Regulation of Oxalate Metabolism in Spinach Revealed by RNA-Seq-Based Transcriptomic Analysis
Abstract
1. Introduction
2. Results
2.1. Validation of Contrasting Oxalate Contents in Spinach Genotypes
2.2. Transcriptome Sequencing, Assembly and Quality Assessment
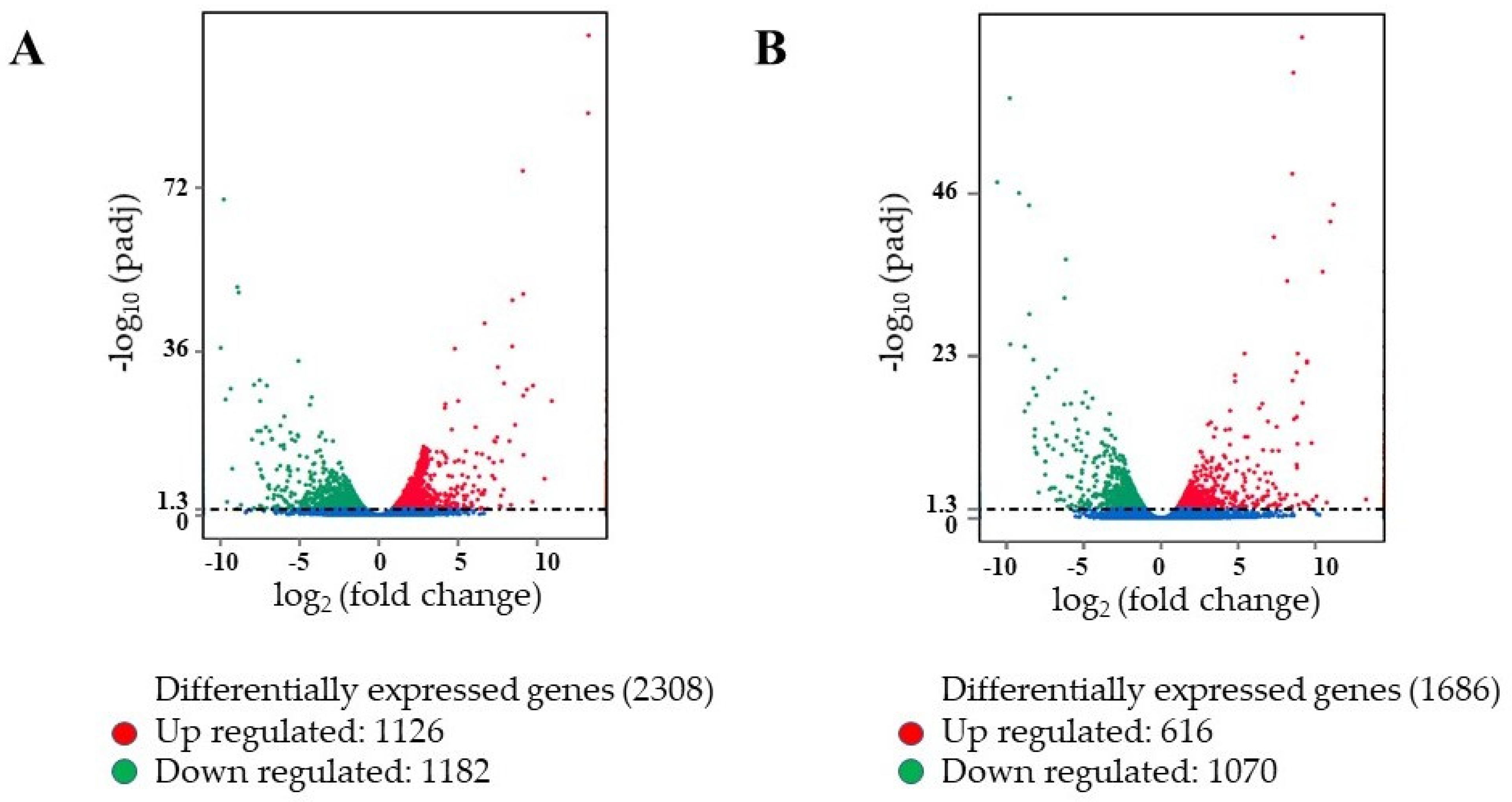
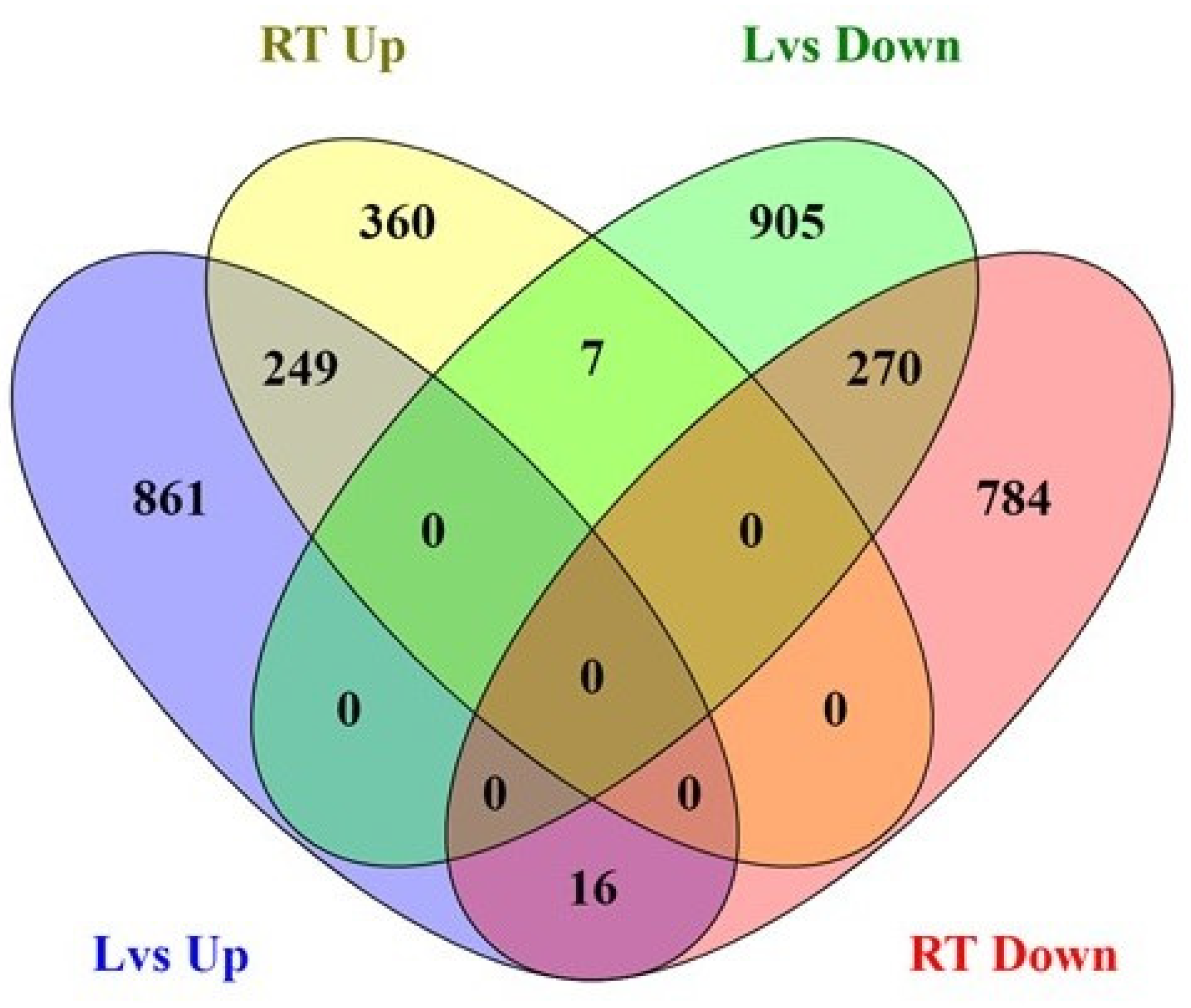
2.3. Analysis of Differentially Expressed Genes (DEGs) in Contrasting Oxalate Genotypes
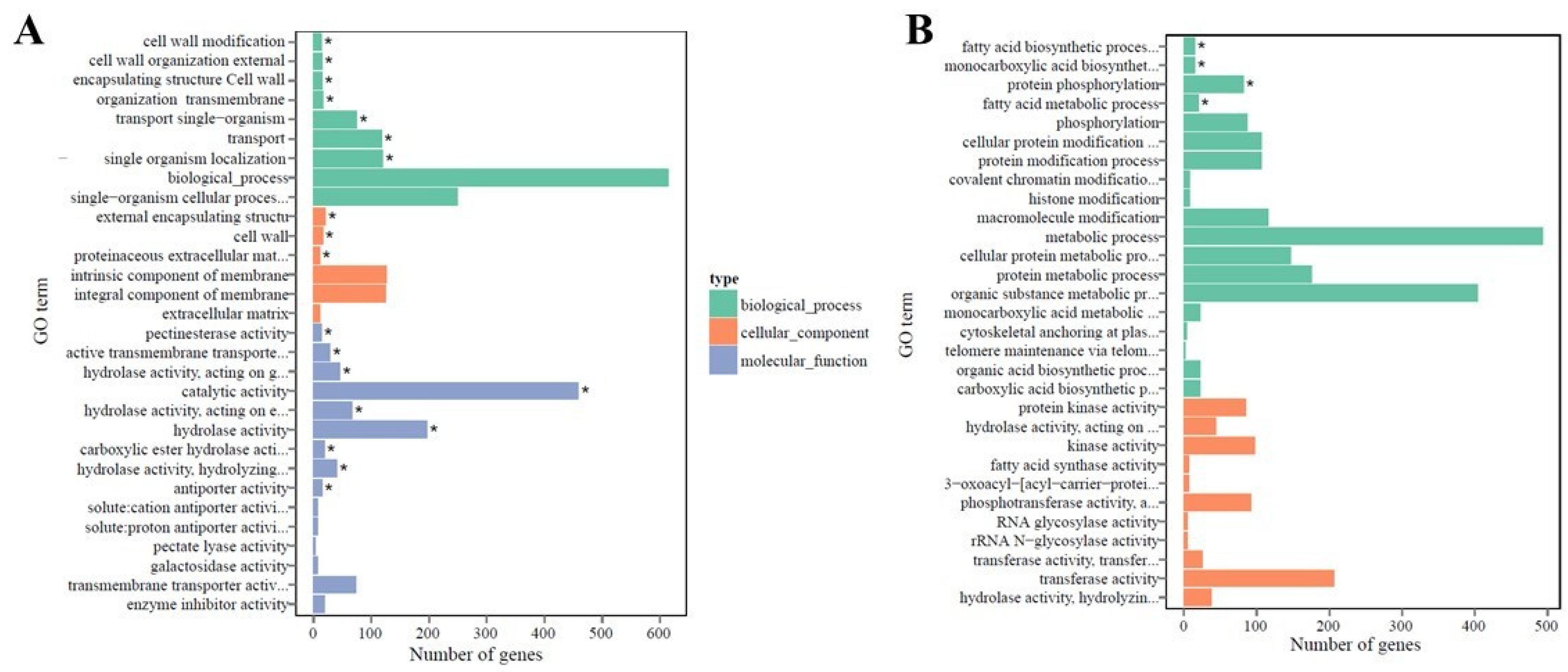
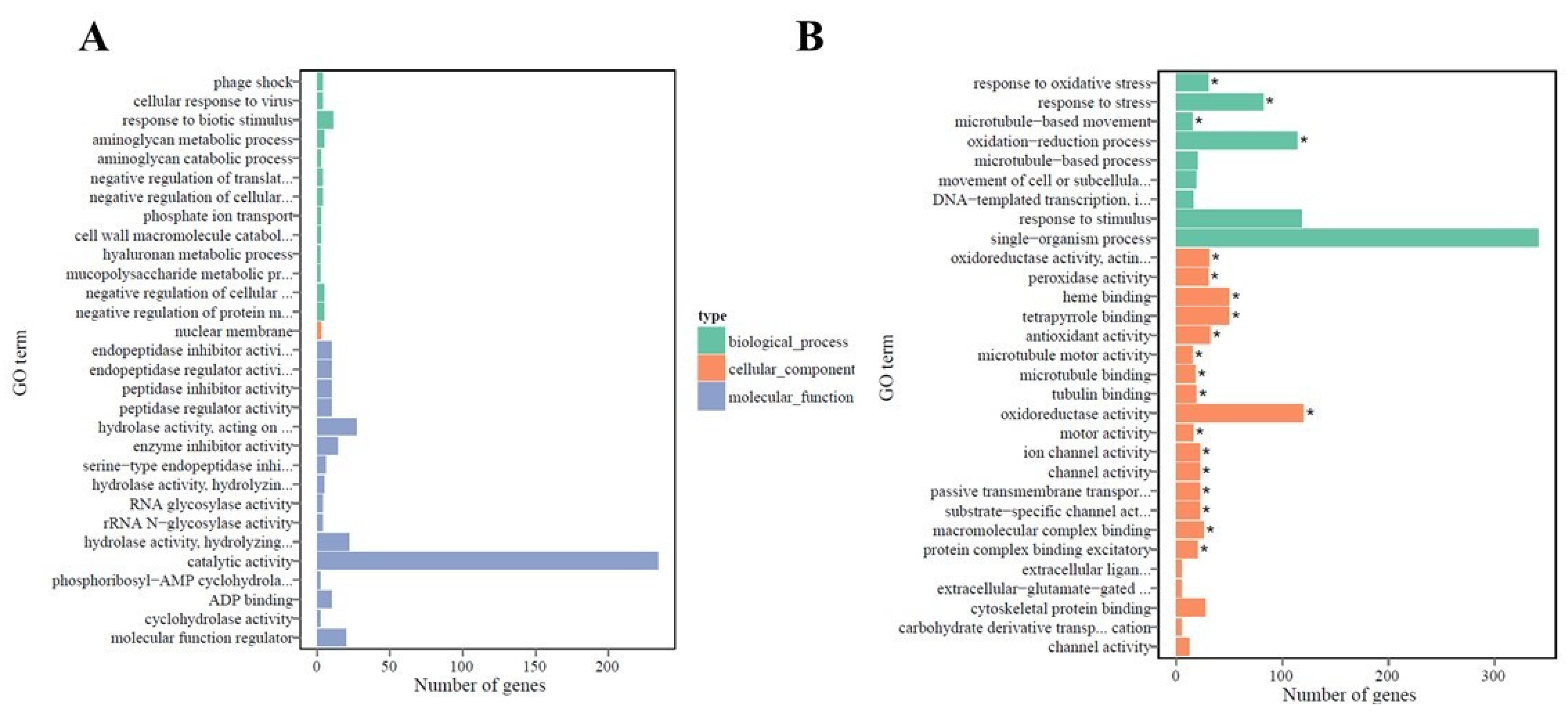
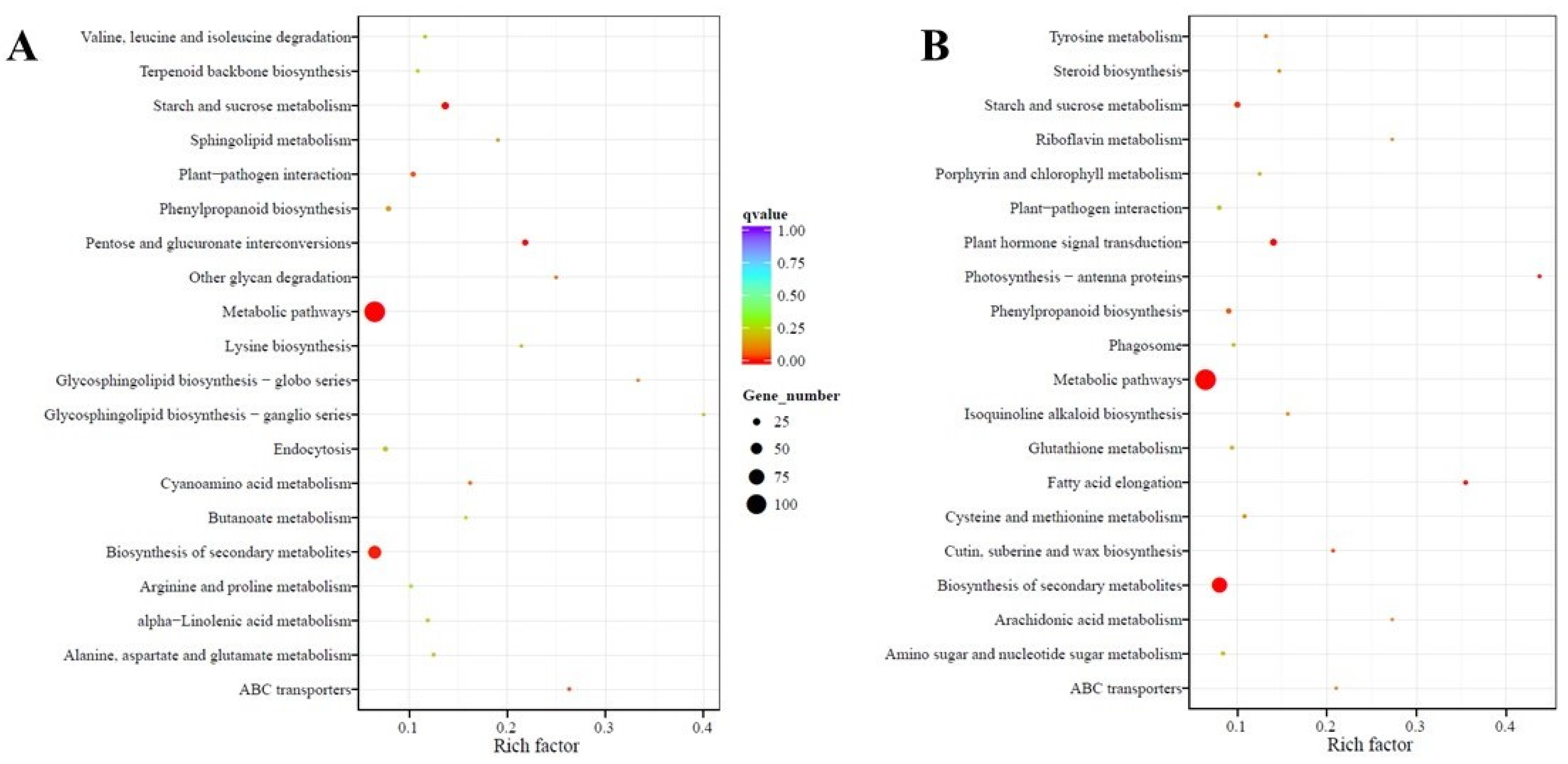
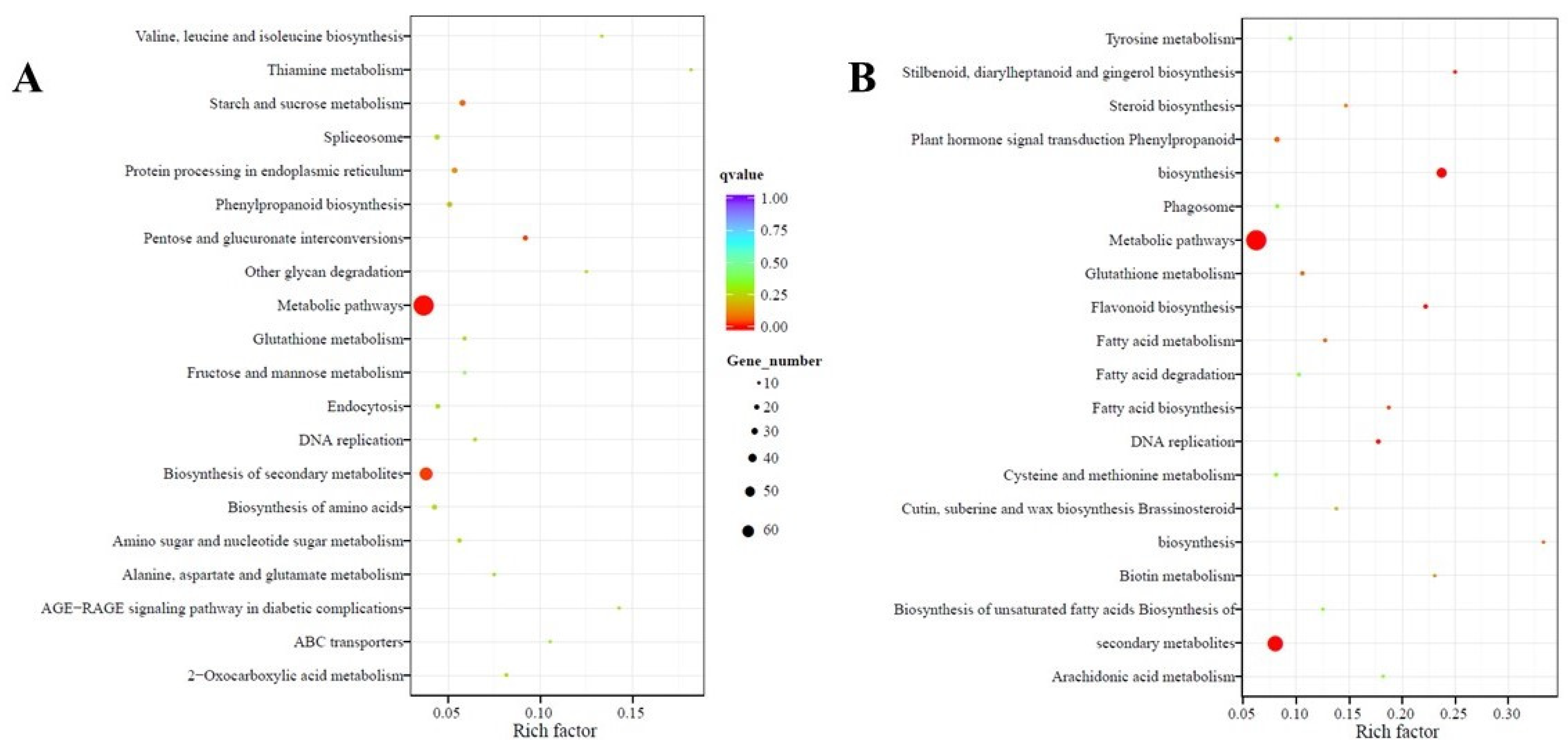
2.4. Functional Annotation and Classification of Transcriptome
2.5. Enrichment Analysis of Transcription Factors (TFs)
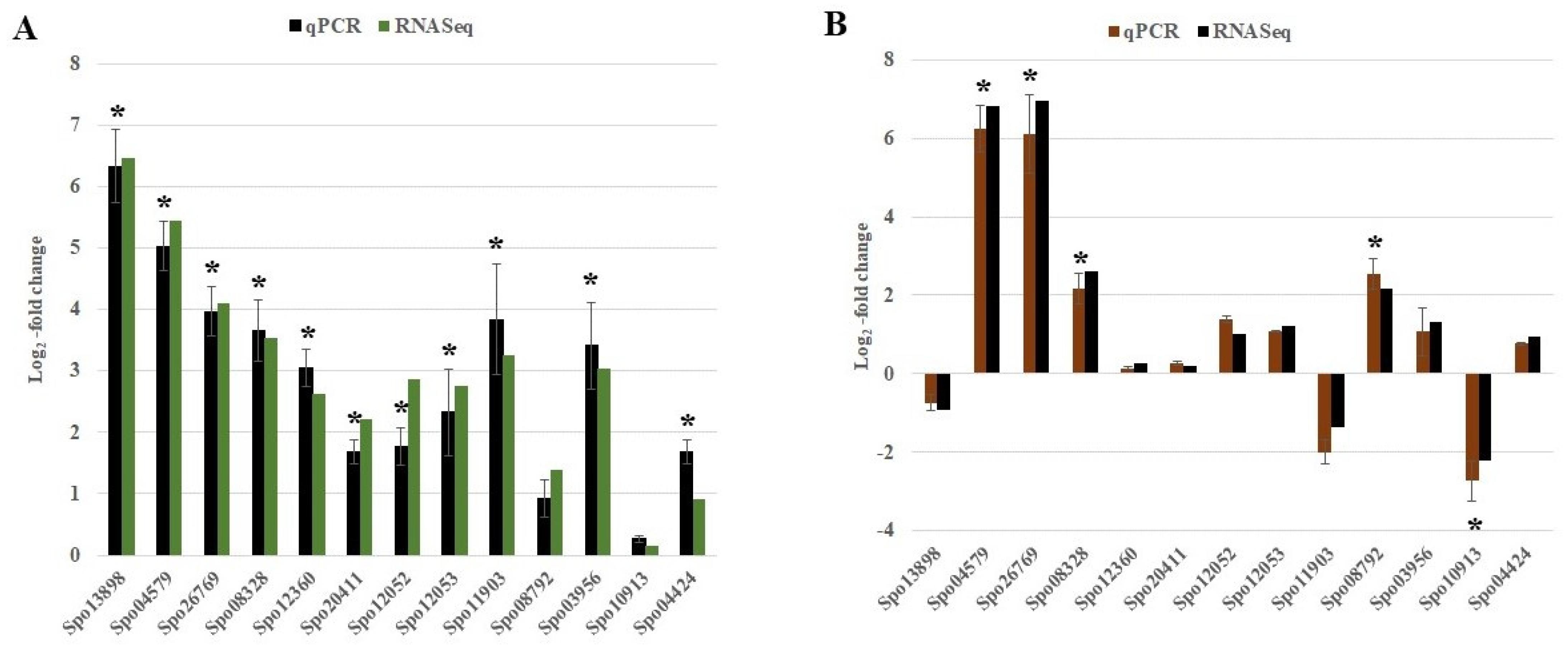
2.6. Quantitative Real-Time PCR Validation of DEGs from RNA-Seq
3. Discussion
3.1. Oxalate Biosynthesis
3.1.1. Cleavage of Isocitrate
3.1.2. Hydrolysis of Oxaloacetate
3.1.3. Oxidation of Glycolate/Glyoxylate
3.1.4. Oxidative Cleavage of Ascorbic Acid
3.2. Oxalate Degradation
4. Conclusions
5. Materials and Methods
5.1. Plant Growth and Estimation of Oxalates in Spinach
5.2. Estimation of Photosynthesis, Minerals, and Quantification of Free Amino Acids
5.3. Oxalic Acid Estimation
5.4. RNA-Seq Analysis of Spinach Leaves and Roots
5.5. Validation by Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siener, R.; Hönow, R.; Seidler, A.; Voss, S.; Hesse, A. Oxalate contents of species of the Polygonaceae, Amaranthaceae and Chenopodiaceae families. Food Chem. 2006, 98, 220–224. [Google Scholar] [CrossRef]
- Fink, S. The micromorphological distribution of bound calcium in needles of Norway spruce [Picea abies (L.) Karst.]. New Phytol. 1991, 119, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, V. Calcium oxalate formation is a rapid and reversible process in Lemna minor L. Protoplasma 1989, 148, 130–137. [Google Scholar] [CrossRef]
- Libert, B.; Franceschi, V.R. Oxalate in crop plants. J. Agric. Food Chem. 1987, 35, 926–938. [Google Scholar] [CrossRef]
- Franceschi, V.R.; Nakata, P.A. Calcium oxalate in plants: Formation and function. Annu. Rev. Plant Biol. 2005, 56, 41–71. [Google Scholar] [CrossRef]
- Mazen, A.M.; Zhang, D.; Franceschi, V.R. Calcium oxalate formation in Lemna minor: Physiological and ultrastructural aspects of high capacity calcium sequestration. New Phytol. 2004, 161, 435–448. [Google Scholar] [CrossRef]
- Webb, M.A. Cell-Mediated Crystallization of Calcium Oxalate in Plants. Plant Cell 1999, 11, 751–761. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ma, J.F.; Zheng, S.J.; Matsumoto, H.; Hiradate, S. Detoxifying aluminium with buckwheat. Nature 1997, 390, 569–570. [Google Scholar] [CrossRef]
- Ma, Z.; Miyasaka, S.C. Oxalate Exudation by Taro in Response to Al. Plant Physiol. 1998, 118, 861–865. [Google Scholar] [CrossRef]
- Franceschi, V.; Schueren, A.M. Incorporation of strontium into plant calcium oxalate crystals. Protoplasma 1986, 130, 199–205. [Google Scholar] [CrossRef]
- Choi, Y.-E.; Harada, E.; Wada, M.; Tsuboi, H.; Morita, Y.; Kusano, T.; Sano, H. Detoxification of cadmium in tobacco plants: Formation and active excretion of crystals containing cadmium and calcium through trichomes. Planta 2001, 213, 45–50. [Google Scholar] [CrossRef]
- Hudgins, J.; Krekling, T.; Franceschi, V.R. Distribution of calcium oxalate crystals in the secondary phloem of conifers: A constitutive defense mechanism? New Phytol. 2003, 159, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Korth, K.L.; Doege, S.J.; Park, S.-H.; Goggin, F.L.; Wang, Q.; Gomez, S.K.; Liu, G.; Jia, L.; Nakata, P.A. Medicago truncatula mutants demonstrate the role of plant calcium oxalate crystals as an effective defense against chewing insects. Plant Physiol. 2006, 141, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, P.A.; Anderson, T.; Lane, B.G.; Davidson, A.L.; Simmonds, D.H. Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol. Mol. Plant Pathol. 2001, 59, 297–307. [Google Scholar] [CrossRef]
- Walz, A.; Zingen-Sell, I.; Loeffler, M.; Sauer, M. Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol. 2008, 57, 453–458. [Google Scholar] [CrossRef]
- Mitchell, T.; Kumar, P.; Reddy, T.; Wood, K.D.; Knight, J.; Assimos, D.G.; Holmes, R.P. Dietary oxalate and kidney stone formation. Am. J. Physiol. Ren. Physiol. 2019, 316, F409–F413. [Google Scholar] [CrossRef]
- Bargagli, M.; Tio, M.C.; Waikar, S.S.; Ferraro, P.M. Dietary Oxalate Intake and Kidney Outcomes. Nutrients 2020, 12, 2673. [Google Scholar] [CrossRef]
- Kawazu, Y.; Okimura, M.; Ishii, T.; Yui, S. Varietal and seasonal differences in oxalate content of spinach. Sci. Hortic. 2003, 97, 203–210. [Google Scholar] [CrossRef]
- Mou, B. Evaluation of Oxalate Concentration in the U.S. Spinach Germplasm Collection. HortScience 2008, 43. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, X.; Zhang, Y.; Zheng, S.J.; Du, S. Effects of nitrogen levels and nitrate/ammonium ratios on oxalate concentrations of different forms in edible parts of spinach. J. Plant Nutr. 2005, 28, 2011–2025. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wei, J.; Sun, M.; Tian, Y.; Li, Z. Effects of Nitrogen and Calcium Nutrition on Oxalate Contents, Forms, and Distribution in Spinach. J. Plant Nutr. 2009, 32, 2123–2139. [Google Scholar] [CrossRef]
- Solberg, S.O.; Yndgaard, F.; Axelsson, J. Nitrate and oxalate in germplasm collections of spinach and other leafy vegetables. Emir. J. Food Agric. 2015, 27, 698–705. [Google Scholar] [CrossRef]
- Tian, H.; Jiang, L.; Liu, E.; Zhang, J.; Liu, F.; Peng, X. Dependence of nitrate-induced oxalate accumulation on nitrate reduction in rice leaves. Physiol. Plant. 2008, 133, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Kagawa, A. Effect of nitrogen nutrients on the oxalate content in spinach plants. J. Jpn. Soc. Hortic. Sci. 1996, 65, 327–332. [Google Scholar] [CrossRef]
- Takebe, M.; Ishihara, T.; Matsuno, K.; Fujimoto, J.; Yoneyama, T. Effect of nitrogen application on the contents of sugars, ascorbic acid, nitrate and oxalic acid in spinach (Spinacia oleracea L.) and komatsuna (Brassica campestris L.). Jpn. J. Soil Sci. Plant Nutr. 1997, 66, 238–246. [Google Scholar]
- Elia, A.; Santamaria, P.; Serio, F. Nitrogen nutrition, yield and quality of spinach. J. Sci. Food Agric. 1998, 76, 341–346. [Google Scholar] [CrossRef]
- Liu, X.; Lu, L.; Chen, Q.; Ding, W.; Dai, P.; Hu, Y.; Yu, Y.; Jin, C.; Lin, X. Ammonium reduces oxalate accumulation in different spinach (Spinacia oleracea L.) genotypes by inhibiting root uptake of nitrate. Food Chem. 2015, 186, 312–318. [Google Scholar] [CrossRef]
- Hodgkinson, A. Oxalic Acid in Biology and Medicine; Academic Press: Cambridge, MA, USA, 1977. [Google Scholar]
- Okutani, I.; Sugiyama, N. Relationship between oxalate concentration and leaf position in various spinach cultivars. HortScience 1994, 29, 1019–1021. [Google Scholar] [CrossRef]
- Maynard, D.; Barker, A.; Minotti, P.; Peck, N. Nitrate Accumulation in Vegetables. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 1976; Volume 28, pp. 71–118. [Google Scholar]
- Cai, X.; Ge, C.; Xu, C.; Wang, X.; Wang, S.; Wang, Q. Expression Analysis of Oxalate Metabolic Pathway Genes Reveals Oxalate Regulation Patterns in Spinach. Molecules 2018, 23, 1286. [Google Scholar] [CrossRef]
- Kitchen, J.W.; Burns, E.E.; Perry, B.A. Calcium oxalate content of spinach (Spinacia oleracea L.). Proc. Am. Soc. Hortic. Sci. 1964, 84, 441–445. [Google Scholar]
- Kaminishi, A.; Kita, N. Seasonal Change of Nitrate and Oxalate Concentration in Relation to the Growth Rate of Spinach Cultivars. Hortsci. Publ. Am. Soc. Hortic. Sci. 2006, 41. [Google Scholar] [CrossRef]
- Murakami, K.; Edamoto, M.; Hata, N.; Itami, Y.; Masuda, M. Low-oxalate Spinach Mutant Induced by Chemical Mutagenesis. J. Jpn. Soc. Hortic. Sci. 2009, 78, 180–184. [Google Scholar] [CrossRef]
- Miyagi, A.; Saimaru, T.; Harigai, N.; Oono, Y.; Hase, Y.; Kawai-Yamada, M. Metabolome analysis of rice leaves to obtain low-oxalate strain from ion beam-mutagenised population. Metabolomics 2020, 16, 94. [Google Scholar] [CrossRef]
- Shi, A.; Mou, B.; Correll, J.C. Association analysis for oxalate concentration in spinach. Euphytica 2016, 212. [Google Scholar] [CrossRef]
- Fujii, N.; Watanabe, M.; Watanabe, Y.; Shimada, N. Kate of oxalate biosynthesis from glycolate and ascorbic acid in spinach leaves. Soil Sci. Plant Nutr. 1993, 39, 627–634. [Google Scholar] [CrossRef]
- Nakata, P.A. Advances in our understanding of calcium oxalate crystal formation and function in plants. Plant Sci. 2003, 164, 901–909. [Google Scholar] [CrossRef]
- Yu, L.; Jiang, J.; Zhang, C.; Jiang, L.; Ye, N.; Lu, Y.; Yang, G.; Liu, E.; Peng, C.; He, Z.; et al. Glyoxylate rather than ascorbate is an efficient precursor for oxalate biosynthesis in rice. J. Exp. Bot. 2010, 61, 1625–1634. [Google Scholar] [CrossRef]
- Franceschi, V.R. Oxalic acid metabolism and calcium oxalate formation in Lemna minor L. Plant Cell Environ. 1987, 10, 397–406. [Google Scholar] [CrossRef]
- Horner, H.T.; Kausch, A.P.; Wagner, B.L. Ascorbic Acid: A Precursor of Oxalate in Crystal Idioblasts of Yucca torreyi in Liquid Root Culture. Int. J. Plant Sci. 2000, 161, 861–868. [Google Scholar] [CrossRef]
- Keates, S.E.; Tarlyn, N.M.; Loewus, F.A.; Franceschi, V.R. L-Ascorbic acid and L-galactose are sources for oxalic acid and calcium oxalate in Pistia stratiotes. Phytochemistry 2000, 53, 433–440. [Google Scholar] [CrossRef]
- Kostman, T.A.; Tarlyn, N.M.; Loewus, F.A.; Franceschi, V.R. Biosynthesis of L-ascorbic acid and conversion of carbons 1 and 2 of L-ascorbic acid to oxalic acid occurs within individual calcium oxalate crystal idioblasts. Plant Physiol. 2001, 125, 634–640. [Google Scholar] [CrossRef]
- Xu, H.-W.; Ji, X.-M.; He, Z.-H.; Shi, W.-P.; Zhu, G.-H.; Niu, J.-K.; Li, B.-S.; Peng, X.-X. Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. J. Exp. Bot. 2006, 57, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.; de la Riva, L.; Garces, F.; Gimenez, R.; Aguilar, J.; Baldoma, L.; Badia, J. The yiaKLX1X2PQRS and ulaABCDEFG gene systems are required for the aerobic utilization of L-ascorbate in Klebsiella pneumoniae strain 13882 with L-ascorbate-6-phosphate as the inducer. J. Bacteriol. 2008, 190, 6615–6624. [Google Scholar] [CrossRef][Green Version]
- Loewus, F.A. Biosynthesis and metabolism of ascorbic acid in plants and of analogs of ascorbic acid in fungi. Phytochemistry 1999, 52, 193–210. [Google Scholar] [CrossRef]
- DeBolt, S.; Cook, D.R.; Ford, C.M. L-tartaric acid synthesis from vitamin C in higher plants. Proc. Natl. Acad. Sci. USA 2006, 103, 5608–5613. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, gkw982. [Google Scholar] [CrossRef] [PubMed]
- Linster, C.L.; Clarke, S.G. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008, 13, 567–573. [Google Scholar] [CrossRef]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; Muñoz-Blanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Hemavathi; Upadhyaya, C.P.; Young, K.E.; Akula, N.; Kim, H.S.; Heung, J.; Oh, O.M.; Aswath, C.R.; Chun, S.C.; Kim, D.H.; et al. Over-expression of strawberry d-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant Sci. 2009, 177, 659–667. [Google Scholar] [CrossRef]
- Ruggieri, V.; Bostan, H.; Barone, A.; Frusciante, L.; Chiusano, M.L. Integrated bioinformatics to decipher the ascorbic acid metabolic network in tomato. Plant Mol. Biol. 2016, 91, 397–412. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Eprintsev, A.T. Organic Acids: The Pools of Fixed Carbon Involved in Redox Regulation and Energy Balance in Higher Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Ji, X.-M.; Peng, X.-X. Oxalate Accumulation as Regulated by Nitrogen Forms and Its Relationship to Photosynthesis in Rice (Oryza sativa L.). J. Integr. Plant Biol. 2005, 47, 831–838. [Google Scholar] [CrossRef]
- Laxa, M.; Fromm, S. Co-expression and regulation of photorespiratory genes in Arabidopsis thaliana: A bioinformatic approach. Curr. Plant Biol. 2018, 14, 2–18. [Google Scholar] [CrossRef]
- Yuenyong, W.; Sirikantaramas, S.; Qu, L.-J.; Buaboocha, T. Isocitrate lyase plays important roles in plant salt tolerance. BMC Plant Biol. 2019, 19, 472. [Google Scholar] [CrossRef] [PubMed]
- Eprintsev, A.; Fedorin, D.; Salnikov, A.; Igamberdiev, A. Expression and properties of the glyoxysomal and cytosolic forms of isocitrate lyase in Amaranthus caudatus L. J. Plant Physiol. 2015, 181. [Google Scholar] [CrossRef] [PubMed]
- Zelitch, I. Synthesis of Glycolate from Pyruvate via Isocitrate Lyase by Tobacco Leaves in Light. Plant Physiol. 1988, 86, 463–468. [Google Scholar] [CrossRef]
- Miyagi, A.; Uchimiya, M.; Kawai-Yamada, M.; Uchimiya, H. An antagonist treatment in combination with tracer experiments revealed isocitrate pathway dominant to oxalate biosynthesis in Rumex obtusifolius L. Metabolomics 2012, 9, 590–598. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, J.; Zeng, J.; Jiang, L.; Liu, E.; Peng, C.; He, Z.; Peng, X. Inducible antisense suppression of glycolate oxidase reveals its strong regulation over photosynthesis in rice. J. Exp. Bot. 2009, 60, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.; Joshi, M.; Penalosa, A. Comparative analysis of tissue-specific transcriptomic responses to nitrogen stress in spinach (Spinacia oleracea). PLoS ONE 2020, 15, e0232011. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liberti, D.; Li, M.; Kim, Y.T.; Hutchens, A.; Wilson, R.; Rollins, J.A. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 2015, 16, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Beevers, H. Biogenesis of oxalate in plant tissues. Plant Physiol. 1968, 43, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Wang, L.; Duan, Q.; Huang, D. Comparative analysis of metabolites profile in spinach (Spinacia oleracea L.) affected by different concentrations of gly and nitrate. Sci. Hortic. 2016, 204, 8–15. [Google Scholar] [CrossRef]
- Mulligan, R.M.; Wilson, B.; Tolbert, N.E. Effects of glyoxylate on photosynthesis by intact chloroplasts. Plant Physiol. 1983, 72, 415–419. [Google Scholar] [CrossRef]
- Kisaki, T.; Tolbert, N.E. Glycolate and glyoxylate metabolism by isolated peroxisomes or chloroplasts. Plant Physiol. 1969, 44, 242–250. [Google Scholar] [CrossRef]
- Allan, W.L.; Clark, S.M.; Hoover, G.J.; Shelp, B.J. Role of plant glyoxylate reductases during stress: A hypothesis. Biochem. J. 2009, 423, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Givan, C.V.; Kleczkowski, L.A. The Enzymic Reduction of Glyoxylate and Hydroxypyruvate in Leaves of Higher Plants. Plant Physiol. 1992, 100, 552–556. [Google Scholar] [CrossRef]
- Hoover, G.; Van Cauwenberghe, O.; Breitkreuz, K.; Clark, S.; Merrill, A.; Shelp, B. Characteristics of an Arabidopsis glyoxylate reductase: General biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Can. J. Bot. 2007, 85, 883–895. [Google Scholar] [CrossRef]
- Simpson, J.P.; Di Leo, R.; Dhanoa, P.K.; Allan, W.L.; Makhmoudova, A.; Clark, S.M.; Hoover, G.J.; Mullen, R.T.; Shelp, B.J. Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: Comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 2008, 59, 2545–2554. [Google Scholar] [CrossRef] [PubMed]
- Esser, C.; Kuhn, A.; Groth, G.; Lercher, M.J.; Maurino, V.G. Plant and animal glycolate oxidases have a common eukaryotic ancestor and convergently duplicated to evolve long-chain 2-hydroxy acid oxidases. Mol. Biol. Evol. 2014, 31, 1089–1101. [Google Scholar] [CrossRef]
- Li, B.-S.; Peng, X.-X.; Li, M.-Q. Relationship between oxalate accumulation and photorespiratory glycolate metabolism in plants. Acta Phytophysiol. Sin. 2000, 26, 148–152. [Google Scholar]
- Guo, Z.; Tan, H.; Zhu, Z.; Lu, S.; Zhou, B. Effect of intermediates on ascorbic acid and oxalate biosynthesis of rice and in relation to its stress resistance. Plant Physiol. Biochem. 2005, 43, 955–962. [Google Scholar] [CrossRef]
- Green, M.A.; Fry, S.C. Vitamin C degradation in plant cells via enzymatic hydrolysis of 4-O-oxalyl-l-threonate. Nature 2005, 433, 83–87. [Google Scholar] [CrossRef]
- Truffault, V.; Fry, S.C.; Stevens, R.G.; Gautier, H. Ascorbate degradation in tomato leads to accumulation of oxalate, threonate and oxalyl threonate. Plant J. 2017, 89, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Fry, S. Apoplastic degradation of ascorbate: Novel enzymes and metabolites permeating the plant cell wall. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2005, 139, 2–7. [Google Scholar] [CrossRef]
- Yang, J.C.; Loewus, F.A. Metabolic Conversion of l-Ascorbic Acid to Oxalic Acid in Oxalate-accumulating Plants. Plant Physiol. 1975, 56, 283–285. [Google Scholar] [CrossRef]
- Dewhirst, R.A.; Clarkson, G.J.; Rothwell, S.D.; Fry, S.C. Novel insights into ascorbate retention and degradation during the washing and post-harvest storage of spinach and other salad leaves. Food Chem. 2017, 233, 237–246. [Google Scholar] [CrossRef]
- Foster, J.; Kim, H.U.; Nakata, P.A.; Browse, J. A Previously Unknown Oxalyl-CoA Synthetase Is Important for Oxalate Catabolism in Arabidopsis. Plant Cell 2012, 24, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An Oxalyl-CoA Synthetase Is Involved in Oxalate Degradation and Aluminum Tolerance. Plant Physiol. 2016, 172, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, N.; Ghosh, R.; Ghosh, S.; Narula, K.; Tayal, R.; Datta, A.; Chakraborty, S. Reduction of Oxalate Levels in Tomato Fruit and Consequent Metabolic Remodeling Following Overexpression of a Fungal Oxalate Decarboxylase. Plant Physiol. 2013, 162, 364–378. [Google Scholar] [CrossRef]
- Foster, J.; Luo, B.; Nakata, P.A. An Oxalyl-CoA Dependent Pathway of Oxalate Catabolism Plays a Role in Regulating Calcium Oxalate Crystal Accumulation and Defending against Oxalate-Secreting Phytopathogens in Medicago truncatula. PLoS ONE 2016, 11, e0149850. [Google Scholar] [CrossRef] [PubMed]
- Lane, B.G.; Dunwell, J.M.; Ray, J.A.; Schmitt, M.R.; Cuming, A.C. Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 1993, 268, 12239–12242. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, D.; Nandy, P.; Ray, A. Study of Germin Like Oxalate Oxidase Enzyme in Monocot Plants. Indian J. Agric. Res. 2020, 54, 10–18. [Google Scholar] [CrossRef]
- Membré, N.; Berna, A.; Neutelings, G.; David, A.; David, H.; Staiger, D.; Vásquez, J.S.; Raynal, M.; Delseny, M.; Bernier, F. cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Mol. Biol. 1997, 35, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Membré, N.; Bernier, F.; Staiger, D.; Berna, A. Arabidopsis thaliana germin-like proteins: Common and specific features point to a variety of functions. Planta 2000, 211, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Moscatello, S.; Famiani, F.; Battistelli, A. Increase of ascorbic acid content and nutritional quality in spinach leaves during physiological acclimation to low temperature. Plant Physiol. Biochem. 2009, 47, 717–723. [Google Scholar] [CrossRef]
- Xu, H.; He, X.; Wang, K.; Chen, L.; Li, K. Identification of Early Nitrate Stress Response Genes in Spinach Roots by Suppression Subtractive Hybridization. Plant Mol. Biol. Rep. 2011, 30. [Google Scholar] [CrossRef]
- Xian, P.; Cai, Z.; Cheng, Y.; Lin, R.; Lian, T.; Ma, Q.; Nian, H. Wild Soybean Oxalyl-CoA Synthetase Degrades Oxalate and Affects the Tolerance to Cadmium and Aluminum Stresses. Int. J. Mol. Sci. 2020, 21, 8869. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Ji, C.; Huang, Y.; Wu, Y. Maize Oxalyl-CoA Decarboxylase1 Degrades Oxalate and Affects the Seed Metabolome and Nutritional Quality. Plant Cell 2018, 30, 2447–2462. [Google Scholar] [CrossRef]
- Foster, J.; Cheng, N.; Paris, V.; Wang, L.; Wang, J.; Wang, X.; Nakata, P.A. An Arabidopsis Oxalyl-CoA Decarboxylase, AtOXC, Is Important for Oxalate Catabolism in Plants. Int. J. Mol. Sci. 2021, 22, 3266. [Google Scholar] [CrossRef]
- Chen, W.W.; Fan, W.; Lou, H.Q.; Yang, J.L.; Zheng, S.J. Regulating cytoplasmic oxalate homeostasis by Acyl activating enzyme3 is critical for plant Al tolerance. Plant Signal. Behav. 2017, 12, e1276688. [Google Scholar] [CrossRef]
- Joshi, V.; Joshi, M.; Silwal, D.; Noonan, K.; Rodriguez, S.; Penalosa, A. Systematized biosynthesis and catabolism regulate citrulline accumulation in watermelon. Phytochemistry 2019, 162, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 4 April 2021).
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
| Fresh wt | Dry Wt. | |||||
|---|---|---|---|---|---|---|
| Tissue | PI 175311 | Bloomsdale | p-Value | PI 175311 | Bloomsdale | p-Value |
| Leaves | 1312.9 ± 85.0 | 785.5 ± 89.2 | 0.0037 | 104.4 ± 5.7 | 54.4 ± 3.1 | 0.0287 |
| Roots | 450.0 ± 11.5 | 287.1 ± 33.3 | 0.0347 | 40.2 ± 3.0 | 20.8 ± 2.6 | 0.0347 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, V.; Penalosa, A.; Joshi, M.; Rodriguez, S. Regulation of Oxalate Metabolism in Spinach Revealed by RNA-Seq-Based Transcriptomic Analysis. Int. J. Mol. Sci. 2021, 22, 5294. https://doi.org/10.3390/ijms22105294
Joshi V, Penalosa A, Joshi M, Rodriguez S. Regulation of Oxalate Metabolism in Spinach Revealed by RNA-Seq-Based Transcriptomic Analysis. International Journal of Molecular Sciences. 2021; 22(10):5294. https://doi.org/10.3390/ijms22105294
Chicago/Turabian StyleJoshi, Vijay, Arianne Penalosa, Madhumita Joshi, and Sierra Rodriguez. 2021. "Regulation of Oxalate Metabolism in Spinach Revealed by RNA-Seq-Based Transcriptomic Analysis" International Journal of Molecular Sciences 22, no. 10: 5294. https://doi.org/10.3390/ijms22105294
APA StyleJoshi, V., Penalosa, A., Joshi, M., & Rodriguez, S. (2021). Regulation of Oxalate Metabolism in Spinach Revealed by RNA-Seq-Based Transcriptomic Analysis. International Journal of Molecular Sciences, 22(10), 5294. https://doi.org/10.3390/ijms22105294







