Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves
Abstract
1. Introduction
2. Results
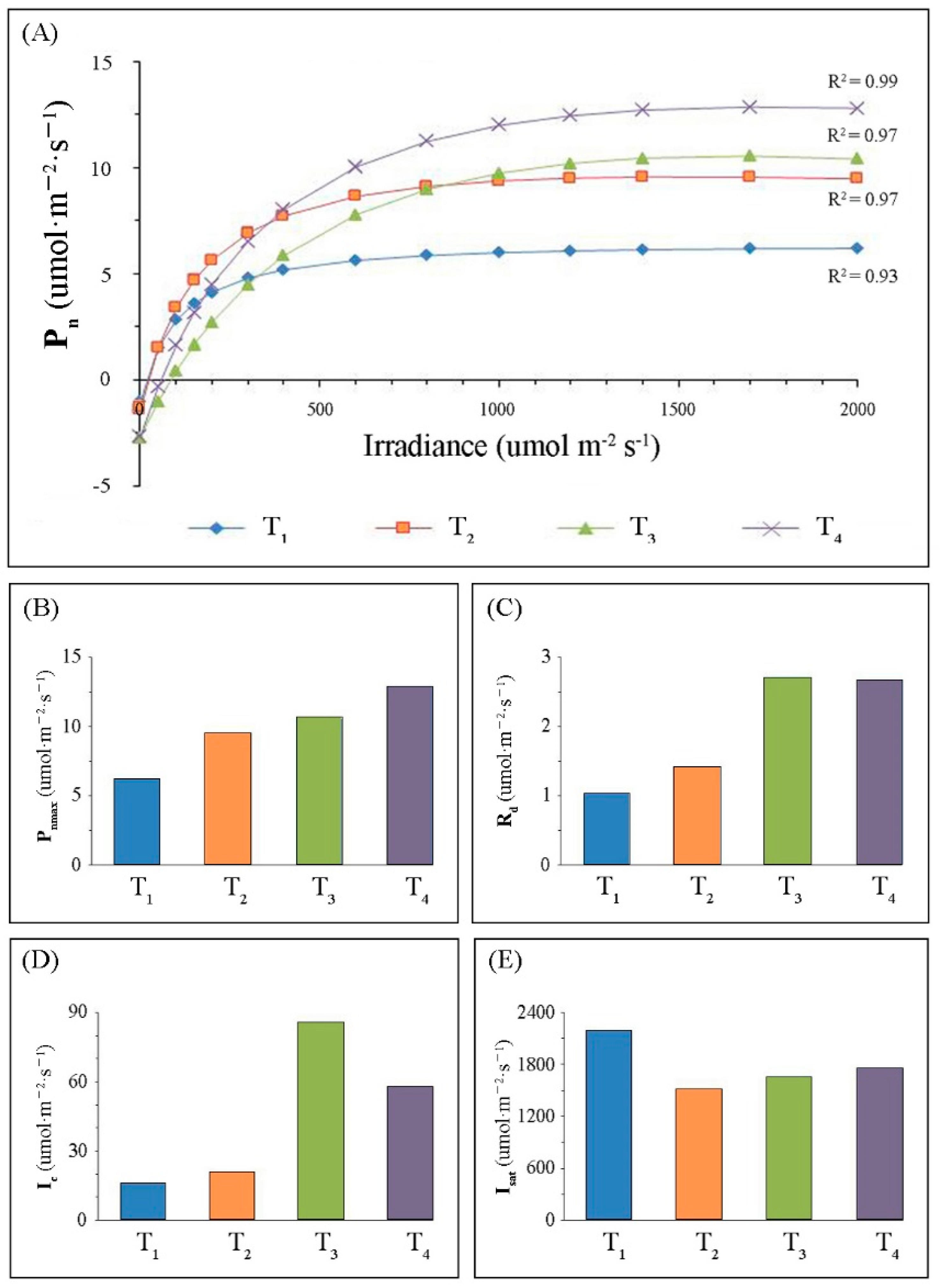
2.1. Variations in Leaf Morphology and Photosynthesis-Related Parameters
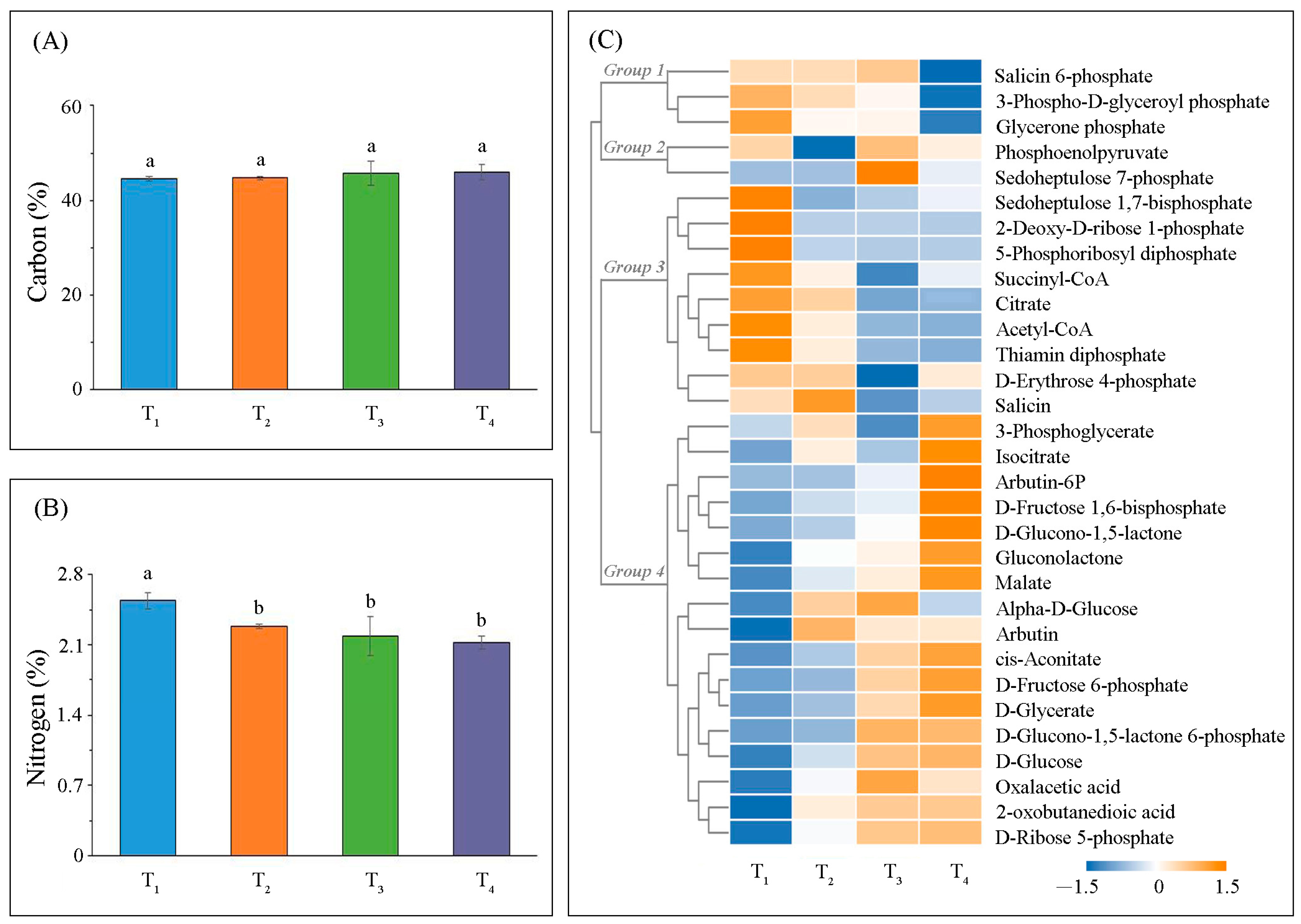
2.2. Variation in Metabolic Levels
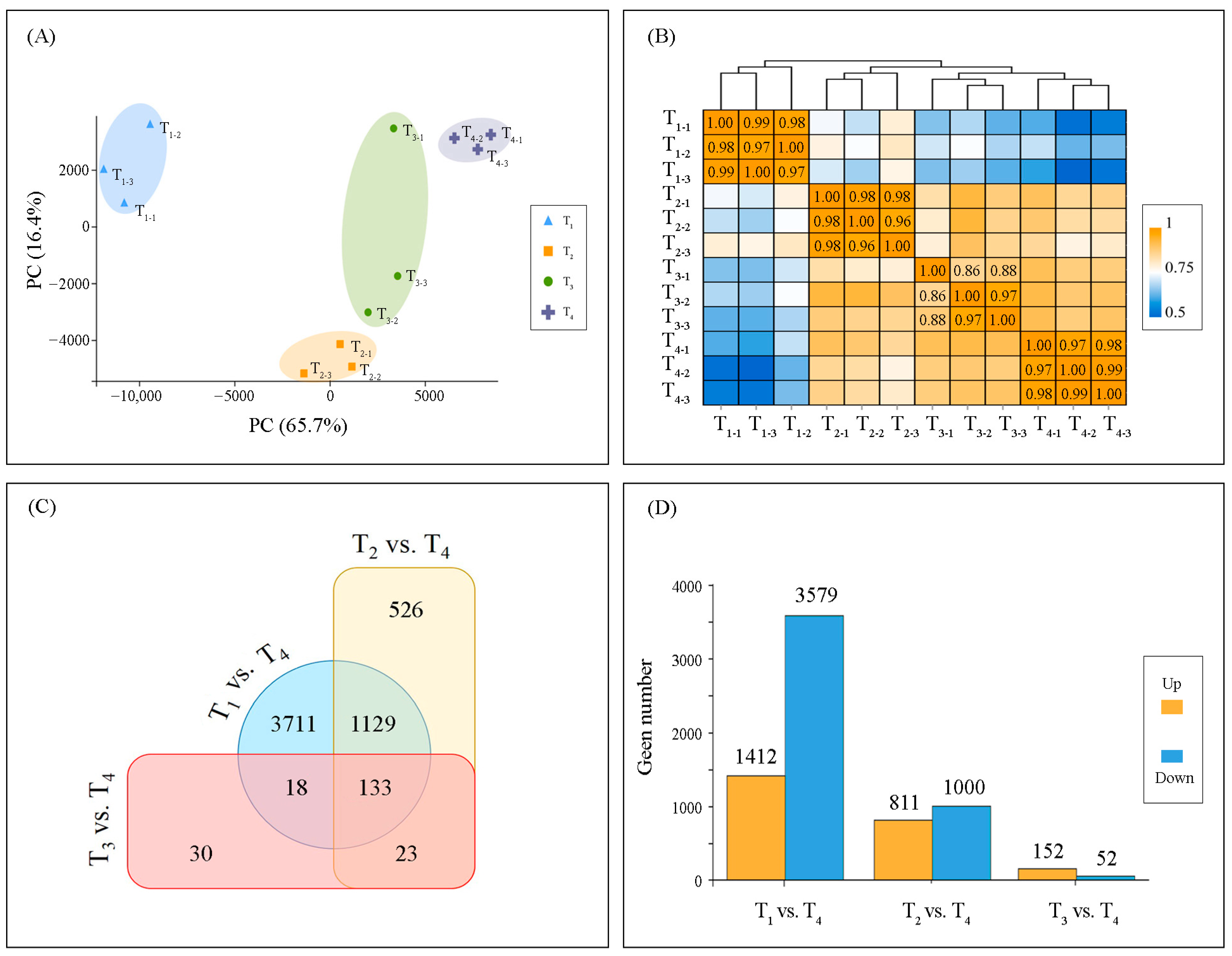
2.3. Transcriptome Reprogramming
2.4. Temporal Dynamics of DEGs Expression
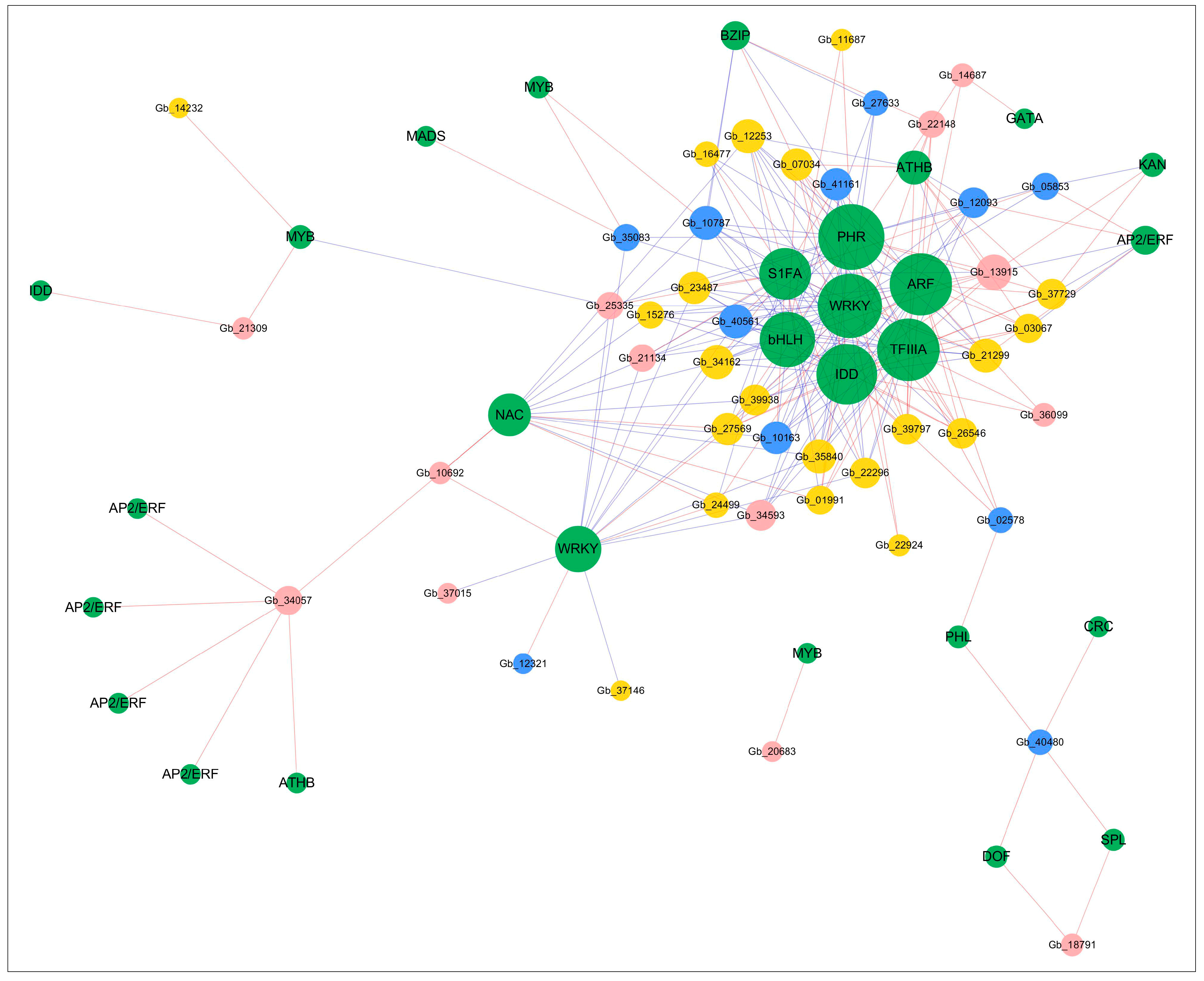
2.5. Gene Co-Expression Network Construction
3. Discussion
3.1. Changes of Photosynthetic Performance of Leaves during Development
3.2. Photosynthetic Carbon Metabolism
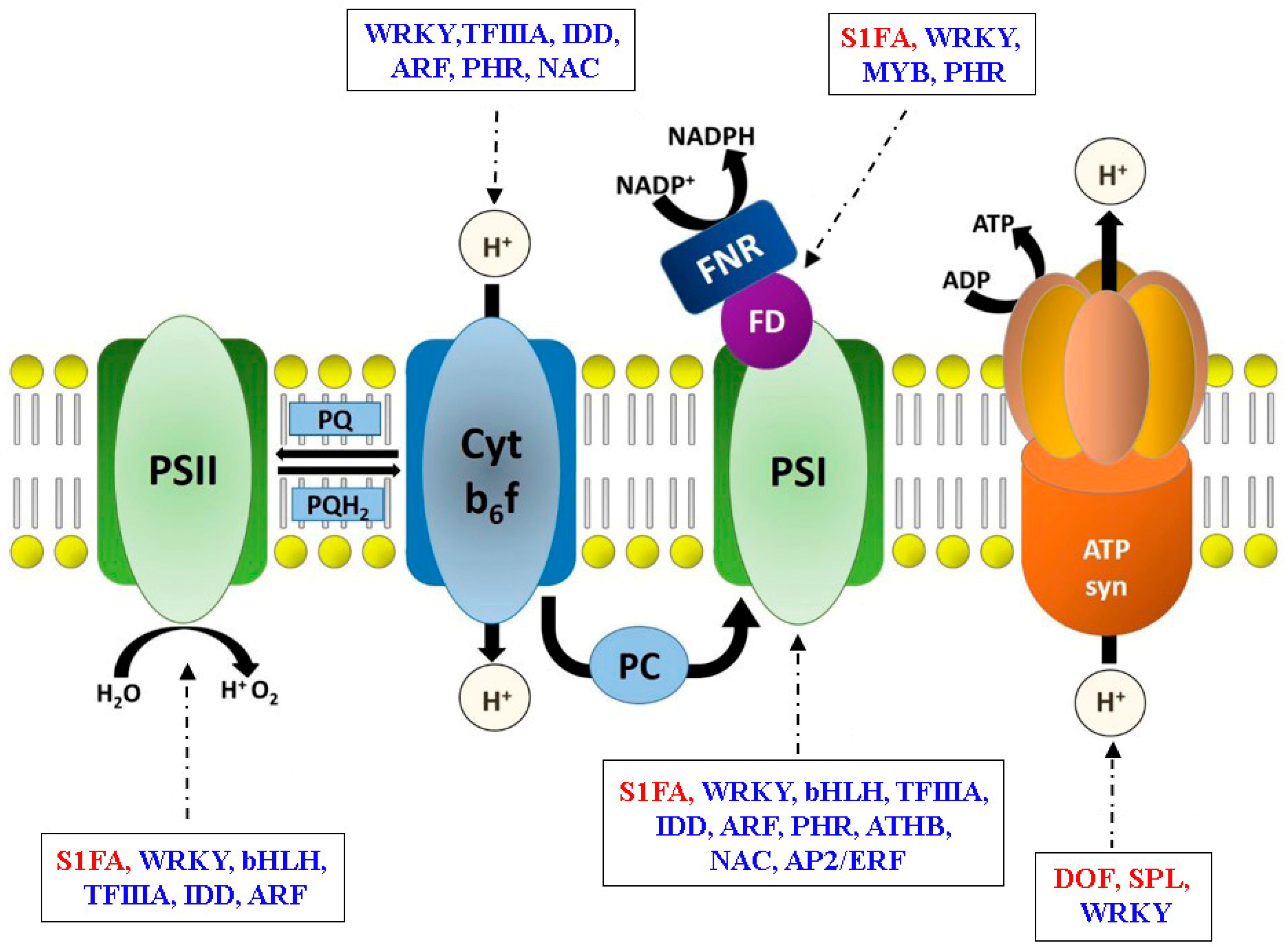
3.3. Transcriptional Regulatory Networks in Photosynthesis Gene Expression
4. Materials and Methods
4.1. Plant Materials
4.2. Observations of Leaf Morphology and Photosynthetic Parameters
4.3. Carbon and Nitrogen Determination
4.4. Metabolomics Analysis
4.5. Transcriptomics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, P.; Hendron, R.W.; Kelly, S. Transcriptional control of photosynthetic capacity: Conservation and divergence from arabidopsis to rice. New Phytol. 2017, 216, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Che, X.; Zhang, Z.; Li, S.; Li, Q.; Li, Y.; Gao, H. Slower development of psi activity limits photosynthesis during euonymus japonicus leaf development. Plant Physiol. Biochem. 2019, 136, 13–21. [Google Scholar] [CrossRef]
- Wang, L.; Du, Q.; Xie, J.; Zhou, D.; Chen, B.; Yang, H.; Zhang, D. Genetic variation in transcription factors and photosynthesis light-reaction genes regulates photosynthetic traits. Tree Physiol. 2018, 38, 1871–1885. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.; Reynolds, M.; Salvucci, M.E.; Raines, C.; Andralojc, P.J.; Zhu, X.-G.; Price, G.D.; Condon, A.G.; Furbank, R.T. Raising yield potential of wheat. Ii. Increasing photosynthetic capacity and efficiency. J. Exp. Bot. 2011, 62, 453–467. [Google Scholar] [CrossRef]
- Tsukaya, H. Controlling size in multicellular organs: Focus on the leaf. PLoS Biol. 2008, 6, e174. [Google Scholar] [CrossRef]
- Maayan, I.; Shaya, F.; Ratner, K.; Mani, Y.; Lavee, S.; Avidan, B.; Shahak, Y.; Ostersetzer-Biran, O. Photosynthetic activity during olive (olea europaea) leaf development correlates with plastid biogenesis and rubisco levels. Physiol. Plant. 2008, 134, 547–558. [Google Scholar] [CrossRef]
- Bertamini, M.; Nedunchezhian, N. Leaf age effects on chlorophyll, rubisco, photosynthetic electron transport activities and thylakoid membrane protein in field grown grapevine leaves. J. Plant Physiol. 2002, 159, 799–803. [Google Scholar] [CrossRef]
- Miyazawa, S.I.; Makino, A.; Terashima, I. Changes in mesophyll anatomy and sink–source relationships during leaf development in quercus glauca, an evergreen tree showing delayed leaf greening. Plant Cell Environ. 2003, 26, 745–755. [Google Scholar] [CrossRef]
- Yoo, S.D.; Greer, D.H.; Laing, W.A.; McManus, M.T. Changes in photosynthetic efficiency and carotenoid composition in leaves of white clover at different developmental stages. Plant Physiol. Biochem. 2003, 41, 887–893. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Yuan, F.; Yang, Z.; Wang, B.; Chen, M. Transcriptome profiling of genes involved in photosynthesis in elaeagnus angustifolia l. Under salt stress. Photosynthetica 2018, 56, 998–1009. [Google Scholar] [CrossRef]
- Zhu, T.; Li, L.; Feng, L.; Ren, M. Stabi5 involved in the regulation of chloroplast development and photosynthesis in potato. Int. J. Mol. Sci. 2020, 21, 1068. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Transcription factors in plants: Physiological functions and regulation of expression. J. Plant Res. 1998, 111, 363–371. [Google Scholar] [CrossRef]
- Stephenson, T.J.; McIntyre, C.L.; Collet, C.; Xue, G.P. Tanf-yb3 is involved in the regulation of photosynthesis genes in triticum aestivum. Funct. Integr. Genom. 2011, 11, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. Glk transcription factors coordinate expression of the photosynthetic apparatus in arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef]
- Oh, S.; Shintani, R.; Koike, S.; Kobayashi, Y. Ginkgo fruit extract as an additive to modify rumen microbiota and fermentation and to mitigate methane production. J. Dairy Sci. 2017, 100, 1923–1934. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Mu, K.; Xu, M.; Ma, X.; Ni, Z.; Wang, J.; Xu, L.-A. Variation in the concentrations of major secondary metabolites in ginkgo leaves from different geographical populations. Forests 2017, 8, 266. [Google Scholar] [CrossRef]
- He, X.; Fu, S.; Chen, W.; Zhao, T.; Xu, S.; Tuba, Z. Changes in effects of ozone exposure on growth, photosynthesis, and respiration of ginkgo biloba in shenyang urban area. Photosynthetica 2007, 45, 555–561. [Google Scholar] [CrossRef]
- Kinoshita, T.; Kume, A.; Hanba, Y.T. Seasonal variations in photosynthetic functions of the urban landscape tree species gingko biloba: Photoperiod is a key trait. Trees 2020, 35, 273–285. [Google Scholar] [CrossRef]
- Jing, J.; Hong, J.; Shuquan, Y.; Guomo, Z. Sex-linked photosynthetic physiologic research and the evolutionary ecological analysis in living fossil plant, ginkgo biloba L. Acta Ecol. Sin. 2008, 28, 1128–1136. [Google Scholar] [CrossRef]
- Shi, D.; Wei, X.; Chen, G.; Xu, Y. Changes in photosynthetic characteristics and antioxidative protection in male and female ginkgo during natural senescence. J. Am. Soc. Hortic. Sci. 2012, 137, 349–360. [Google Scholar] [CrossRef]
- Kagotani, Y.; Nishida, K.; Kiyomizu, T.; Sasaki, K.; Kume, A.; Hanba, Y.T. Photosynthetic responses to soil water stress in summer in two japanese urban landscape tree species (ginkgo biloba and prunus yedoensis): Effects of pruning mulch and irrigation management. Trees 2016, 30, 697–708. [Google Scholar] [CrossRef]
- Pandey, S.; Kumar, S.; Nagar, P. Photosynthetic performance of ginkgo biloba L. Grown under high and low irradiance. Photosynthetica 2003, 41, 505–511. [Google Scholar] [CrossRef]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Zhu, X.G.; Naidu, S.L.; Ort, D.R. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006, 29, 315–330. [Google Scholar] [CrossRef]
- Herrmann, H.A.; Schwartz, J.-M.; Johnson, G.N. From empirical to theoretical models of light response curves-linking photosynthetic and metabolic acclimation. Photosynth. Res. 2020, 145, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.A.; Johnson, D. Changes in photosynthetic characteristics during leaf development in apple. Photosynth. Res. 1981, 2, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Reich, P.; Walters, M.; Ellsworth, D. Leaf life-span in relation to leaf, plant, and stand characteristics among diverse ecosystems. Ecol. Monogr. 1992, 62, 365–392. [Google Scholar] [CrossRef]
- Lobo, F.d.A.; De Barros, M.; Dalmagro, H.; Dalmolin, Â.; Pereira, W.; de Souza, É.; Vourlitis, G.; Ortíz, C.R. Erratum to: Fitting net photosynthetic light-response curves with microsoft excel—A critical look at the models. Photosynthetica 2013, 52, 445–456. [Google Scholar] [CrossRef]
- Zhu, M.; Xu, W.; Wen, J.; Zhu, Y.; Li, Y.; Su, Y.; Zhang, Q.; Sun, R. Dynamic changes of photosynthetic properties and chemical compositions of eucommia ulmoides oliver under two planting models. Ind. Crop. Prod. 2017, 96, 46–56. [Google Scholar] [CrossRef]
- Cai, Z.; Slot, M.; Fan, Z. Leaf development and photosynthetic properties of three tropical tree species with delayed greening. Photosynthetica 2005, 43, 91–98. [Google Scholar] [CrossRef]
- Reich, P.B.; Walters, M.B.; Ellsworth, D.S.; Vose, J.M.; Volin, J.C.; Gresham, C.; Bowman, W.D. Relationships of leaf dark respiration to leaf nitrogen, specific leaf area and leaf life-span: A test across biomes and functional groups. Oecologia 1998, 114, 471–482. [Google Scholar] [CrossRef]
- Dodd, I.; Critchley, C.; Woodall, G.; Stewart, G. Photoinhibition in differently coloured juvenile leaves of syzygium species. J. Exp. Bot. 1998, 49, 1437–1445. [Google Scholar] [CrossRef]
- Manetas, Y.; Drinia, A.; Petropoulou, Y. High contents of anthocyanins in young leaves are correlated with low pools of xanthophyll cycle components and low risk of photoinhibition. Photosynthetica 2002, 40, 349–354. [Google Scholar] [CrossRef]
- Stirling, C.; Aguilera, C.; Baker, N.; Long, S. Changes in the photosynthetic light response curve during leaf development of field grown maize with implications for modelling canopy photosynthesis. Photosynth. Res. 1994, 42, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Thiele, A.; Herold, M.; Lenk, I.; Quail, P.H.; Gatz, C. Heterologous expression of arabidopsis phytochrome b in transgenic potato influences photosynthetic performance and tuber development. Plant Physiol. 1999, 120, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Pellny, T.K. Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 2003, 54, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Ensminger, I.; Busch, F.; Huner, N.P. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant. 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Ding, F.; Wang, M.; Zhang, S.; Ai, X. Changes in sbpase activity influence photosynthetic capacity, growth, and tolerance to chilling stress in transgenic tomato plants. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ölçer, H.; Lloyd, J.C.; Raines, C.A. Photosynthetic capacity is differentially affected by reductions in sedoheptulose-1, 7-bisphosphatase activity during leaf development in transgenic tobacco plants. Plant Physiol. 2001, 125, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Heyneke, E.; Fernie, A.R. Metabolic regulation of photosynthesis. Biochem. Soc. Trans. 2018, 46, 321–328. [Google Scholar] [CrossRef]
- Krahmer, J.; Ganpudi, A.; Abbas, A.; Romanowski, A.; Halliday, K.J. Phytochrome, carbon sensing, metabolism, and plant growth plasticity. Plant Physiol. 2018, 176, 1039–1048. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the tca cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, D.B.; Martins, S.C.; Cavalcanti, J.H.F.; Daloso, D.M.; Martinoia, E.; Nunes-Nesi, A.; DaMatta, F.M.; Fernie, A.R.; Araújo, W.L. Enhanced photosynthesis and growth in atquac1 knockout mutants are due to altered organic acid accumulation and an increase in both stomatal and mesophyll conductance. Plant Physiol. 2016, 170, 86–101. [Google Scholar] [CrossRef]
- Bechtold, U.; Penfold, C.A.; Jenkins, D.J.; Legaie, R.; Moore, J.D.; Lawson, T.; Matthews, J.S.; Vialet-Chabrand, S.R.; Baxter, L.; Subramaniam, S. Time-series transcriptomics reveals that agamous-like22 affects primary metabolism and developmental processes in drought-stressed arabidopsis. Plant Cell 2016, 28, 345–366. [Google Scholar] [CrossRef]
- Waters, M.T.; Langdale, J.A. The making of a chloroplast. EMBO J. 2009, 28, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E. Pifs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Juarez, M.T.; Twigg, R.W.; Timmermans, M.C. Specification of adaxial cell fate during maize leaf development. Development 2004, 131, 4533–4544. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ohashi-Ito, K.; Bergmann, D.C. Orthologs of arabidopsis thaliana stomatal bhlh genes and regulation of stomatal development in grasses. Development 2009, 136, 2265–2276. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Bian, Y.; Lv, Y.; Xie, Q. Genome-wide analysis of primary auxin-responsive aux/iaa gene family in maize (zea mays. L.). Mol. Biol. Rep. 2010, 37, 3991–4001. [Google Scholar] [CrossRef]
- Qi, S.; Lin, Q.; Feng, X.; Han, H.; Liu, J.; Zhang, L.; Wu, S.; Le, J.; Blumwald, E.; Hua, X. Idd 16 negatively regulates stomatal initiation via trans-repression of spch in arabidopsis. Plant Biotechnol. J. 2019, 17, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Hall, D.A.; Last, R.L. A small zinc finger thylakoid protein plays a role in maintenance of photosystem ii in arabidopsis thaliana. Plant Cell 2011, 23, 1861–1875. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. Wrky transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F.; de Paula, W.B.; Puthiyaveetil, S.; Nield, J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011, 16, 645–655. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Sheen, J. Involvement of maize dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998, 10, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yu, L.; Li, Z.; Yan, Y.; Han, R.; Liu, H.; Ma, M. Genome-wide analysis of the spl family transcription factors and their responses to abiotic stresses in maize. Plant Gene 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Ye, Z. A new model for relationship between irradiance and the rate of photosynthesis in oryza sativa. Photosynthetica 2007, 45, 637–640. [Google Scholar] [CrossRef]
- Guo, Y.; Gao, C.; Wang, M.; Fu, F.-F.; El-Kassaby, Y.A.; Wang, T.; Wang, G. Metabolome and transcriptome analyses reveal flavonoids biosynthesis differences in ginkgo biloba associated with environmental conditions. Ind. Crop. Prod. 2020, 158, 112963. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Wang, T.; Fu, F.-F.; El-Kassaby, Y.A.; Wang, G. Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves. Int. J. Mol. Sci. 2021, 22, 2601. https://doi.org/10.3390/ijms22052601
Guo Y, Wang T, Fu F-F, El-Kassaby YA, Wang G. Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves. International Journal of Molecular Sciences. 2021; 22(5):2601. https://doi.org/10.3390/ijms22052601
Chicago/Turabian StyleGuo, Ying, Tongli Wang, Fang-Fang Fu, Yousry A. El-Kassaby, and Guibin Wang. 2021. "Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves" International Journal of Molecular Sciences 22, no. 5: 2601. https://doi.org/10.3390/ijms22052601
APA StyleGuo, Y., Wang, T., Fu, F.-F., El-Kassaby, Y. A., & Wang, G. (2021). Metabolome and Transcriptome Analyses Reveal the Regulatory Mechanisms of Photosynthesis in Developing Ginkgo biloba Leaves. International Journal of Molecular Sciences, 22(5), 2601. https://doi.org/10.3390/ijms22052601







