Remodeler Catalyzed Nucleosome Repositioning: Influence of Structure and Stability
Abstract
1. Introduction
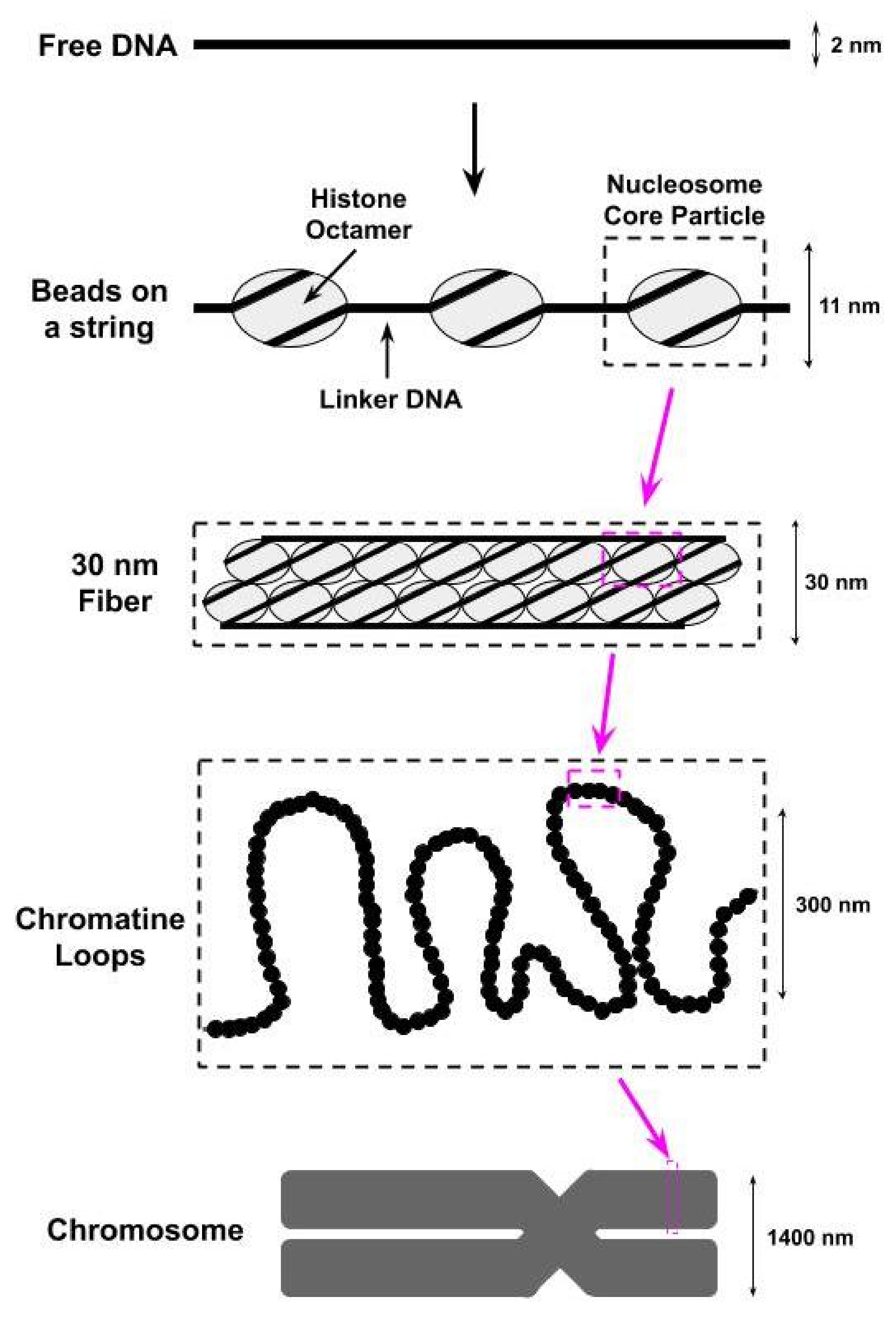
2. Structure
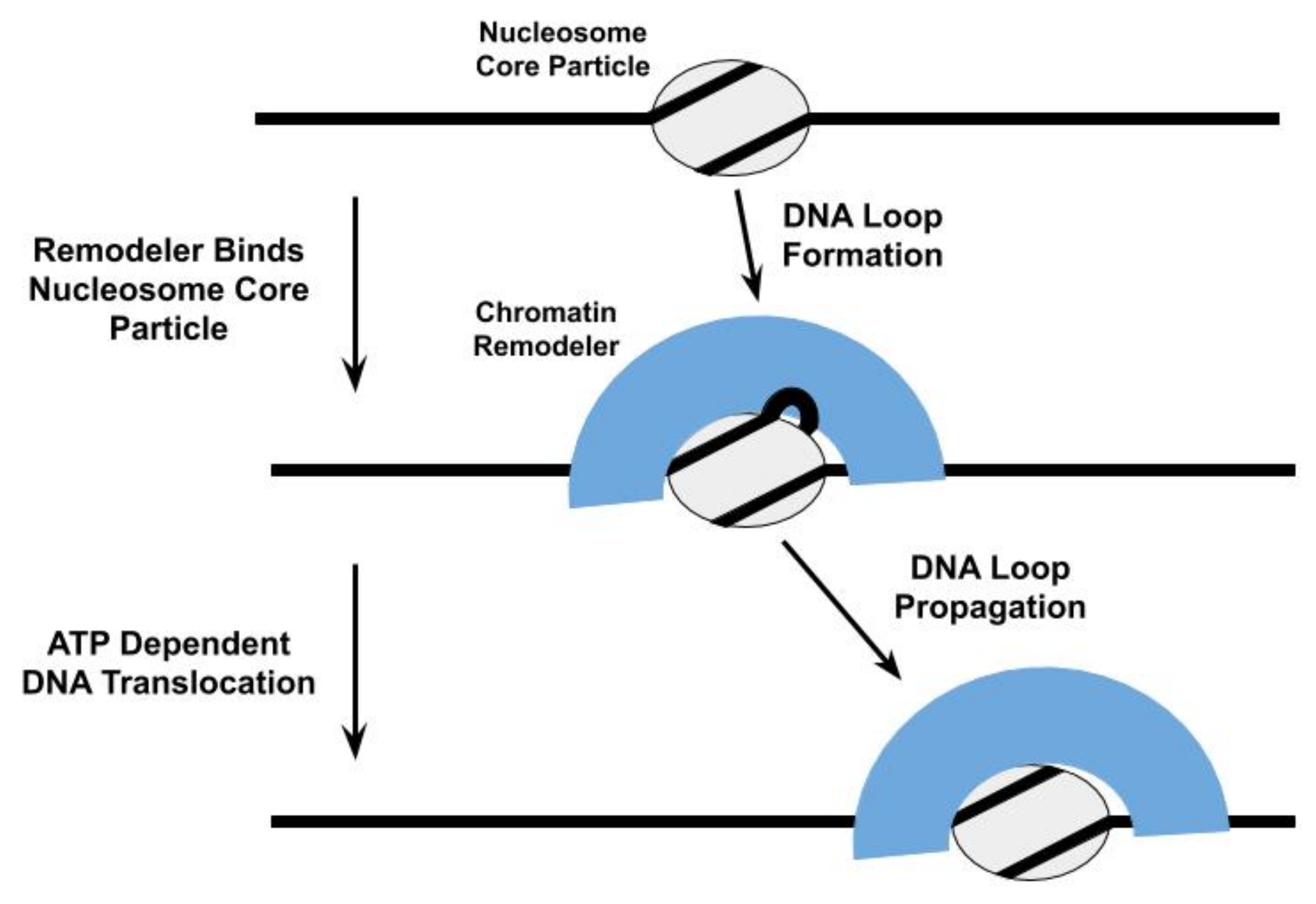
3. Mechanisms of Nucleosome Repositioning
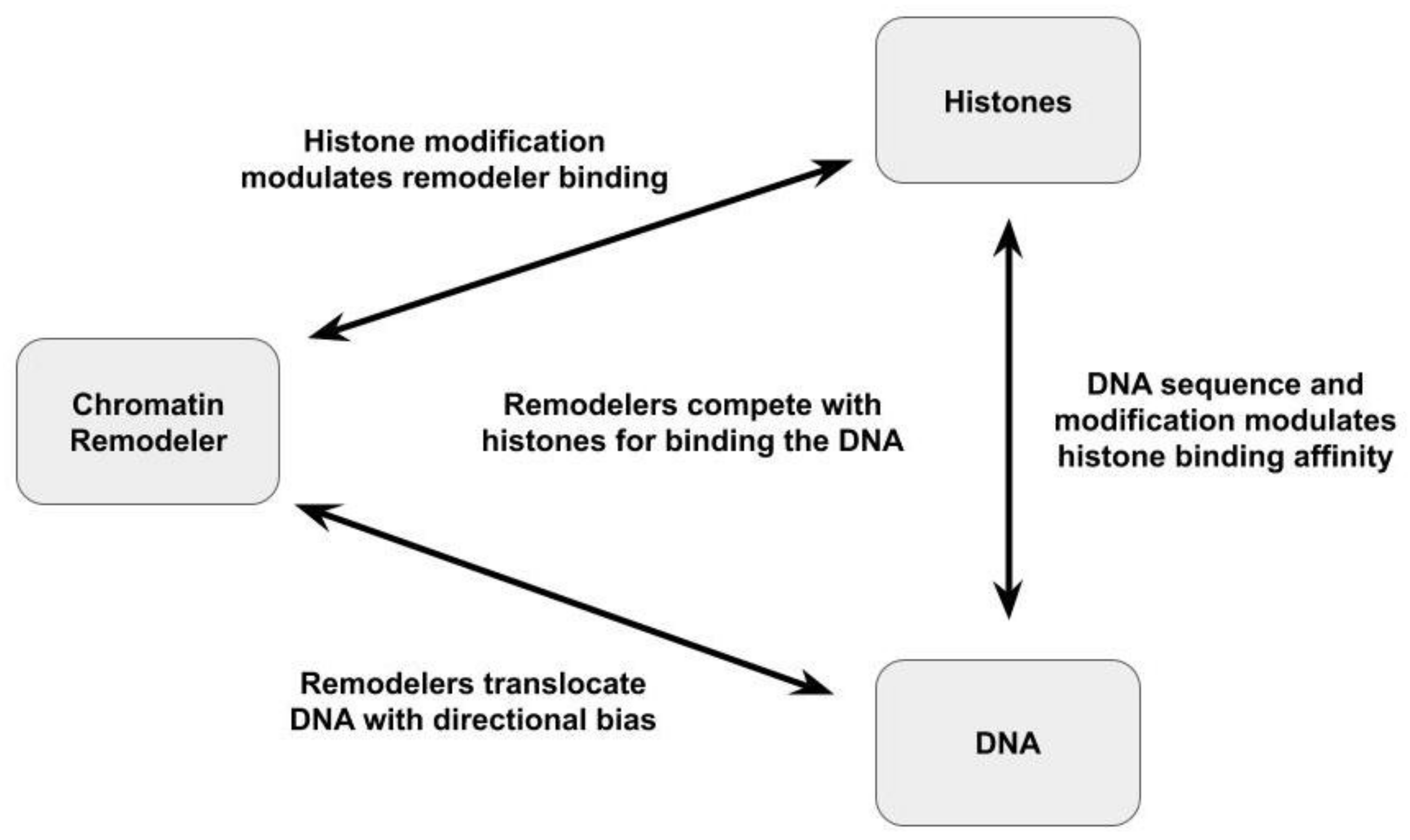
4. Effects of Substrate Modification On NCP Binding and Mobilization
5. Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 angstrom resolution. Nature 1997, 389, 251. [Google Scholar] [CrossRef] [PubMed]
- Davey, C.A.; Sargent, D.F.; Luger, K.; Maeder, A.W.; Richmond, T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002, 319, 1097–1113. [Google Scholar] [CrossRef]
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145. [Google Scholar] [CrossRef] [PubMed]
- Frouws, T.D.; Duda, S.C.; Richmond, T.J. X-ray structure of the MMTV-A nucleosome core. Proc. Natl. Acad. Sci. USA 2016, 113, 1214–1219. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Luger, K.; Tan, S.; Richmond, T.J. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. USA 1996, 93, 1370–1375. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Tan, S. Nucleosome structure and function. Chem. Rev. 2015, 115, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Szerlong, H.J.; Hansen, J.C. Nucleosome distribution and linker DNA: connecting nuclear function to dynamic chromatin structure. Biochem. Cell Biol. 2011, 89, 24–34. [Google Scholar] [CrossRef]
- Dann, G.P.; Liszczak, G.P.; Bagert, J.D.; Müller, M.M.; Nguyen, U.T.; Wojcik, F.; Brown, Z.Z.; Bos, J.; Panchenko, T.; Pihl, R.; et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548, 607–611. [Google Scholar] [CrossRef]
- Lowary, P.; Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998, 276, 19–42. [Google Scholar] [CrossRef]
- Thåström, A.; Lowary, P.; Widom, J. Measurement of histone–DNA interaction free energy in nucleosomes. Methods 2004, 33, 33–44. [Google Scholar] [CrossRef]
- Battistini, F.; Hunter, C.A.; Moore, I.K.; Widom, J. Structure-based identification of new high-affinity nucleosome binding sequences. J. Mol. Biol. 2012, 420, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.T.; Yoo, J.; Dai, Q.; Zhang, Q.; He, C.; Aksimentiev, A.; Ha, T. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tóth, K.; Böhm, V.; Sellmann, C.; Danner, M.; Hanne, J.; Berg, M.; Barz, I.; Gansen, A.; Langowski, J. Histone-and DNA sequence-dependent stability of nucleosomes studied by single-pair FRET. Cytom. Part A 2013, 83, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Widom, J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001, 34, 269–324. [Google Scholar] [CrossRef] [PubMed]
- Polach, K.; Widom, J. Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995, 254, 130–149. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Widom, J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000, 296, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Widom, J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004, 11, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Azmi, I.F.; Watanabe, S.; Maloney, M.F.; Kang, S.; Belsky, J.A.; MacAlpine, D.M.; Peterson, C.L.; Bell, S.P. Nucleosomes influence multiple steps during replication initiation. eLife 2017, 6, e22512. [Google Scholar] [CrossRef]
- Kadonaga, J.T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 1998, 92, 307–313. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Bowman, G.D. Mechanisms of ATP-dependent nucleosome sliding. Curr. Opin. Struct. Biol. 2010, 20, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Eisen, J.A.; Sweder, K.S.; Hanawalt, P.C. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995, 23, 2715–2723. [Google Scholar] [CrossRef] [PubMed]
- Lusser, A.; Kadonaga, J.T. Chromatin remodeling by ATP-dependent molecular machines. Bioessays 2003, 25, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Imam, N.; Verma, K.; Patel, A.K. Chromatin remodelers: We are the drivers! Nucleus 2016, 7, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Markert, J.; Luger, K. Nucleosomes Meet Their Remodeler Match. Trends Biochem. Sci. 2020. [Google Scholar] [CrossRef]
- Peng, A.Y.T.; Kolhe, J.A.; Behrens, L.D.; Freeman, B.C. Genome organization: Tag it, move it, place it. Current Opinion in Cell Biology 2020, 68, 90–97. [Google Scholar] [CrossRef]
- Mueller-Planitz, F.; Klinker, H.; Becker, P.B. Nucleosome sliding mechanisms: new twists in a looped history. Nat. Struct. Mol. Biol. 2013, 20, 1026–1032. [Google Scholar] [CrossRef]
- Singleton, M.R.; Dillingham, M.S.; Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007, 76, 23–50. [Google Scholar] [CrossRef]
- Marmorstein, R.; Berger, S.L. Structure and function of bromodomains in chromatin-regulating complexes. Gene 2001, 272, 1–9. [Google Scholar] [CrossRef]
- Olave, I.A.; Reck-Peterson, S.L.; Crabtree, G.R. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 2002, 71, 755–781. [Google Scholar] [CrossRef] [PubMed]
- Szerlong, H.; Saha, A.; Cairns, B.R. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003, 22, 3175–3187. [Google Scholar] [CrossRef] [PubMed]
- Zofall, M.; Persinger, J.; Bartholomew, B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol. Cell. Biol. 2004, 24, 10047–10057. [Google Scholar] [CrossRef] [PubMed]
- Petty, E.; Pillus, L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013, 29, 621–629. [Google Scholar] [CrossRef]
- Wang, S.S.; Zhou, B.O.; Zhou, J.Q. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol. Cell. Biol. 2011, 31, 3171–3181. [Google Scholar] [CrossRef][Green Version]
- Becker, P.B.; Hörz, W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 2002, 71, 247–273. [Google Scholar] [CrossRef]
- Cairns, B.R.; Lorch, Y.; Li, Y.; Zhang, M.; Lacomis, L.; Erdjument-Bromage, H.; Tempst, P.; Du, J.; Laurent, B.; Kornberg, R.D. RSC, an essential, abundant chromatin-remodeling complex. Cell 1996, 87, 1249–1260. [Google Scholar] [CrossRef]
- Toto, M.; D’Angelo, G.; Corona, D.F. Regulation of ISWI chromatin remodelling activity. Chromosoma 2014, 123, 91–102. [Google Scholar] [CrossRef]
- Tran, H.G.; Steger, D.J.; Iyer, V.R.; Johnson, A.D. The chromo domain protein Chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000, 19, 2323–2331. [Google Scholar] [CrossRef]
- Ebbert, R.; Birkmann, A.; Schüller, H.J. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 1999, 32, 741–751. [Google Scholar] [CrossRef]
- Klopf, E.; Paskova, L.; Solé, C.; Mas, G.; Petryshyn, A.; Posas, F.; Wintersberger, U.; Ammerer, G.; Schüller, C. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2009, 29, 4994–5007. [Google Scholar] [CrossRef] [PubMed]
- Varga-Weisz, P.D.; Wilm, M.; Bonte, E.; Dumas, K.; Mann, M.; Becker, P.B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature 1997, 388, 598–602. [Google Scholar] [CrossRef]
- Vary, J.C., Jr.; Gangaraju, V.K.; Qin, J.; Landel, C.C.; Kooperberg, C.; Bartholomew, B.; Tsukiyama, T. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 2003, 23, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Jónsson, Z.O.; Jha, S.; Wohlschlegel, J.A.; Dutta, A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 2004, 16, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Bouazoune, K.; Brehm, A. ATP-dependent chromatin remodeling complexes in Drosophila. Chromosome Res. 2006, 14, 433–449. [Google Scholar] [CrossRef]
- Hall, J.A.; Georgel, P.T. CHD proteins: A diverse family with strong ties. Biochem. Cell Biol. 2007, 85, 463–476. [Google Scholar] [CrossRef]
- Marfella, C.G.; Imbalzano, A.N. The Chd family of chromatin remodelers. Mutat. Res. Mol. Mech. Mutagen. 2007, 618, 30–40. [Google Scholar] [CrossRef]
- Chittori, S.; Hong, J.; Bai, Y.; Subramaniam, S. Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome. Nucleic Acids Res. 2019, 47, 9400–9409. [Google Scholar] [CrossRef]
- Yan, L.; Wu, H.; Li, X.; Gao, N.; Chen, Z. Structures of the ISWI–nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 2019, 26, 258–266. [Google Scholar] [CrossRef]
- Al-Ani, G.; Briggs, K.; Malik, S.S.; Conner, M.; Azuma, Y.; Fischer, C.J. Quantitative determination of binding of ISWI to nucleosomes and DNA shows allosteric regulation of DNA binding by nucleotides. Biochemistry 2014, 53, 4334–4345. [Google Scholar] [CrossRef]
- Hota, S.K.; Bhardwaj, S.K.; Deindl, S.; Lin, Y.c.; Zhuang, X.; Bartholomew, B. Nucleosome mobilization by ISW2 requires the concerted action of the ATPase and SLIDE domains. Nat. Struct. Mol. Biol. 2013, 20, 222. [Google Scholar] [CrossRef]
- Liu, X.; Li, M.; Xia, X.; Li, X.; Chen, Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 2017, 544, 440–445. [Google Scholar] [CrossRef]
- Li, M.; Xia, X.; Tian, Y.; Jia, Q.; Liu, X.; Lu, Y.; Li, M.; Li, X.; Chen, Z. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 2019, 567, 409–413. [Google Scholar] [CrossRef]
- Han, Y.; Reyes, A.A.; Malik, S.; He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 2020, 579, 452–455. [Google Scholar] [CrossRef]
- Lohman, T.M.; Hsieh, J.; Maluf, N.K.; Cheng, W.; Lucius, A.L.; Fischer, C.J.; Brendza, K.M.; Korolev, S.; Waksman, G. DNA helicases, motors that move along nucleic acids: lessons from the SF1 helicase superfamily. In The Enzymes; Elsevier: Amsterdam, The Netherlands, 2003; Volume 23, pp. 303–311. [Google Scholar]
- Fischer, C.J.; Wooten, L.; Tomko, E.J.; Lohman, T.M. Kinetics of motor protein translocation on single-stranded DNA. In Helicases; Springer: Berlin/Heidelberg, Germany, 2009; pp. 45–56. [Google Scholar]
- Blosser, T.R.; Yang, J.G.; Stone, M.D.; Narlikar, G.J.; Zhuang, X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature 2009, 462, 1022. [Google Scholar] [CrossRef]
- Racki, L.R.; Yang, J.G.; Naber, N.; Partensky, P.D.; Acevedo, A.; Purcell, T.J.; Cooke, R.; Cheng, Y.; Narlikar, G.J. The chromatin remodeler ACF acts as a dimeric motor to space nucleosomes. Nature 2009, 462, 1016. [Google Scholar] [CrossRef]
- Brehm, A.; Längst, G.; Kehle, J.; Clapier, C.R.; Imhof, A.; Eberharter, A.; Müller, J.; Becker, P.B. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 2000, 19, 4332–4341. [Google Scholar] [CrossRef]
- Wagner, F.R.; Dienemann, C.; Wang, H.; Stützer, A.; Tegunov, D.; Urlaub, H.; Cramer, P. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 2020, 579, 448–451. [Google Scholar] [CrossRef]
- Farnung, L.; Vos, S.M.; Wigge, C.; Cramer, P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature 2017, 550, 539–542. [Google Scholar] [CrossRef]
- Armache, J.P.; Gamarra, N.; Johnson, S.L.; Leonard, J.D.; Wu, S.; Narlikar, G.J.; Cheng, Y. Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome. Elife 2019, 8, e46057. [Google Scholar] [CrossRef]
- Patel, A.B.; Moore, C.M.; Greber, B.J.; Luo, J.; Zukin, S.A.; Ranish, J.; Nogales, E. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. Elife 2019, 8, e54449. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef]
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002, 16, 2120–2134. [Google Scholar] [CrossRef]
- Whitehouse, I.; Stockdale, C.; Flaus, A.; Szczelkun, M.D.; Owen-Hughes, T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 2003, 23, 1935–1945. [Google Scholar] [CrossRef]
- Zhang, Y.; Smith, C.L.; Saha, A.; Grill, S.W.; Mihardja, S.; Smith, S.B.; Cairns, B.R.; Peterson, C.L.; Bustamante, C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell 2006, 24, 559–568. [Google Scholar] [CrossRef]
- Lia, G.; Indrieri, M.; Owen-Hughes, T.; Finzi, L.; Podesta, A.; Milani, P.; Dunlap, D. ATP-dependent looping of DNA by ISWI. J. Biophotonics 2008, 1, 280–286. [Google Scholar] [CrossRef]
- Eastlund, A.; Malik, S.S.; Fischer, C.J. Kinetic mechanism of DNA translocation by the RSC molecular motor. Arch. Biochem. Biophys. 2013, 532, 73–83. [Google Scholar] [CrossRef][Green Version]
- Eastlund, A.; Al-Ani, G.; Fischer, C.J. Low processivity for DNA translocation by the ISWI molecular motor. Biochim. Biophys. Acta-(Bba)-Proteins Proteom. 2015, 1854, 1487–1493. [Google Scholar] [CrossRef]
- Côté, J.; Peterson, C.L.; Workman, J.L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. USA 1998, 95, 4947–4952. [Google Scholar] [CrossRef]
- Dürr, H.; Körner, C.; Müller, M.; Hickmann, V.; Hopfner, K.P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell 2005, 121, 363–373. [Google Scholar] [CrossRef]
- Mueller-Planitz, F.; Klinker, H.; Ludwigsen, J.; Becker, P.B. The ATPase domain of ISWI is an autonomous nucleosome remodeling machine. Nat. Struct. Mol. Biol. 2013, 20, 82–89. [Google Scholar] [CrossRef]
- Kim, D.E.; Narayan, M.; Patel, S.S. T7 DNA helicase: a molecular motor that processively and unidirectionally translocates along single-stranded DNA. J. Mol. Biol. 2002, 321, 807–819. [Google Scholar] [CrossRef]
- Khaki, A.R.; Field, C.; Malik, S.; Niedziela-Majka, A.; Leavitt, S.A.; Wang, R.; Hung, M.; Sakowicz, R.; Brendza, K.M.; Fischer, C.J. The macroscopic rate of nucleic acid translocation by hepatitis C virus helicase NS3h is dependent on both sugar and base moieties. J. Mol. Biol. 2010, 400, 354–378. [Google Scholar] [CrossRef]
- Tomko, E.J.; Fischer, C.J.; Lohman, T.M. Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J. Mol. Biol. 2012, 418, 32–46. [Google Scholar] [CrossRef][Green Version]
- Sirinakis, G.; Clapier, C.R.; Gao, Y.; Viswanathan, R.; Cairns, B.R.; Zhang, Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011, 30, 2364–2372. [Google Scholar] [CrossRef]
- Malik, S.S.; Rich, E.; Viswanathan, R.; Cairns, B.R.; Fischer, C.J. Allosteric interactions of DNA and nucleotides with S. cerevisiae RSC. Biochemistry 2011, 50, 7881–7890. [Google Scholar] [CrossRef]
- Cairns, B.R.; Erdjument-Bromage, H.; Tempst, P.; Winston, F.; Kornberg, R.D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 1998, 2, 639–651. [Google Scholar] [CrossRef]
- Fischer, C.J.; Maluf, N.K.; Lohman, T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004, 344, 1287–1309. [Google Scholar] [CrossRef]
- Brendza, K.M.; Cheng, W.; Fischer, C.J.; Chesnik, M.A.; Niedziela-Majka, A.; Lohman, T.M. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. USA 2005, 102, 10076–10081. [Google Scholar] [CrossRef]
- Eberharter, A.; Vetter, I.; Ferreira, R.; Becker, P.B. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD–histone contacts. EMBO J. 2004, 23, 4029–4039. [Google Scholar] [CrossRef]
- Strohner, R.; Wachsmuth, M.; Dachauer, K.; Mazurkiewicz, J.; Hochstatter, J.; Rippe, K.; Längst, G. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat. Struct. Mol. Biol. 2005, 12, 683. [Google Scholar] [CrossRef]
- Zofall, M.; Persinger, J.; Kassabov, S.R.; Bartholomew, B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006, 13, 339–346. [Google Scholar] [CrossRef]
- Deindl, S.; Hwang, W.L.; Hota, S.K.; Blosser, T.R.; Prasad, P.; Bartholomew, B.; Zhuang, X. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 2013, 152, 442–452. [Google Scholar] [CrossRef]
- Dechassa, M.L.; Zhang, B.; Horowitz-Scherer, R.; Persinger, J.; Woodcock, C.L.; Peterson, C.L.; Bartholomew, B. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 2008, 28, 6010–6021. [Google Scholar] [CrossRef]
- Lorch, Y.; Maier-Davis, B.; Kornberg, R.D. Mechanism of chromatin remodeling. Proc. Natl. Acad. Sci. USA 2010, 107, 3458–3462. [Google Scholar] [CrossRef]
- Saha, A.; Wittmeyer, J.; Cairns, B.R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 2005, 12, 747–755. [Google Scholar] [CrossRef]
- Kulić, I.; Schiessel, H. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys. Rev. Lett. 2003, 91, 148103. [Google Scholar] [CrossRef]
- Brandani, G.B.; Takada, S. Chromatin remodelers couple inchworm motion with twist-defect formation to slide nucleosomal DNA. PLoS Comput. Biol. 2018, 14, e1006512. [Google Scholar] [CrossRef]
- Fischer, C.J.; Saha, A.; Cairns, B.R. Kinetic model for the ATP-dependent translocation of Saccharomyces cerevisiae RSC along double-stranded DNA. Biochemistry 2007, 46, 12416–12426. [Google Scholar] [CrossRef]
- Al-Ani, G.; Malik, S.S.; Eastlund, A.; Briggs, K.; Fischer, C.J. ISWI remodels nucleosomes through a random walk. Biochemistry 2014, 53, 4346–4357. [Google Scholar] [CrossRef]
- Morgan, A.M.; LeGresley, S.E.; Briggs, K.; Al-Ani, G.; Fischer, C.J. Effects of nucleosome stability on remodeler-catalyzed repositioning. Phys. Rev. E 2018, 97, 032422. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature 2012, 492, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Ludwigsen, J.; Klinker, H.; Mueller-Planitz, F. No need for a power stroke in ISWI-mediated nucleosome sliding. EMBO Rep. 2013, 14, 1092–1097. [Google Scholar] [CrossRef] [PubMed]
- Udugama, M.; Sabri, A.; Bartholomew, B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol. 2011, 31, 662–673. [Google Scholar] [CrossRef]
- Fyodorov, D.V.; Kadonaga, J.T. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 2002, 418, 896–900. [Google Scholar] [CrossRef]
- He, X.; Fan, H.Y.; Narlikar, G.J.; Kingston, R.E. Human ACF1 alters the remodeling strategy of SNF2h. J. Biol. Chem. 2006, 281, 28636–28647. [Google Scholar] [CrossRef]
- Partensky, P.D.; Narlikar, G.J. Chromatin remodelers act globally, sequence positions nucleosomes locally. J. Mol. Biol. 2009, 391, 12–25. [Google Scholar] [CrossRef]
- Rippe, K.; Schrader, A.; Riede, P.; Strohner, R.; Lehmann, E.; Längst, G. DNA sequence-and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl. Acad. Sci. USA 2007, 104, 15635–15640. [Google Scholar] [CrossRef]
- Manelyte, L.; Strohner, R.; Gross, T.; Längst, G. Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin remodeling complex NoRC. PLoS Genet. 2014, 10, e1004157. [Google Scholar] [CrossRef]
- Winger, J.; Bowman, G.D. The Sequence of Nucleosomal DNA Modulates Sliding by the Chd1 Chromatin Remodeler. J. Mol. Biol. 2017, 429, 808–822. [Google Scholar] [CrossRef]
- Lequieu, J.; Schwartz, D.C.; de Pablo, J.J. In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. USA 2017, 114, E9197–E9205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Johnson, S.L.; Lee, L.J.; Longhurst, A.D.; Beckwith, S.L.; Johnson, M.J.; Morrison, A.J.; Narlikar, G.J. The yeast INO80 complex operates as a tunable DNA length-sensitive switch to regulate nucleosome sliding. Mol. Cell 2018, 69, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Blossey, R.; Schiessel, H. Kinetic proofreading of gene activation by chromatin remodeling. HFSP J. 2008, 2, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J. A proposal for kinetic proof reading by ISWI family chromatin remodeling motors. Curr. Opin. Chem. Biol. 2010, 14, 660–665. [Google Scholar] [CrossRef][Green Version]
- Blossey, R.; Schiessel, H. Histone mark recognition controls nucleosome translocation via a kinetic proofreading mechanism: Confronting theory and high-throughput experiments. Phys. Rev. E 2019, 99, 060401. [Google Scholar] [CrossRef]
- Gregory, P.D.; Wagner, K.; Hörz, W. Histone acetylation and chromatin remodeling. Exp. Cell Res. 2001, 265, 195–202. [Google Scholar] [CrossRef]
- Clapier, C.R.; Nightingale, K.P.; Becker, P.B. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002, 30, 649–655. [Google Scholar] [CrossRef]
- Längst, G.; Becker, P.B. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta-(Bba)-Gene Struct. Expr. 2004, 1677, 58–63. [Google Scholar] [CrossRef]
- Grunstein, M. Histone acetylation in chromatin structure and transcription. Nature 1997, 389, 349–352. [Google Scholar] [CrossRef]
- Turner, B.M. Histone acetylation and an epigenetic code. Bioessays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Kouzarides, T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar] [CrossRef]
- Bannister, A.J.; Schneider, R.; Kouzarides, T. Histone methylation: dynamic or static? Cell 2002, 109, 801–806. [Google Scholar] [CrossRef]
- Hamiche, A.; Kang, J.G.; Dennis, C.; Xiao, H.; Wu, C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. USA 2001, 98, 14316–14321. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Längst, G.; Corona, D.F.; Becker, P.B.; Nightingale, K.P. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 2001, 21, 875–883. [Google Scholar] [CrossRef]
- Dang, W.; Kagalwala, M.N.; Bartholomew, B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol. Cell. Biol. 2006, 26, 7388–7396. [Google Scholar] [CrossRef]
- Ferreira, H.; Flaus, A.; Owen-Hughes, T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J. Mol. Biol. 2007, 374, 563–579. [Google Scholar] [CrossRef]
- Racki, L.R.; Naber, N.; Pate, E.; Leonard, J.D.; Cooke, R.; Narlikar, G.J. The histone H4 tail regulates the conformation of the ATP-binding pocket in the SNF2h chromatin remodeling enzyme. J. Mol. Biol. 2014, 426, 2034–2044. [Google Scholar] [CrossRef][Green Version]
- Makowski, M.; Grawe, C.; Foster, B.; Nguyen, N.; Bartke, T.; Michiel, V. Global profiling of protein–DNA and protein–nucleosome binding affinities using quantitative mass spectrometry. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Barbera, A.J.; Chodaparambil, J.V.; Kelley-Clarke, B.; Joukov, V.; Walter, J.C.; Luger, K.; Kaye, K.M. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 2006, 311, 856–861. [Google Scholar] [CrossRef]
- Kalashnikova, A.A.; Porter-Goff, M.E.; Muthurajan, U.M.; Luger, K.; Hansen, J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 2013, 10, 20121022. [Google Scholar] [CrossRef]
- McGinty, R.K.; Tan, S. Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 2016, 37, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Dao, H.T.; Dul, B.E.; Dann, G.P.; Liszczak, G.P.; Muir, T.W. A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol. 2020, 16, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Gamarra, N.; Johnson, S.L.; Trnka, M.J.; Burlingame, A.L.; Narlikar, G.J. The nucleosomal acidic patch relieves auto-inhibition by the ISWI remodeler SNF2h. Elife 2018, 7, e35322. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Shen, X. Chromatin Remodeling: INO80 and SWR1. Cell 2011, 144, 158–158.e2. [Google Scholar] [CrossRef]
- Turinetto, V.; Giachino, C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015, 43, 2489–2498. [Google Scholar] [CrossRef]
- Rudnizky, S.; Bavly, A.; Malik, O.; Pnueli, L.; Melamed, P.; Kaplan, A. H2A. Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Giaimo, B.D.; Ferrante, F.; Herchenröther, A.; Hake, S.B.; Borggrefe, T. The histone variant H2A. Z in gene regulation. Epigenetics Chromatin 2019, 12, 1–22. [Google Scholar] [CrossRef]
- Hada, A.; Hota, S.K.; Luo, J.; Lin, Y.C.; Kale, S.; Shaytan, A.K.; Bhardwaj, S.K.; Persinger, J.; Ranish, J.; Panchenko, A.R.; et al. Histone Octamer Structure Is Altered Early in ISW2 ATP-Dependent Nucleosome Remodeling. Cell Rep. 2019, 28, 282–294. [Google Scholar] [CrossRef]
- Moshkin, Y.M.; Chalkley, G.E.; Kan, T.W.; Reddy, B.A.; Ozgur, Z.; van Ijcken, W.F.; Dekkers, D.H.; Demmers, J.A.; Travers, A.A.; Verrijzer, C.P. Remodelers organize cellular chromatin by counteracting intrinsic histone-DNA sequence preferences in a class-specific manner. Mol. Cell. Biol. 2012, 32, 675–688. [Google Scholar] [CrossRef]
- Smeenk, G.; Wiegant, W.W.; Vrolijk, H.; Solari, A.P.; Pastink, A.; van Attikum, H. The NuRD chromatin–remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010, 190, 741–749. [Google Scholar] [CrossRef]
- Polo, S.E.; Kaidi, A.; Baskcomb, L.; Galanty, Y.; Jackson, S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010, 29, 3130–3139. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E. Reshaping chromatin after DNA damage: The choreography of histone proteins. J. Mol. Biol. 2015, 427, 626–636. [Google Scholar] [CrossRef] [PubMed]
- LeGresley, S.E.; Wilt, J.; Antonik, M. DNA damage may drive nucleosomal reorganization to facilitate damage detection. Phys. Rev. E 2014, 89, 032708. [Google Scholar] [CrossRef] [PubMed]

| Subfamily | Principle Activity | Additional Domains | Domain Function |
|---|---|---|---|
| ISWI | Nucleosome assembly and spacing Transcription regulation | HAND, SANT, and SLIDE | DNA and nucleosome binding |
| SWI/SNF | Transcription regulation | HSA BROMO | Binds nuclear actin-related proteins Recognizes acetylated histones |
| CHD | Interacts with promoter DNA sequences Transcription regulation | CHROMO | Recognizes methylated histones |
| INO80 | Inositol-responsive gene expression Deposition of histone variant H2AZ | HSA | Binds actin and actin-related proteins |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morgan, A.; LeGresley, S.; Fischer, C. Remodeler Catalyzed Nucleosome Repositioning: Influence of Structure and Stability. Int. J. Mol. Sci. 2021, 22, 76. https://doi.org/10.3390/ijms22010076
Morgan A, LeGresley S, Fischer C. Remodeler Catalyzed Nucleosome Repositioning: Influence of Structure and Stability. International Journal of Molecular Sciences. 2021; 22(1):76. https://doi.org/10.3390/ijms22010076
Chicago/Turabian StyleMorgan, Aaron, Sarah LeGresley, and Christopher Fischer. 2021. "Remodeler Catalyzed Nucleosome Repositioning: Influence of Structure and Stability" International Journal of Molecular Sciences 22, no. 1: 76. https://doi.org/10.3390/ijms22010076
APA StyleMorgan, A., LeGresley, S., & Fischer, C. (2021). Remodeler Catalyzed Nucleosome Repositioning: Influence of Structure and Stability. International Journal of Molecular Sciences, 22(1), 76. https://doi.org/10.3390/ijms22010076





