MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coli and Pseudomonas aeruginosa
Abstract
1. Introduction
2. Results
2.1. Estimation of the Expected PG Interacting Partners
2.2. Composition Analysis of the PG from E. coli and P. aeruginosa
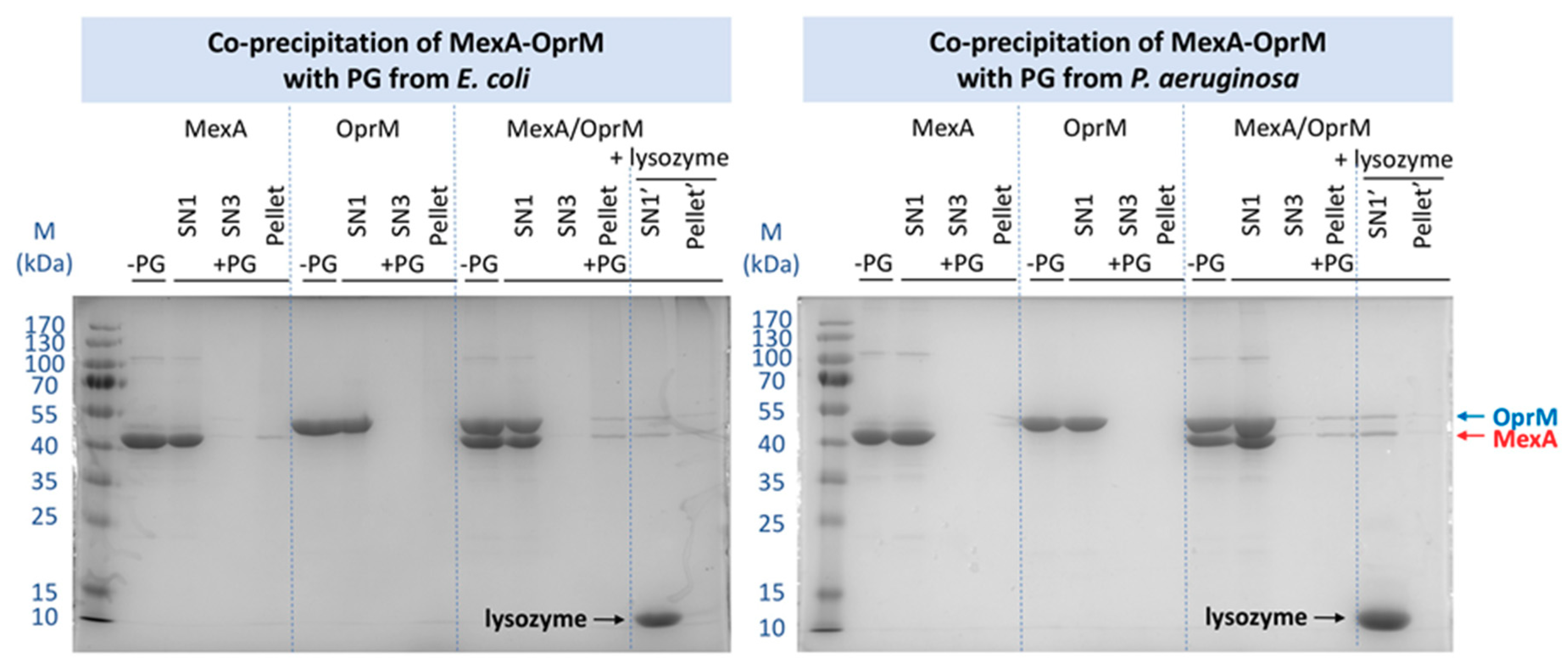
2.3. Pull-Down Analysis of MexA and OprM with Purified PG
2.3.1. Comparison of the PG from E. coli and P. aeruginosa
2.3.2. Effects of Different Parameters on the Co-Precipitation of MexA-OprM with PG from P. aeruginosa
3. Discussion
4. Materials and Methods
4.1. Purification of the Peptidoglycan
4.2. Cloning and Purification of the Analyzed Proteins
4.3. Pull Down with PG
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PG | peptidoglycan |
| NAM | N-acetylmuramic acid |
| NAG | N-acetylglucosamine |
| DAP | diaminopimelic acid |
References
- Delmar, J.A.; Su, C.C.; Yu, E.W. Bacterial Multidrug Efflux Transporters. Annu. Rev. Biophys. 2014, 43, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Du, D.J.; van Veen, H.W.; Luisi, B.F. Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol. 2015, 23, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Matsumoto, T.; Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006, 443, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Seeger, M.A.; Schiefner, A.; Eicher, T.; Verrey, F.; Diederichs, K.; Pos, K.M. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 2006, 313, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Mikolosko, J.; Bobyk, K.; Zgurskaya, H.I.; Ghosh, P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 2006, 14, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Koronakis, V.; Sharff, A.; Koronakis, E.; Luisi, B.; Hughes, C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 2000, 405, 914–919. [Google Scholar] [CrossRef] [PubMed]
- Sennhauser, G.; Bukowska, M.A.; Briand, C.; Grutter, M.G. Crystal Structure of the Multidrug Exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009, 389, 134–145. [Google Scholar] [CrossRef]
- Symmons, M.F.; Bokma, E.; Koronakis, E.; Hughes, C.; Koronakis, V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA 2009, 106, 7173–7178. [Google Scholar] [CrossRef] [PubMed]
- Akama, H.; Kanemaki, M.; Yoshimura, M.; Tsukihara, T.; Kashiwagi, T.; Yoneyama, H.; Narita, S.; Nakagawa, A.; Nakae, T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa—Dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004, 279, 52816–52819. [Google Scholar] [CrossRef]
- Phan, G.; Benabdelhak, H.; Lascombe, M.B.; Benas, P.; Rety, S.; Picard, M.; Ducruix, A.; Etchebest, C.; Broutin, I. Structural and Dynamical Insights into the Opening Mechanism of P. aeruginosa OprM Channel. Structure 2010, 18, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Monlezun, L.; Phan, G.; Benabdelhak, H.; Lascombe, M.B.; Enguene, V.Y.N.; Picard, M.; Broutin, I. New OprM structure highlighting the nature of the N-terminal anchor. Front. Microbiol. 2015, 6, 667. [Google Scholar] [CrossRef]
- Du, D.J.; Wang, Z.; James, N.R.; Voss, J.E.; Klimont, E.; Ohene-Agyei, T.; Venter, H.; Chiu, W.; Luisi, B.F. Structure of the AcrAB-TolC multidrug efflux pump. Nature 2014, 509, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, G.Z.; Hryc, C.F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.; Du, D.J. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife 2017, 6, e24905. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, K.; Yonehara, R.; Ishizaka-Ikeda, E.; Miyazaki, N.; Maeda, S.; Iwasaki, K.; Nakagawa, A.; Yamashita, E. Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism. Nat. Commun. 2019, 10, 1520. [Google Scholar] [CrossRef] [PubMed]
- Glavier, M.; Puvanendran, D.; Salvador, D.; Decossas, M.; Phan, G.; Garnier, C.; Frezza, E.; Cece, Q.; Schoehn, G.; Picard, M.; et al. Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex. Nat. Commun. 2020, 11, 4948. [Google Scholar] [CrossRef] [PubMed]
- Enguene, V.Y.N.; Verchere, A.; Phan, G.; Broutin, I.; Picard, M. Catch me if you can: A biotinylated proteoliposome affinity assay for the investigation of assembly of the MexA-MexB-OprM efflux pump from Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 541. [Google Scholar]
- Verchere, A.; Dezi, M.; Adrien, V.; Broutin, I.; Picard, M. In vitro transport activity of the fully assembled MexAB-OprM efflux pump from Pseudomonas aeruginosa. Nat. Commun. 2015, 6, 6890. [Google Scholar] [CrossRef]
- Sakurai, K.; Yamasaki, S.; Nakao, K.; Nishino, K.; Yamaguchi, A.; Nakashima, R. Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds. Sci. Rep. 2019, 9, 4359. [Google Scholar] [CrossRef]
- Kobylka, J.; Kuth, M.S.; Muller, R.T.; Geertsma, E.R.; Pos, K.M. AcrB: A mean, keen, drug efflux machine. Ann. N. Y. Acad. Sci. 2020, 1459, 38–68. [Google Scholar] [CrossRef]
- Reading, E.; Ahdash, Z.; Fais, C.; Ricci, V.; Xuan, W.K.; Grimsey, E.; Stone, J.; Malloci, G.; Lau, A.M.; Findlay, H.; et al. Perturbed structural dynamics underlie inhibition and altered efflux of the multidrug resistance pump AcrB. Nat. Commun. 2020, 11, 5565. [Google Scholar] [CrossRef]
- Simsir, M.; Broutin, I.; Mus-Veteau, I.; Cazals, F. Studying dynamics without explicit dynamics: A structure-based study of the export mechanism by AcrB. Proteins 2021, 89, 259–275. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Nakashima, R.; Sakurai, K. Structural basis of RND-type multidrug exporters. Front. Microbiol. 2015, 6, 327. [Google Scholar] [CrossRef] [PubMed]
- Zwama, M.; Yamaguchi, A. Molecular mechanisms of AcrB-mediated multidrug export. Res. Microbiol. 2018, 169, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Zwama, M.; Yamasaki, S.; Nakashima, R.; Sakurai, K.; Nishino, K.; Yamaguchi, A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat. Commun. 2018, 9, 124. [Google Scholar] [CrossRef] [PubMed]
- Atzori, A.; Malviya, V.N.; Malloci, G.; Dreier, J.; Pos, K.M.; Vargiu, A.V.; Ruggerone, P. Identification and characterization of carbapenem binding sites within the RND-transporter AcrB. Biochim. Biophys. Acta Biomembr. 2019, 1861, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Tam, H.K.; Malviya, V.N.; Foong, W.E.; Herrmann, A.; Malloci, G.; Ruggerone, P.; Vargiu, A.V.; Pos, K.M. Binding and Transport of Carboxylated Drugs by the Multidrug Transporter AcrB. J. Mol. Biol. 2020, 432, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Atzori, A.; Malloci, G.; Cardamone, F.; Bosin, A.; Vargiu, A.V.; Ruggerone, P. Molecular Interactions of Carbapenem Antibiotics with the Multidrug Efflux Transporter AcrB of Escherichia coli. Int. J. Mol. Sci. 2020, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Vargiu, A.V.; Ramaswamy, V.K.; Malvacio, I.; Malloci, G.; Kleinekathofer, U.; Ruggerone, P. Water-mediated interactions enable smooth substrate transport in a bacterial efflux pump. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Hazel, A.J.; Abdali, N.; Leus, I.V.; Parks, J.M.; Smith, J.C.; Zgurskaya, H.I.; Gumbart, J.C. Conformational Dynamics of AcrA Govern Multidrug Efflux Pump Assembly. ACS Infect. Dis. 2019, 5, 1926–1935. [Google Scholar] [CrossRef]
- Schulz, R.; Vargiu, A.V.; Collu, F.; Kleinekathofer, U.; Ruggerone, P. Functional Rotation of the Transporter AcrB: Insights into Drug Extrusion from Simulations. PLoS Comput. Biol. 2010, 6, e1000806. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Ramaswamy, V.K.; Malloci, G.; Malvacio, I.; Atzori, A.; Ruggerone, P. Computer simulations of the activity of RND efflux pumps. Res. Microbiol. 2018, 169, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Bavro, V.N.; Pietras, Z.; Furnham, N.; Perez-Cano, L.; Fernandez-Recio, J.; Pei, X.Y.; Misra, R.; Luisi, B. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell 2008, 30, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.A.; Travers, T.; Pos, K.M.; Zgurskaya, H.I.; Gnanakaran, S. Dynamics of Intact MexAB-OprM Efflux Pump: Focusing on the MexA-OprM Interface. Sci. Rep. 2017, 7, 16521. [Google Scholar] [CrossRef]
- Fabre, L.; Ntreh, A.T.; Yazidi, A.; Leus, I.V.; Weeks, J.W.; Bhattacharyya, S.; Ruickoldt, J.; Rouiller, I.; Zgurskaya, H.I.; Sygusch, J.A. “Drug Sweeping” State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa. Structure 2021, 29, 261–274.E6. [Google Scholar] [CrossRef]
- Matias, V.R.F.; Al-Amoudi, A.; Dubochet, J.; Beveridge, T.J. Cryo-transmission electron Microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 6112–6118. [Google Scholar] [CrossRef] [PubMed]
- Quintela, J.C.; Caparros, M.; De Pedro, M.A. Variability of Peptidoglycan Structural Parameters in Gram-Negative Bacteria. FEMS Microbiol. Lett. 1995, 125, 95–100. [Google Scholar] [CrossRef]
- Barry, A.L.; Reller, L.B.; Miller, G.H.; Washington, J.A.; Schoenknect, F.D.; Peterson, L.R.; Hare, R.S.; Knapp, C. Revision of Standards for Adjusting the Cation Content of Mueller-Hinton Broth for Testing Susceptibility of Pseudomonas aeruginosa to Aminoglycosides. J. Clin. Microbiol. 1992, 30, 585–589. [Google Scholar] [CrossRef]
- Glauner, B. Separation and Quantification of Muropeptides with High-Performance Liquid-Chromatography. Anal. Biochem. 1988, 172, 451–464. [Google Scholar] [CrossRef]
- Yanai, A.; Kato, K.; Beppu, T.; Arima, K. Peptidoglycan of Pseudomonas aeruginosa. Agr. Biol. Chem. 1976, 40, 1505–1508. [Google Scholar] [CrossRef]
- Crump, G.M.; Zhou, J.H.; Mashayekh, S.; Grimes, C.L. Revisiting peptidoglycan sensing: Interactions with host immunity and beyond. Chem. Commun. 2020, 56, 13313–13322. [Google Scholar] [CrossRef] [PubMed]
- Ferrandez, Y.; Monlezun, L.; Phan, G.; Benabdelhak, H.; Benas, P.; Ulryck, N.; Falson, P.; Ducruix, A.; Picard, M.; Broutin, I. Stoichiometry of the MexA-OprM binding, as investigated by blue native gel electrophoresis. Electrophoresis 2012, 33, 1282–1287. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.D.; Chen, M.Y.; Yu, Z.L.; Bell, J.M.; Wang, H.; Forrester, I.; Villarreal, H.; Jakana, J.; Du, D.J.; Luisi, B.; et al. In situ structure and assembly of the multidrug efflux pump AcrAB-TolC. Nat. Commun. 2019, 10, 2635. [Google Scholar] [CrossRef] [PubMed]
- Boags, A.T.; Samsudin, F.; Khalid, S. Binding from Both Sides: TolR and Full-Length OmpA Bind and Maintain the Local Structure of the E-coli Cell Wall. Structure 2019, 27, 713–724. [Google Scholar] [CrossRef]
- Xu, Y.B.; Moeller, A.; Jun, S.Y.; Le, M.; Yoon, B.Y.; Kim, J.S.; Lee, K.; Ha, N.C. Assembly and Channel Opening of Outer Membrane Protein in Tripartite Drug Efflux Pumps of Gram-negative Bacteria. J. Biol. Chem. 2012, 287, 11740–11750. [Google Scholar] [CrossRef]
- Meroueh, S.O.; Bencze, K.Z.; Hesek, D.; Lee, M.; Fisher, J.F.; Stemmler, T.L.; Mobashery, S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA 2006, 103, 4404–4409. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; van Heijenoort, J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J. Bacteriol. 1985, 163, 208–212. [Google Scholar] [CrossRef]
- Pisabarro, A.G.; De Pedro, M.A.; Vazquez, D. Structural Modifications in the Peptidoglycan of Escherichia coli Associated with Changes in the State of Growth of the Culture. J. Bacteriol. 1985, 161, 238–242. [Google Scholar] [CrossRef]
- Vollmer, W.; Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta Biomembr. 2008, 1778, 1714–1734. [Google Scholar] [CrossRef]
- Barreteau, H.; Patin, D.; Bouhss, A.; Blanot, D.; Mengin-Lecreulx, D.; Touzé, T. CbrA mediates colicin M resistance in Escherichia coli through modification of undecaprenyl-phosphate-linked peptidoglycan precursors. J. Bacteriol. 2020, 202, e00436-20. [Google Scholar] [CrossRef]
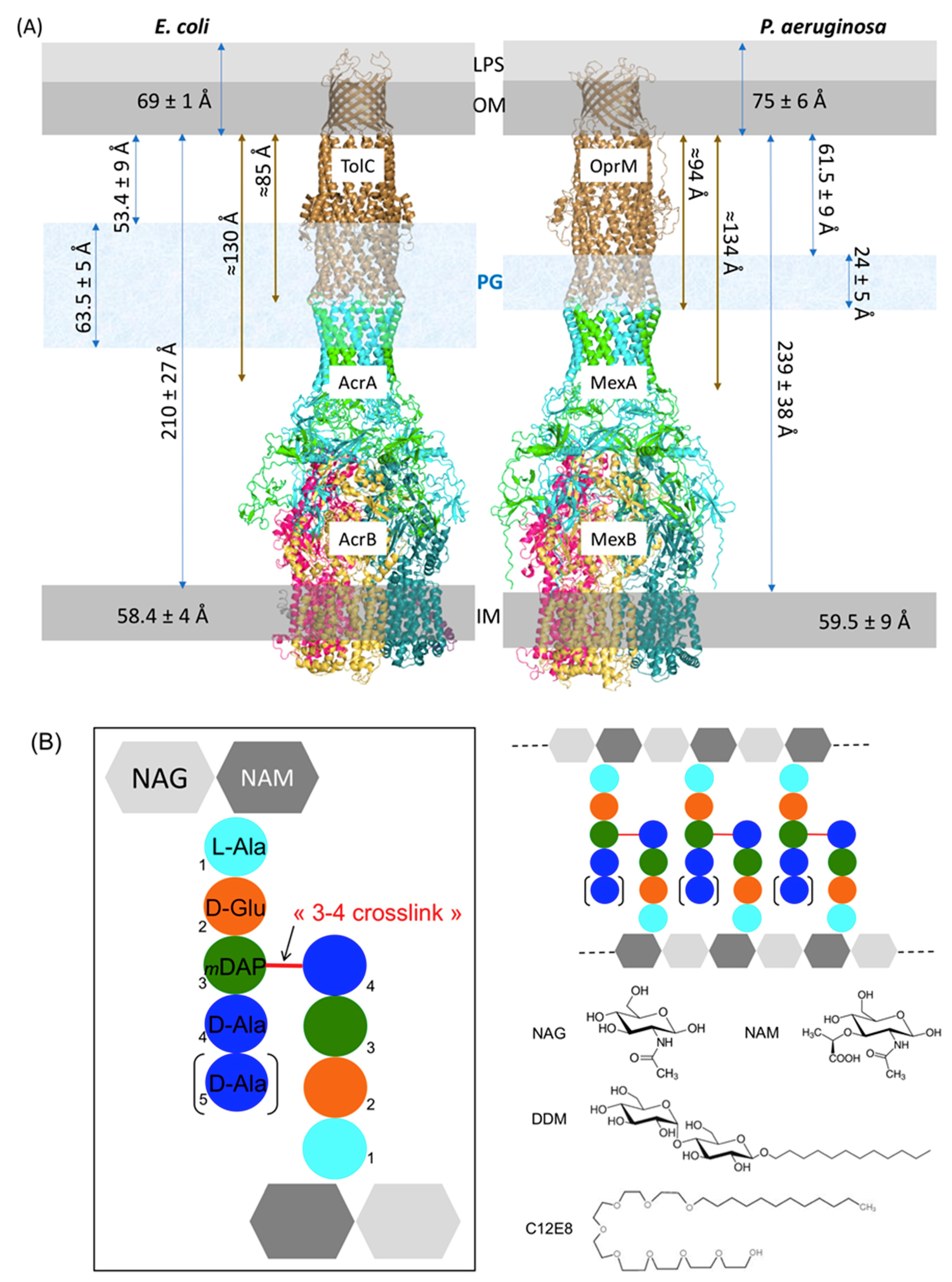

| NAG | NAM | Ala | Glu | DAP | Gly | |
|---|---|---|---|---|---|---|
| E. coli | ||||||
| crude PG extract | 1 | 0.93 | 6.35 | 5.81 | 0.98 | 3.19 |
| purified PG | 1 | 0.98 | 1.82 | 1.13 | 1.03 | 0.19 |
| P. aeruginosa | ||||||
| LB medium crude PG extract | 1 | 1.02 | 2.38 | 1.78 | 1.11 | 0.31 |
| LB medium PG treated by E1 | 1 | 0.98 | 1.57 | 1.06 | 0.99 | 0.04 |
| LB medium PG treated by E1+E2 | 1 | 0.98 | 1.60 | 1.16 | 1.00 | 0.08 |
| MH medium crude PG extract | 1 | 0.94 | 2.70 | 2.21 | 1.11 | 0.63 |
| MH medium PG treated by E1 | 1 | 1.03 | 1.58 | 1.09 | 1.02 | 0.03 |
| MH medium PG treated by E1+E2 | 1 | 0.98 | 1.60 | 1.25 | 1.05 | 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Lustig, M.; Salem, M.; Mengin-Lecreulx, D.; Phan, G.; Broutin, I. MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coli and Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 5328. https://doi.org/10.3390/ijms22105328
Ma M, Lustig M, Salem M, Mengin-Lecreulx D, Phan G, Broutin I. MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coli and Pseudomonas aeruginosa. International Journal of Molecular Sciences. 2021; 22(10):5328. https://doi.org/10.3390/ijms22105328
Chicago/Turabian StyleMa, Miao, Margaux Lustig, Michèle Salem, Dominique Mengin-Lecreulx, Gilles Phan, and Isabelle Broutin. 2021. "MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coli and Pseudomonas aeruginosa" International Journal of Molecular Sciences 22, no. 10: 5328. https://doi.org/10.3390/ijms22105328
APA StyleMa, M., Lustig, M., Salem, M., Mengin-Lecreulx, D., Phan, G., & Broutin, I. (2021). MexAB-OprM Efflux Pump Interaction with the Peptidoglycan of Escherichia coli and Pseudomonas aeruginosa. International Journal of Molecular Sciences, 22(10), 5328. https://doi.org/10.3390/ijms22105328








