudry, B.; Saadaoui, M.; Graba, Y. Classification of sequence signatures: A guide to Hox protein function. BioEssays 2009, 31, 500–511. [Google Scholar] [CrossRef]
- Johnson, F.B.; Parker, E.; Krasnow, M.A. Extradenticle protein is a selective cofactor for the Drosophila homeotics: Role of the homeodomain and YPWM amino acid motif in the interaction. Proc. Natl. Acad. Sci. USA 1995, 92, 739–743. [Google Scholar] [CrossRef]
- Joshi, R.; Passner, J.M.; Rohs, R.; Jain, R.; Sosinsky, A.; Crickmore, M.A.; Jacob, V.; Aggarwal, A.K.; Honig, B.; Mann, R.S. Functional Specificity of a Hox Protein Mediated by the Recognition of Minor Groove Structure. Cell 2007, 131, 530–543. [Google Scholar] [CrossRef]
- Passner, J.M.; Ryoo, H.D.; Shen, L.; Mann, R.S.; Aggarwal, A.K. Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature 1999, 397, 714–719. [Google Scholar] [CrossRef]
- Shen, W.-F.; Chang, C.-P.; Rozenfeld, S.; Sauvageau, G.; Humphries, R.K.; Lu, M.; Lawrence, H.J.; Cleary, M.L.; Largman, C. Hox Homeodomain Proteins Exhibit Selective Complex Stabilities with Pbx and DNA. Nucleic Acids Res. 1996, 24, 898–906. [Google Scholar] [CrossRef]
- Joshi, R.; Sun, L.; Mann, R. Dissecting the functional specificities of two Hox proteins. Genes Dev. 2010, 24, 1533–1545. [Google Scholar] [CrossRef]
- Chang, C.P.; Brocchieri, L.; Shen, W.F.; Largman, C.; Cleary, M.L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 1996, 16, 1734–1745. [Google Scholar] [CrossRef]
- Lu, Q.; Kamps, M.P. Structural determinants within Pbx1 that mediate cooperative DNA binding with pentapeptide-containing Hox proteins: Proposal for a model of a Pbx1-Hox-DNA complex. Mol. Cell. Biol. 1996, 16, 1632–1640. [Google Scholar] [CrossRef]
- Peltenburg, L.T.; Murre, C. Specific residues in the Pbx homeodomain differentially modulate the DNA-binding activity of Hox and Engrailed proteins. Development 1997, 124, 1089–1098. [Google Scholar] [CrossRef]
- Chang, C.P.; Shen, W.F.; Rozenfeld, S.; Lawrence, H.J.; Largman, C.; Cleary, M.L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995, 9, 663–674. [Google Scholar] [CrossRef]
- Perricone, U.; Gulotta, M.R.; Lombino, J.; Parrino, B.; Cascioferro, S.; Diana, P.; Cirrincione, G.; Padova, A. An overview of recent molecular dynamics applications as medicinal chemistry tools for the undruggable site challenge. MedChemComm 2018, 9, 920–936. [Google Scholar] [CrossRef]
- Gulotta, M.R.; Vittorio, S.; Gitto, R.; Perricone, U.; De Luca, L. Exploring Molecular Contacts of MUC1 at CIN85 Binding Interface to Address Future Drug Design Efforts. Int. J. Mol. Sci. 2021, 22, 2208. [Google Scholar] [CrossRef]
- Ji, T.; Lee, M.; Pruitt, S.C.; Hangauer, D.G. Privileged scaffolds for blocking protein–protein interactions: 1,4-disubstituted naphthalene antagonists of transcription factor complex HOX–PBX/DNA. Bioorg. Med. Chem. Lett. 2004, 14, 3875–3879. [Google Scholar] [CrossRef]
- Morgan, R.G.L.; Sohal, J. HOX peptides as PBX modulators for the treatment of cancer. European Patent EP1909811B1, 2009. Available online: https://patentimages.storage.googleapis.com/09/d8/95/6ccfb3d2ef2f38/EP1909811B1.pdf (accessed on 21 May 2020).
- Morgan, R.; Pirard, P.M.; Shears, L.; Sohal, J.; Pettengell, R.; Pandha, H.S. Antagonism of HOX/PBX Dimer Formation Blocks the In vivo Proliferation of Melanoma. Cancer Res. 2007, 67, 5806–5813. [Google Scholar] [CrossRef]
- Plowright, L.; Harrington, K.J.; Pandha, H.S.; Morgan, R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer). Br. J. Cancer 2009, 100, 470–475. [Google Scholar] [CrossRef]
- Morgan, R.; Boxall, A.; Harrington, K.J.; Simpson, G.R.; Gillett, C.; Michael, A.; Pandha, H.S. Targeting the HOX/PBX dimer in breast cancer. Breast Cancer Res. Treat. 2012, 136, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Boxall, A.; Harrington, K.J.; Simpson, G.R.; Michael, A.; Pandha, H.S. Targeting HOX transcription factors in prostate cancer. BMC Urol. 2014, 14, 17. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Simpson, G.; Gray, S.; Gillett, C.; Tabi, Z.; Spicer, J.; Harrington, K.J.; Pandha, H.S. HOX transcription factors are potential targets and markers in malignant mesothelioma. BMC Cancer 2016, 16, 85. [Google Scholar] [CrossRef]
- Errico, M.C.; Felicetti, F.; Bottero, L.; Mattia, G.; Boe, A.; Felli, N.; Petrini, M.; Bellenghi, M.; Pandha, H.S.; Calvaruso, M.; et al. The abrogation of the HOXB7/PBX2 complex induces apoptosis in melanoma through the miR-221&222-c-FOS pathway. Int. J. Cancer 2013, 133, 879–892. [Google Scholar] [PubMed]
- Ando, H.; Natsume, A.; Senga, T.; Watanabe, R.; Ito, I.; Ohno, M.; Iwami, K.; Ohka, F.; Motomura, K.; Kinjo, S.; et al. Peptide-based inhibition of the HOXA9/PBX interaction retards the growth of human meningioma. Cancer Chemother. Pharmacol. 2014, 73, 53–60. [Google Scholar] [CrossRef]
- Primon, M.; Hoffman, E.; Pandha, H.S.; Morgan, R. HTL001, a novel inhibitor of HOX/PBX binding, is highly cytotoxic to prostate and breast cancer cells. Eur. J. Cancer 2016, 69, S133. [Google Scholar] [CrossRef]
- Shears, L.; Plowright, L.; Harrington, K.; Pandha, H.S.; Morgan, R. Disrupting the Interaction Between HOX and PBX Causes Necrotic and Apoptotic Cell Death in the Renal Cancer Lines CaKi-2 and 769-P. J. Urol. 2008, 180, 2196–2201. [Google Scholar] [CrossRef]
- Protein Data Bank. Available online: https://pdb101.rcsb.org (accessed on 21 May 2020).
- Perricone, U.; Wieder, M.; Seidel, T.; Langer, T.; Padova, A.; Almerico, A.M.; Tutone, M. A Molecular Dynamics–Shared Pharmacophore Approach to Boost Early-Enrichment Virtual Screening: A Case Study on Peroxisome Proliferator-Activated Receptor α. ChemMedChem 2017, 12, 1399–1407. [Google Scholar] [CrossRef]
- Database of Non-Standard Amino Acids. Available online: https://www.swisssidechain.ch/ (accessed on 27 July 2020).
- Gfeller, D.; Michielin, O.; Zoete, V. Expanding molecular modeling and design tools to non-natural sidechains. J. Comput. Chem. 2012, 33, 1525–1535. [Google Scholar] [CrossRef]
- Gulotta, M.R.; Lombino, J.; Perricone, U.; De Simone, G.; Mekni, N.; De Rosa, M.; Diana, P.; Padova, A. Targeting SARS-CoV-2 RBD Interface: A Supervised Computational Data-Driven Approach to Identify Potential Modulators. ChemMedChem 2020, 15, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- Beard, H.; Cholleti, A.; Pearlman, D.; Sherman, W.; Loving, K.A. Applying Physics-Based Scoring to Calculate Free Energies of Binding for Single Amino Acid Mutations in Protein-Protein Complexes. PLoS ONE 2013, 8, e82849. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Matsson, P.; Kihlberg, J. How Big Is Too Big for Cell Permeability? J. Med. Chem. 2017, 60, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Duffy, E.M.; Jorgensen, W.L. Prediction of Properties from Simulations: Free Energies of Solvation in Hexadecane, Octanol, and Water. J. Am. Chem. Soc. 2000, 122, 2878–2888. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, J.R.; Calkins, D.; Sullivan, A.P.; Shelley, J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010, 24, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical p K a Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the Role of the Crystal Environment in Determining Protein Side-chain Conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, November 2006; Association for Computing Machinery: New York, NY, USA. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]

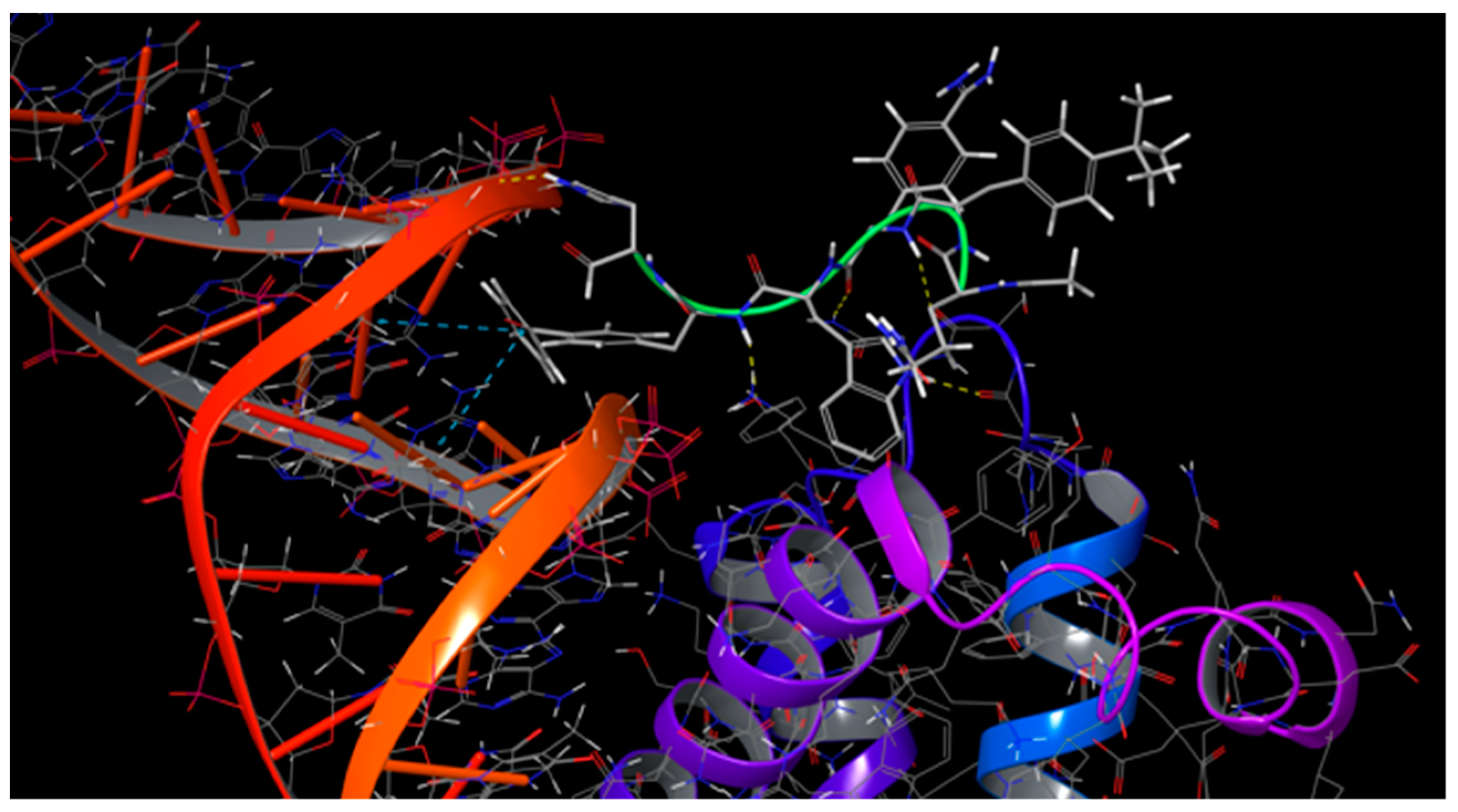
| HOXA9 Residue | PBX1 Residue | Interaction Type |
|---|---|---|
| Trp199 | Ser257 | 1 H-bond |
| Trp199 | Leu256 | 1 H-bond |
| Trp199 | Tyr291 | π-stacking |
| Trp199 | Tyr260 | π-stacking |
| Leu200 | Tyr260 | 1 H-bond |
| Ala197 | Asn258 | 1 H-bond |
| Peptide Involved | HOXA9 Hexapeptide | HTL001 Hexapeptide |
|---|---|---|
| ΔGbinding average (kcal/mol) | −58.1922 | −53.6882 |
| ΔGbinding Std. Dev. | 8.99 | 8.53 |
| ΔGbinding range (kcal/mol) | −84.6286 to −34.1107 | −78.0904 to −28.9169 |
| Corresponding HOXA9 aa | Non-natural Amino Acid | Non-natural Amino Acid Structure | ΔΔGaffinity | ΔΔGstability |
|---|---|---|---|---|
| ALA196 | CIR | 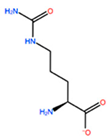 | −45.128 kcal/mol | −0.816 kcal/mol |
| ALC | 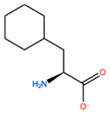 | −18.704 kcal/mol | −4.095 kcal/mol | |
| MTR |  | −17.088 kcal/mol | −3.208 kcal/mol | |
| CTE | 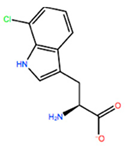 | −15.008 kcal/mol | −5.778 kcal/mol | |
| ALA197 | BIF | 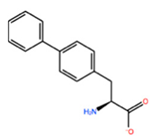 | −12.892 kcal/mol | −3.615 kcal/mol |
| TBP | 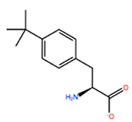 | −8.133 kcal/mol | −3.556 kcal/mol | |
| HRG | 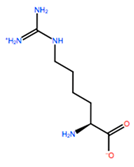 | −6.681 kcal/mol | −9.341 kcal/mol | |
| CIR |  | −5.075 kcal/mol | −2.606 kcal/mol | |
| ASN198 | MOT | 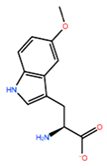 | −7.505 kcal/mol | −13.303 kcal/mol |
| 0BN |  | −6.051 kcal/mol | −10.151 kcal/mol | |
| KYN | 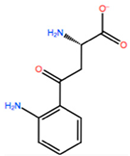 | −6.041 kcal/mol | −5.511 kcal/mol | |
| GBU | 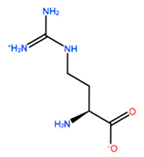 | −5.867 kcal/mol | −0.688 kcal/mol | |
| LEU200 | PBF | 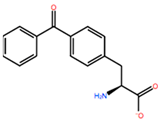 | −51.368 kcal/mol | −3.302 kcal/mol |
| CP3 | 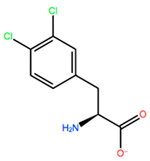 | −11.929 kcal/mol | −1.106 kcal/mol | |
| QU4 |  | −11.415 kcal/mol | −4.372 kcal/mol | |
| ANT | 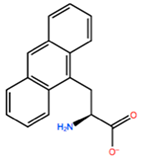 | −11.134 kcal/mol | −1.353 kcal/mol | |
| HIS201 | ILX | 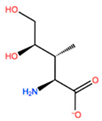 | −11.562 kcal/mol | −1.371 kcal/mol |
| HIL | 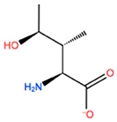 | −10.330 kcal/mol | −2.198 kcal/mol | |
| DPP |  | −4.195 kcal/mol | −1.029 kcal/mol | |
| HRG |  | −4.018 kcal/mol | −5.686 kcal/mol |
| X1 = ALA196 Mutation | X2 = ALA197 Mutation | X3 = ASN198 Mutation | ||||
| ΔGbinding average (kcal/mol) | CIR | −68.1064 | BIF | −52.5114 | MOT | −51.3925 |
| ΔGbinding Std. Dev. | 9.19 | 8.23 | 9.77 | |||
| ΔGbinding range (kcal/mol) | −91.4424 to −33.6024 | −77.3444 to −25.9087 | −79.7187 to −21.3466 | |||
| ΔGbinding average (kcal/mol) | MTR | −58.3419 | TBP | −59.0603 | 0BN | −59.1051 |
| ΔGbinding Std. Dev. | 7.37 | 9.34 | 8.71 | |||
| ΔGbinding range (kcal/mol) | −85.6739 to −35.5824 | −80.7314 to −31.1314 | −82.4310 to −27.3523 | |||
| ΔGbinding average (kcal/mol) | ALC | −59.6952 | HRG | −54.3707 | KYN | −56.4406 |
| ΔGbinding Std. Dev. | 7.39 | 9.34 | 7.35 | |||
| ΔGbinding range (kcal/mol) | −78.3738 to −26.9511 | −85.1600 to −24.8071 | −75.4170 to −28.6690 | |||
| ΔGbinding average (kcal/mol) | CTE | −56.3011 | CIR | −57.6592 | GBU | −55.1339 |
| ΔGbinding Std. Dev. | 7.55 | 8.88 | 7.97 | |||
| ΔGbinding range (kcal/mol) | −80.4156 to −29.7472 | −81.7311 to −26.2628 | −77.5916 to −30.5304 | |||
| X4= LEU200 Mutation | X5 = HIS201 Mutation | |||||
| ΔGbinding average (kcal/mol) | PBF | −68.1857 | ILX | −48.9087 | ||
| ΔGbinding Std. Dev. | 8.44 | 8.81 | ||||
| ΔGbinding range (kcal/mol) | −95.1687 to −39.7321 | −74.3783 to −20.2857 | ||||
| ΔGbinding average (kcal/mol) | CP3 | −64.6802 | HIL | −50.3148 | ||
| ΔGbinding Std. Dev. | 9.49 | 9.03 | ||||
| ΔGbinding range (kcal/mol) | −89.3192 to−34.0239 | −79.2197 to −22.2265 | ||||
| ΔGbinding average (kcal/mol) | QU4 | −61.8016 | DPP | −55.6169 | ||
| ΔGbinding Std. Dev. | 10.53 | 11.48 | ||||
| ΔGbinding range (kcal/mol) | −88.4144 to −33.9895 | −91.1993 to −22.9813 | ||||
| ΔGbinding average (kcal/mol) | ANT | −63.3043 | HRG | −57.0861 | ||
| ΔGbinding Std. Dev. | 8.50 | 9.46 | ||||
| ΔGbinding range (kcal/mol) | −87.5841 to −35.9672 | −86.8681 to −27.0434 | ||||
| PEPTIDE INVOLVED | First Peptide | Second Peptide | Third Peptide |
| ΔGbinding average | −79.6771 kcal/mol | −61.8602 kcal/mol | −68.0795 kcal/mol |
| ΔGbinding Std. Dev. | 10.18 | 12.72 | 10.63 |
| ΔGbinding range | −104.585 to −38.2615 kcal/mol | −99.0013 to −30.7190 kcal/mol | −98.3690 to −29.5313 kcal/mol |
| PEPTIDE INVOLVED | Fourth Peptide | Fifth Peptide | Sixth Peptide |
| ΔGbinding average | −64.6664 kcal/mol | −81.8766 kcal/mol | −55.1927 kcal/mol |
| ΔGbinding Std. Dev. | 7.53 | 7.44 | 10.09 |
| ΔGbinding range | −87.5689 to −30.1013 kcal/mol | −101.5164 to −45.3623 kcal/mol | −85.5158 to −22.5652 kcal/mol |
| PEPTIDE INVOLVED | Seventh Peptide | Eighth Peptide | Ninth Peptide |
| ΔGbinding average | −62.8885 kcal/mol | −71.9163 kcal/mol | −74.0909 kcal/mol |
| ΔGbinding Std. Dev. | 9.19 | 9.19 | 11.42 |
| ΔGbinding range | −89.1247 to −19.4438 kcal/mol | −101.6790 to −44.4808 kcal/mol | −105.5444 to −32.2303 kcal/mol |
| PEPTIDE INVOLVED | Tenth Peptide | Eleventh Peptide | Twelfth Peptide |
| ΔGbinding average | −60.2167 kcal/mol | −65.0198 kcal/mol | −68.3222 kcal/mol |
| ΔGbinding Std. Dev. | 9.56 | 8.24 | 8.13 |
| ΔGbinding range | −89.0633 to −28.6783 kcal/mol | −89.3709 to −36.2812 kcal/mol | −95.3406 to −42.1593 kcal/mol |
| PEPTIDES | PSA | logPo/w | |
|---|---|---|---|
| 1 | CIR-TBP-0BN-TRP-PBF-HIS | 302.15 | 3.2 |
| 2 | CIR-TBP-0BN-TRP-CP3-HIS | 310.38 | 2.6 |
| 3 | CIR-TBP-0BN-TRP-QU4-HIS | 322.80 | 2.0 |
| 4 | CIR-TBP-0BN-TRP-ANT-HIS | 307.47 | 3.3 |
| 5 | ALC-TBP-0BN-TRP-PBF-HIS | 300.04 | 6.1 |
| 6 | ALC-TBP-0BN-TRP-CP3-HIS | 284.52 | 6.0 |
| 7 | ALC-TBP-0BN-TRP-QU4-HIS | 297.08 | 5.4 |
| 8 | ALC-TBP-0BN-TRP-ANT-HIS | 250.48 | 5.9 |
| 9 | MTR-TBP-0BN-TRP-PBF-HIS | 276.41 | 4.3 |
| 10 | MTR-TBP-0BN-TRP-CP3-HIS | 268.43 | 5.4 |
| 11 | MTR-TBP-0BN-TRP-QU4-HIS | 280.80 | 4.8 |
| 12 | MTR-TBP-0BN-TRP-ANT-HIS | 260.87 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulotta, M.R.; De Simone, G.; John, J.; Perricone, U.; Brancale, A. A Computer-Based Methodology to Design Non-Standard Peptides Potentially Able to Prevent HOX-PBX1-Associated Cancer Diseases. Int. J. Mol. Sci. 2021, 22, 5670. https://doi.org/10.3390/ijms22115670
Gulotta MR, De Simone G, John J, Perricone U, Brancale A. A Computer-Based Methodology to Design Non-Standard Peptides Potentially Able to Prevent HOX-PBX1-Associated Cancer Diseases. International Journal of Molecular Sciences. 2021; 22(11):5670. https://doi.org/10.3390/ijms22115670
Chicago/Turabian StyleGulotta, Maria Rita, Giada De Simone, Justin John, Ugo Perricone, and Andrea Brancale. 2021. "A Computer-Based Methodology to Design Non-Standard Peptides Potentially Able to Prevent HOX-PBX1-Associated Cancer Diseases" International Journal of Molecular Sciences 22, no. 11: 5670. https://doi.org/10.3390/ijms22115670
APA StyleGulotta, M. R., De Simone, G., John, J., Perricone, U., & Brancale, A. (2021). A Computer-Based Methodology to Design Non-Standard Peptides Potentially Able to Prevent HOX-PBX1-Associated Cancer Diseases. International Journal of Molecular Sciences, 22(11), 5670. https://doi.org/10.3390/ijms22115670






