Implication of a Key Region of Six Bacillus cereus Genes Involved in Siroheme Synthesis, Nitrite Reductase Production and Iron Cluster Repair in the Bacterial Response to Nitric Oxide Stress
Abstract
1. Introduction
2. Material and Method
2.1. Bacterial Strains
2.2. Growth and Stress Conditions
2.3. RNAseq Library Preparation and Data Analysis
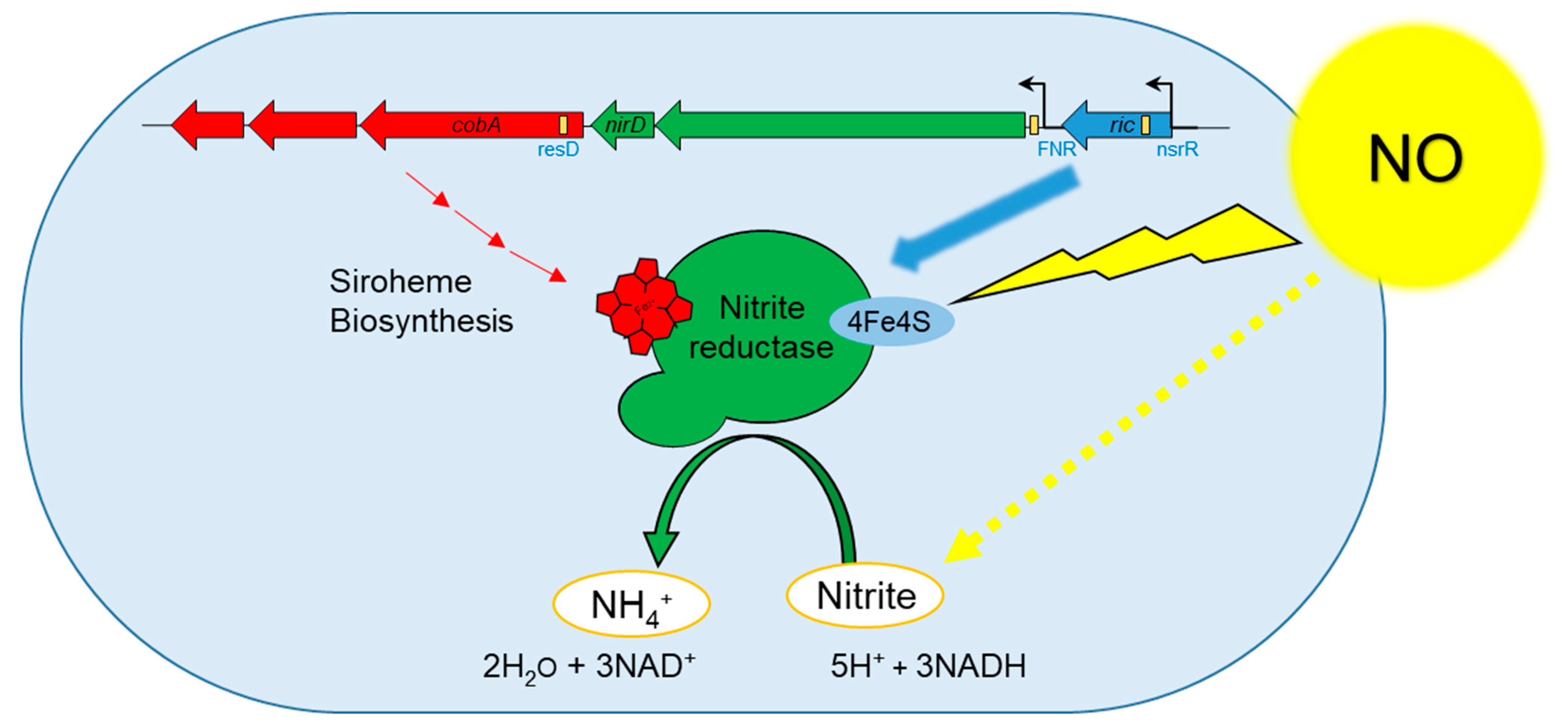
2.4. Bio-Informatic Analysis of the Six Genes Region
2.5. Reverse Transcription Quantitative PCR (RT-qPCR)
3. Results
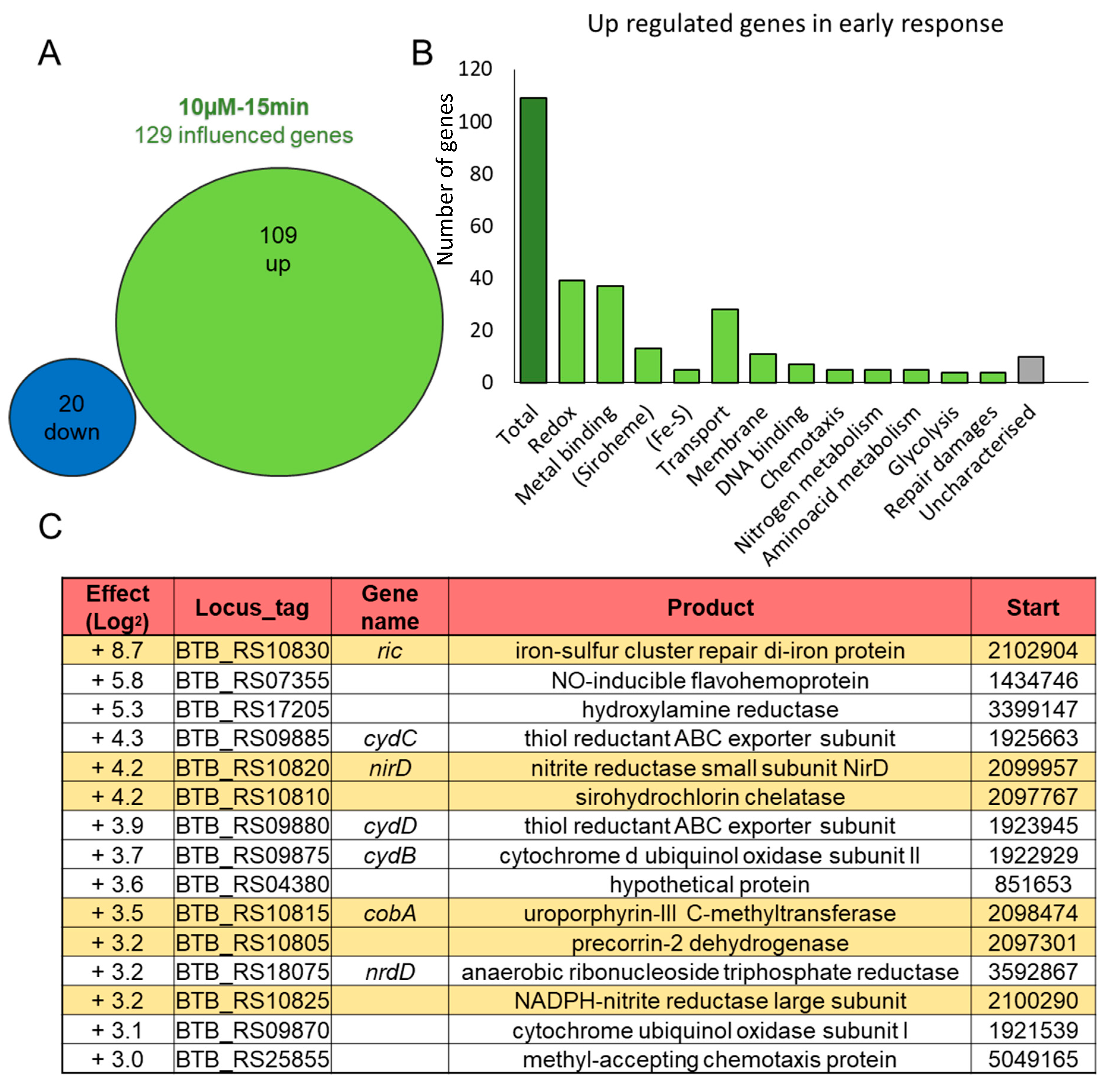
3.1. B. cereus Transcriptomic Response to NO Stress
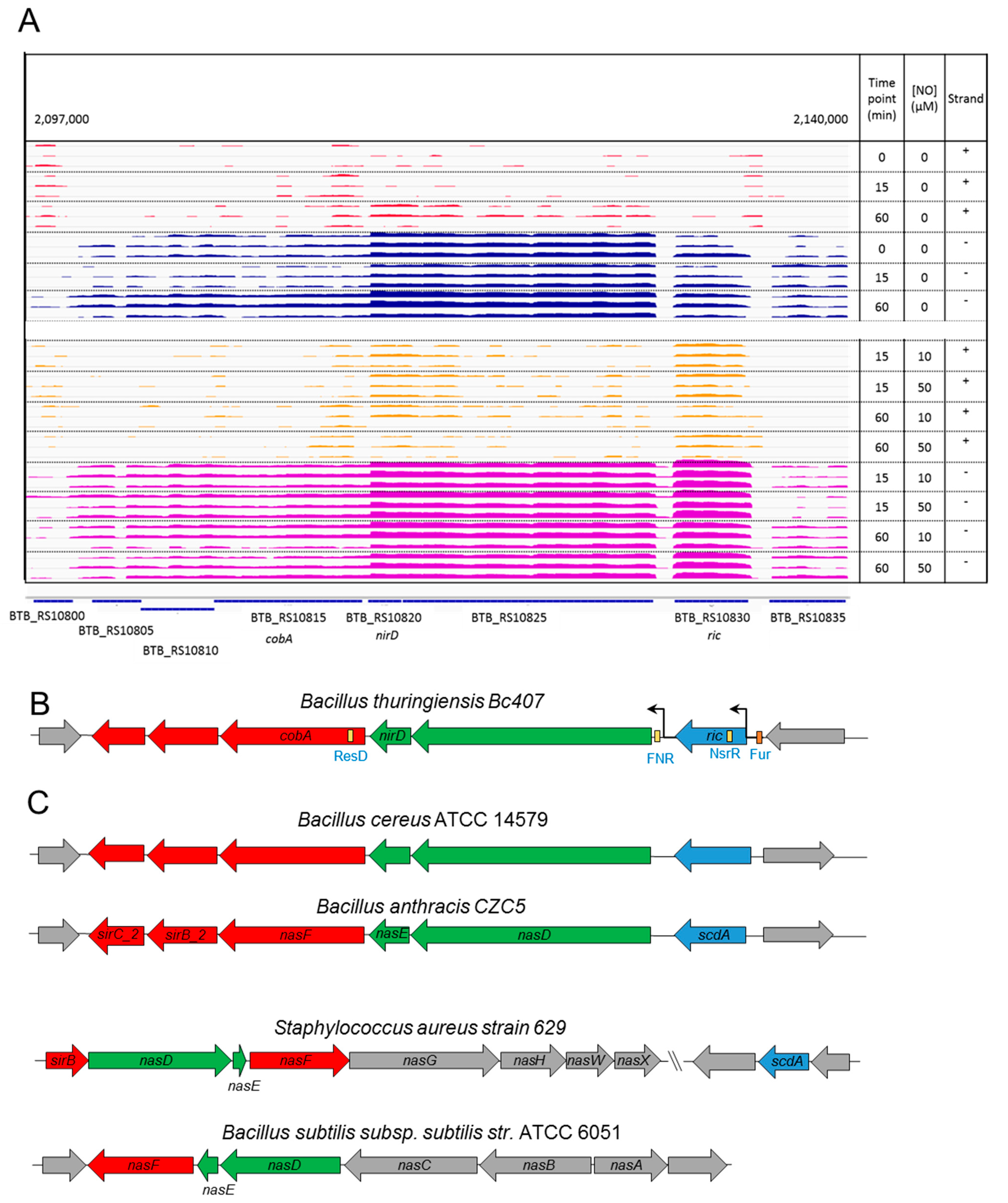
3.2. A Chromosomal Region of 6 Genes over Activated during Early Response to NO Stress
3.3. Specific Response to NO Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Porrini, C.; Ramarao, N.; Tran, S.L. Dr. NO and Mr. Toxic-the versatile role of nitric oxide. Biol. Chem. 2020, 401, 547–572. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.H.; Akuta, T.; Akaike, T. Nitric Oxide-Induced Nitrative Stress Involved in Microbial Pathogenesis. J. Pharmacol. Sci. 2005, 98, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Justino, M.C. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 2006, 257, 278–284. [Google Scholar] [CrossRef]
- Overton, T.W. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and ni-trosative damage to iron-sulfur centers. J. Bacteriol. 2008, 190, 2004–2013. [Google Scholar] [CrossRef] [PubMed]
- Darrigo, C. The Bacterial Mfd Protein Prevents DNA Damage Induced by the Host Nitrogen Immune Response in a NER-Independent but RecBC-Dependent Pathway. PLoS ONE 2016, 11, e0163321. [Google Scholar]
- Guillemet, E. The bacterial repair protein Mfd confers resistance to the host nitric-oxide response. Sci. Rep. 2016, 6, 29349. [Google Scholar] [CrossRef]
- Justino, M.C.; Vicente, J.B.; Teixeira, M.; Saraiva, L.M. New Genes Implicated in the Protection of Anaerobically Grown Escherichia coli against Nitric Oxide*. J. Biol. Chem. 2005, 280, 2636–2643. [Google Scholar] [CrossRef] [PubMed]
- Bauer, C.E.; Elsen, S.; Bird, T.H. Mechanisms for Redox Control of Gene Expression. Annu. Rev. Microbiol. 1999, 53, 495–523. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.M. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 2006, 188, 5878–5887. [Google Scholar] [CrossRef]
- Hantke, K. Regulation of ferric iron transport in Escherichia coli K12: Isolation of a constitutive mutant. Mol. Genet. Genom. 1981, 182, 288–292. [Google Scholar] [CrossRef]
- Stenfors Arnesen, L.P.; Fagerlund, A.; Granum, P.E. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 2008, 32, 579–606. [Google Scholar] [CrossRef] [PubMed]
- RamaRao, N.; Sanchis, V. The Pore-Forming Haemolysins of Bacillus Cereus: A Review. Toxins 2013, 5, 1119–1139. [Google Scholar] [CrossRef]
- Glasset, B. Large-scale survey of Bacillus cereus-induced food-borne outbreaks: Epidemiologic and genetic characterization. Euro Surveill. 2016, 21, 30413. [Google Scholar] [CrossRef] [PubMed]
- Veysseyre, F.; Fourcade, C.; Lavigne, J.-P.; Sotto, A. Bacillus cereus infection: 57 case patients and a literature review. Médecine Mal. Infect. 2015, 45, 436–440. [Google Scholar] [CrossRef]
- Ramarao, N.; Belotti, L.; Deboscker, S.; Ennahar-Vuillemin, M.; de Launay, J.; Lavigne, T.; Koebel, C.; Escande, B.; Guinebretière, M.H. Two unrelated episodes of Bacillus cereus bacteremia in a neonatal intensive care unit. Am. J. Infect. Control. 2014, 42, 694–695. [Google Scholar] [CrossRef] [PubMed]
- Glasset, B.; Herbin, S.; Granier, S.A.; Cavalié, L.; Lafeuille, E.; Guérin, C.; Ruimy, R.; Casagrande-Magne, F.; Levast, M.; Chautemps, N.; et al. Bacillus cereus, a serious cause of nosocomial infections: Epidemiologic and genetic survey. PLoS ONE 2018, 13, e0194346. [Google Scholar] [CrossRef] [PubMed]
- Cormontagne, D.; Rigourd, V.; Vidic, J.; Rizzotto, F.; Bille, E.; Ramarao, N. Bacillus cereus Induces Severe Infections in Preterm Neonates: Implication at the Hospital and Human Milk Bank Level. Toxins 2021, 13, 123. [Google Scholar] [CrossRef] [PubMed]
- Haydar, A. InhA1-Mediated Cleavage of the Metalloprotease NprA Allows Bacillus cereus to Escape from Macrophages. Front. Microbiol. 2018, 23, 1063. [Google Scholar] [CrossRef]
- Ramarao, N.; Lereclus, D. The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell. Microbiol. 2005, 7, 1357–1364. [Google Scholar] [CrossRef]
- Tran, S.-L.; Guillemet, E.; Ngo-Camus, M.; Clybouw, C.; Puhar, A.; Moris, A.; Gohar, M.; Lereclus, D.; Ramarao, N. Hemolysin II is a Bacillus cereus virulence factor that induces apoptosis of macrophages. Cell. Microbiol. 2011, 13, 92–108. [Google Scholar] [CrossRef]
- Tran, S.-L.; Puhar, A.; Ngo-Camus, M.; Ramarao, N. Trypan Blue Dye Enters Viable Cells Incubated with the Pore-Forming Toxin HlyII of Bacillus cereus. PLoS ONE 2011, 6, e22876. [Google Scholar] [CrossRef]
- Tran, S.L.; Ramarao, N. Bacillus cereus immune escape: A journey within macrophages. FEMS Microbiol. Lett. 2013, 347, 1–6. [Google Scholar] [CrossRef]
- Lereclus, D. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 1989, 51, 7–211. [Google Scholar]
- Tourasse, N.; Helgason, E.; Okstad, O.; Hegna, I.; Kolsto, A.-B. The Bacillus cereus group: Novel aspects of population structure and genome dynamics. J. Appl. Microbiol. 2006, 101, 579–593. [Google Scholar] [CrossRef] [PubMed]
- Kaes, C.; Katz, A.; Hosseini, M.W. Bipyridine: The Most Widely Used Ligand. A Review of Molecules Comprising at Least Two 2,2‘-Bipyridine Units. Chem. Rev. 2000, 100, 3553–3590. [Google Scholar] [CrossRef] [PubMed]
- Tran, S.-L.; Guillemet, E.; Lereclus, D.; Ramarao, N. Iron regulates Bacillus thuringiensis haemolysin hlyII gene expression during insect infection. J. Invertebr. Pathol. 2013, 113, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Fouquier d’Herouel, A. A simple and efficient method to search for selected primary transcripts: Non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011, 39, e46. [Google Scholar] [CrossRef]
- Cadot, C.; Tran, S.-L.; Vignaud, M.-L.; De Buyser, M.-L.; Kolstø, A.-B.; Brisabois, A.; Nguyen-Thé, C.; Lereclus, D.; Guinebretière, M.-H.; Ramarao, N. InhA1, NprA and HlyII as candidates to differentiate pathogenic from non-pathogenic Bacillus cereus strains. J. Clin. Microbiol. 2010, 48, 65–1358. [Google Scholar] [CrossRef]
- Joshi, N.; Fass, J. A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33). 2011. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Strimmer, K. A unified approach to false discovery rate estimation. BMC Bioinform. 2008, 9, 1–14. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences. In Meta-Genomics and Its Applications in Agriculture, Biomedicine and Environmental Studies; Nova Science Publishers: Haupauge, NY, USA, 2011; pp. 61–78. [Google Scholar]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Reents, H.; Münch, R.; Dammeyer, T.; Jahn, D.; Härtig, E. The Fnr Regulon of Bacillus subtilis. J. Bacteriol. 2006, 188, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Partridge, J.D.; Bodenmiller, D.M.; Humphrys, M.S.; Spiro, S. NsrR targets in the Escherichia coli genome: New insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009, 73, 94–680. [Google Scholar] [CrossRef] [PubMed]
- Chumsakul, O.; Anantsri, D.P.; Quirke, T.; Oshima, T.; Nakamura, K.; Ishikawa, S.; Nakano, M.M. Genome-Wide Analysis of ResD, NsrR, and Fur Binding in Bacillus subtilis during Anaerobic Fermentative Growth by In Vivo Footprinting. J. Bacteriol. 2017, 199, e00086-17. [Google Scholar] [CrossRef]
- Harvie, D.R.; Vílchez, S.; Steggles, J.R.; Ellar, D.J. Bacillus cereus Fur regulates iron metabolism and is required for full virulence. Microbiology 2005, 151, 569–577. [Google Scholar] [CrossRef]
- Glasset, B.; Sperry, M.; Dervyn, R.; Herbin, S.; Brisabois, A.; Ramarao, N. The cytotoxic potential of Bacillus cereus strains of various origins. Food Microbiol. 2021, 98, 103759. [Google Scholar] [CrossRef]
- Pacher, P.; Beckman, J.S.; Liaudet, L. Nitric Oxide and Peroxynitrite in Health and Disease. Physiol. Rev. 2007, 87, 315–424. [Google Scholar] [CrossRef]
- Carter, C.W., Jr. Structure of the iron-sulfur cluster in the Chromatius iron protein at 2.25 Angstrom resolution. Cold Spring Harb. Symp. Quant. Biol. 1972, 36, 5–381. [Google Scholar] [CrossRef]
- Fitzpatrick, J.; Kim, E. Synthetic Modeling Chemistry of Iron–Sulfur Clusters in Nitric Oxide Signaling. Acc. Chem. Res. 2015, 48, 2453–2461. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.R.; Megson, I.L. Non-Heme Iron Nitrosyls in Biology. Chem. Rev. 2002, 102, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Kinkel, T.L.; Roux, C.M.; Dunman, P.M.; Fang, F.C. The Staphylococcus aureus SrrAB Two-Component System Promotes Resistance to Nitrosative Stress and Hypoxia. mBio 2013, 4, e00696-13. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gunsalus, R.P. The nrfA and nirB Nitrite Reductase Operons in Escherichia coli Are Expressed Differently in Response to Nitrate than to Nitrite. J. Bacteriol. 2000, 182, 5813–5822. [Google Scholar] [CrossRef] [PubMed]
- Colandene, J.D.; Garrett, R.H. Functional Dissection and Site-directed Mutagenesis of the Structural Gene for NAD(P)H-Nitrite Reductase in Neurospora crassa. J. Biol. Chem. 1996, 271, 24096–24104. [Google Scholar] [CrossRef]
- Green, J. Activation of FNR-dependent transcription by iron: An in vitro switch for FNR. FEMS Microbiol. Lett. 1993, 113, 22–219. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.B.; Stolowich, N.J.; Roessner, C.A.; Scott, A. The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 1993, 335, 57–60. [Google Scholar] [CrossRef]
- Dietl, A.-M.; Binder, U.; Shadkchan, Y.; Osherov, N.; Haas, H. Siroheme Is Essential for Assimilation of Nitrate and Sulfate as Well as Detoxification of Nitric Oxide but Dispensable for Murine Virulence of Aspergillus fumigatus. Front. Microbiol. 2018, 9, 2615. [Google Scholar] [CrossRef] [PubMed]
- Cammack, R.; Jackson, R.H.; Cornish-Bowden, A.; Cole, J.A. Electron-spin-resonance studies of the NADH-dependent nitrite reductase from Escherichia coli K12. Biochem. J. 1982, 207, 333–339. [Google Scholar] [CrossRef]



| Regulators | Consensus Sequences | Sources |
|---|---|---|
| FNR | TGTGANNNNNNTCACA | [37] |
| NsrR | AANATGCATTT | [38] |
| ResD | TNTNANNANTNTNTGACAANT | [39] |
| Fur | GATAATGATAATCATTCT | [40] |
| Locus Tag | Primer Forward | Primer Reverse |
|---|---|---|
| BTB_RS10830 | GAAAATGAACATAACCACGCTG | TGTAAACAAGTCGATAAGTGCC |
| BTB_RS10825 | GACCAAATACGCAAATAGCAAG | TTCATAAAGAGGGGCAACAAG |
| BTB_RS10820 | TCATAAAAACGGACCATTAGCC | TGTCACCATCAATGACTTCTAC |
| BTB_RS10815 | CGACAAGCAGGAAAAAAAGAAG | GTAATGAAACGACATCACCAAC |
| BTB_RS10805 | AGAATCATCGTTTCACACACC | GTTCCTGCTTCATACGCTTC |
| BTB_RS00795 | TAACTCCTTACGTCGTATTC | ATTTCCAACGTCTTCTCTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porrini, C.; Guérin, C.; Tran, S.-L.; Dervyn, R.; Nicolas, P.; Ramarao, N. Implication of a Key Region of Six Bacillus cereus Genes Involved in Siroheme Synthesis, Nitrite Reductase Production and Iron Cluster Repair in the Bacterial Response to Nitric Oxide Stress. Int. J. Mol. Sci. 2021, 22, 5079. https://doi.org/10.3390/ijms22105079
Porrini C, Guérin C, Tran S-L, Dervyn R, Nicolas P, Ramarao N. Implication of a Key Region of Six Bacillus cereus Genes Involved in Siroheme Synthesis, Nitrite Reductase Production and Iron Cluster Repair in the Bacterial Response to Nitric Oxide Stress. International Journal of Molecular Sciences. 2021; 22(10):5079. https://doi.org/10.3390/ijms22105079
Chicago/Turabian StylePorrini, Constance, Cyprien Guérin, Seav-Ly Tran, Rozenn Dervyn, Pierre Nicolas, and Nalini Ramarao. 2021. "Implication of a Key Region of Six Bacillus cereus Genes Involved in Siroheme Synthesis, Nitrite Reductase Production and Iron Cluster Repair in the Bacterial Response to Nitric Oxide Stress" International Journal of Molecular Sciences 22, no. 10: 5079. https://doi.org/10.3390/ijms22105079
APA StylePorrini, C., Guérin, C., Tran, S.-L., Dervyn, R., Nicolas, P., & Ramarao, N. (2021). Implication of a Key Region of Six Bacillus cereus Genes Involved in Siroheme Synthesis, Nitrite Reductase Production and Iron Cluster Repair in the Bacterial Response to Nitric Oxide Stress. International Journal of Molecular Sciences, 22(10), 5079. https://doi.org/10.3390/ijms22105079







