Abstract
Polycomb repressive complexes (PRCs) are epigenetic regulators that mediate repressive histone modifications. PRCs play a pivotal role in the maintenance of hematopoietic stem cells through repression of target genes involved in cell proliferation and differentiation. Next-generation sequencing technologies have revealed that various hematologic malignancies harbor mutations in PRC2 genes, such as EZH2, EED, and SUZ12, and PRC1.1 genes, such as BCOR and BCORL1. Except for the activating EZH2 mutations detected in lymphoma, most of these mutations compromise PRC function and are frequently associated with resistance to chemotherapeutic agents and poor prognosis. Recent studies have shown that mutations in PRC genes are druggable targets. Several PRC2 inhibitors, including EZH2-specific inhibitors and EZH1 and EZH2 dual inhibitors have shown therapeutic efficacy for tumors with and without activating EZH2 mutations. Moreover, EZH2 loss-of-function mutations appear to be attractive therapeutic targets for implementing the concept of synthetic lethality. Further understanding of the epigenetic dysregulation associated with PRCs in hematological malignancies should improve treatment outcomes.
1. Introduction
Polycomb repressive complexes (PRCs) are composed of polycomb group (PcG) proteins and are epigenetic regulators that mediate repressive histone modifications [1]. There are two major types of PRCs, PRC1 and PRC2. Canonical PRC1 consists of polycomb group ring finger (PCGF) 4/BMI1 or PCGF2/MEL18, RING1A or RING1B, CBX, and PHC. RING1A/B is a ubiquitin ligase (E3 ligase), and PRC1 monoubiquitinates histone H2A at lysine residue 119 (H2AK119ub1). PRC2 mainly consists of EZH1/2, EED, SUZ12, and RBBP4/7. EZH1/2 has specific histone methylation activity and mediates the mono-, di-, and trimethylation of H3 at lysine 27 (H3K27me1/me2/me3) (Figure 1) [2]. Generally, PRC2 is recruited to target gene loci, which typically contains non-methylated CpG islands. Then, PRC1 binds to H3K27me3 through one of its components, CBX, and monoubiquitinates H2AK119. These histone modifications inhibit transcription and induce chromatin aggregation, which maintains transcriptional repression of target genes. Recently, several variant PRC1 complexes were identified that can monoubiquitinate H2AK119 independent of PRC2 or H3K27me3. Among them, PRC 1.1, comprising PCGF1, RING1A/B, BCOR or BCORL1, and KDM2B, is of particular interest because of its distinct functions in hematopoietic stem cells and hematologic malignancies [3].
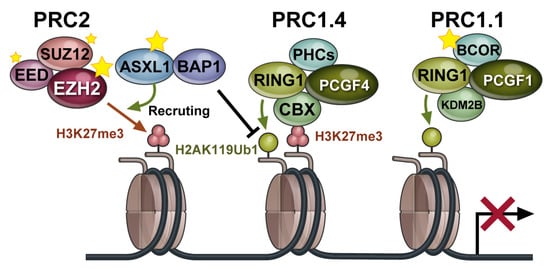
Figure 1.
Polycomb repressive complexes (PRCs) are multiprotein complexes that play important roles in the repression of target genes through chromatin modifications. PRC components that are targeted by somatic gene mutations are indicated by stars. Black T arrow indicates inhibition of ubiquitination at H2K119.
PRCs have been shown to function in many aspects of hematopoiesis, and next-generation sequencing has revealed gain-of-function and loss-of-function mutations in PRCs and related genes. In this review, we present the current knowledge on PRC aberrations in hematologic malignancies and their prognostic significance, as well as potential therapeutic approaches targeting PRCs.
2. Physiological Functions of PRCs in Hematopoiesis
Among the various PRC components, BMI1 (also known as PCGF4) has been extensively investigated in hematopoiesis. In Bmi1 deficient mice, Cdkn2a (Ink4a/Arf), which encodes Ink4a and Arf tumor suppressors, was de-repressed [4]. As a result, Bmi1 knockout mice showed a significantly lower frequency of hematopoietic stem cells (HSCs) due to impaired self-renewal [5]. In addition, the forced expression of Bmi1 enhanced the self-renewal of HSCs [6]. Bmi1 also suppresses the commitment and differentiation of HSCs into B cells by repressing Ebf1 and Pax5, which encode key transcription factors that are essential for B-cell differentiation. Consequently, Bmi1-deficient mice showed premature expression of Ebf1 and Pax5 in HSCs and multipotent progenitors (MPPs), which was accompanied by accelerated lymphoid specification and a marked reduction in HSC/MPPs [7]. Bmi1 is also required to maintain leukemic stem cells as well as normal HSCs [8].
The functions of EZH1 and EZH2 in hematopoiesis have also been well characterized. Ezh2-deficient HSCs retained their reconstitution capacity [9] because Ezh1 can compensate for Ezh2 [10,11]. Conditional deletion of Ezh1 in adult mice impaired the self-renewal capacity of HSCs through de-repression of Cdkn2a [12], while HSCs in Ezh1-null mice retained almost normal repopulation capacity. Ezh1 and Ezh2 double knockout mice completely lost repopulation capacity [10]. With respect to other PRC2 components, loss of Eed impaired differentiation of HSCs and led to HSC exhaustion [13]. Suz12 has also been shown to be required for the maintenance of HSC [14].
Several studies have revealed the physiological roles of PRC1.1 in hematopoiesis. KDM2B binds to nonmethylated CpG islands through its zinc finger-CxxC (ZF-CxxC) DNA-binding domain, thereby recruiting other components of PRC1.1. Kdm2b-deficient mice had significantly decreased numbers of hematopoietic stem and progenitor cells (HSPCs) [15] which showed impaired repopulation of hematopoietic cells, particularly lymphoid cells [15,16]. In contrast, forced expression of Kdm2b prevented exhaustion of the long-term repopulating potential of HSCs following serial transplantation [17]. BCOR, a co-repressor of BCL6, played an important role in restricting differentiation toward the myeloid lineage, partly by repressing Cebp and HoxA family genes. As a consequence, Bcor knockout mice showed myeloid-skewed differentiation [18,19]. Depletion of PCGF1 also led to myeloid-skewing [20] and de-repressed expression of HoxA family genes [21]. Taken altogether, these data show that PRC1.1 regulates the functions of HSCs and restricts their differentiation toward the myeloid lineage by repressing the transcription of genes required for myeloid differentiation, such as C/EBP and HOXA family genes.
3. Functions of PRCs in Hematologic Malignancies
Dysregulated function of epigenetic regulators is frequently involved in the pathogenesis of solid and hematological malignancies. PRCs play a pivotal role in the maintenance of HSCs and hematopoiesis, and dysregulation of PRC function has been implicated in the pathogenesis of hematological malignancies. Overexpression of PcG genes generally promotes tumorigenesis, partly through their ability to transcriptionally repress tumor suppressor genes, such as the CDKN2A locus (encoding p16INK4A and p14ARF), and developmental regulator genes [22]. The expression of BMI1/PCGF4 has correlated with disease progression and the prognosis of myelodysplastic syndrome (MDS) [23], the prognoses of acute myeloid leukemia (AML) and chronic myeloid leukemia [24]. In contrast, loss-of-function mutations in PRC genes, such as EZH2, EED, SUZ12, BCOR, and BCORL1, have been found in various hematological malignancies, suggesting their tumor suppressor functions (Table 1) [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51]. Therefore, PcG genes may have dual roles as oncogenes and tumor suppressor genes, depending on the situation and underlying disease. Among the observed PcG gene mutations, genetic abnormalities in EZH2 and BCOR are of particular interest owing to their relatively high frequencies, pathological significance, and potential as therapeutic targets.

Table 1.
Frequencies of mutations in PRC genes in hematologic malignancies.
3.1. EZH2
EZH2 is frequently overexpressed and/or amplified in prostate, breast, bladder, and colon cancers [52], and its expression is correlated with metastasis [53] and poor prognosis [54,55]. We and other groups have shown that Ezh2 exerts an oncogenic function during the maintenance phase of MLL-AF9 AML in mice and could be therapeutically targeted. In contrast, Ezh2 acts as a tumor suppressor during the induction phase of AML [56,57,58]. EZH2 is strongly expressed in germinal center (GC) B cells and acts with BCL6 to recruit a noncanonical PRC1-BCOR complex containing CBX8 in a GC B-cell–specific manner to repress the expression of differentiation genes [59]. Correspondingly, gain-of-function mutations in EZH2 are frequently found in GC B-cell–type lymphoma [47] in which H3K27me3 levels are significantly elevated [47]. Mutant EZH2 contributes to lymphomagenesis partly by repressing Prdm1 and/or Irf4, which are essential for B-cell differentiation [60,61,62]. Mutant EZH2 not only increases the abundance of H3K27me3 but also causes the widespread redistribution of this repressive mark, suggesting that mutant EZH2 induces persistent repression and aberrant activation of EZH2 target genes [62].
Loss-of-function mutations in EZH2 are frequently found in patients with MDS and myeloproliferative neoplasms (MPN). Although EZH2 mutations are rare in de novo AML patients, they are frequently found in patients with secondary AML transformed from MDS [31]. Abnormalities of chromosome 7, including -7 and -7q, are frequently found in patients with MDS, and they involve EZH2, which is located in the long arm of chromosome 7 (7q36). In patients with MDS, EZH2 mutations are associated with poor prognosis when compared to that of MDS patients with wild-type EZH2. Furthermore, patients with bi-allelic EZH2 mutations showed worse overall survival than those with mono-allelic EZH2 mutations [47]. In MDS and AML, EZH2 expression is also regulated by mutations in spliceosome genes, such as SRSF2 and U2AF1, which cause mis-splicing of EZH2, leading to degradation of variant EZH2 mRNAs via nonsense-mediated decay [63,64]. EZH2 mutations frequently co-occur with TET2, RUNX1, and ASXL1 mutations [63]. We have demonstrated that loss of Ezh2 cooperated with a Tet2 hypomorph or a RUNX1 mutant to induce MDS and MDS/MPN in mice [10,65,66]. The deletion of Ezh2 in mice conferred a growth advantage to HSCs and promoted myeloid-biased repopulation. The deletion of Ezh2 resulted in an MDS/MPN-like disease with myeloproliferative features such as the enhanced repopulating capacity of HSCs and extramedullary hematopoiesis in the spleen, and myelodysplastic features such as anemia accompanied by enhanced apoptosis in erythroblasts and dysplasia in myeloid cells. Concurrent depletion of Ezh2 and Tet2 established more advanced myelodysplasia and markedly accelerated the development of myelodysplastic disorders, including both MDS and MDS/MPN [9,57].
These findings clearly indicate that EZH2 plays a tumor-suppressive role in myelodysplastic disorders and enhances transformation triggered by other driver mutations. In combination with the RUNX1S291fs mutant, loss of Ezh2 also promoted development of MDS in mice. In this MDS model, we found that RUNX1S291fs/Ezh2-null cells compromise normal HSPCs function [65,66]. In MDS patients, mutant cells propagate in the bone marrow (BM) despite impaired proliferative capacity and are thought to eventually outcompete normal hematopoietic cells. RUNX1S291fs/Ezh2-null MDS cells appeared to be defective in proliferative capacity. Of interest, inflammatory cytokine pathways such as IL-6 pathway, which are inhibitory towards HSC functions were significantly activated in residing normal HSPCs, but not in RUNX1S291fs/Ezh2-null HSPCs. Expression of IL-6 was significantly elevated in RUNX1S291fs/Ezh2-null BM cells compared with Rx291 BM cells. Excessive IL-6 production by MDS cells has also been shown to promote leukopenia and thrombocytosis in a 5q- MDS mouse model [67]. These findings suggest that RUNX1S291fs/Ezh2-null cells outcompete normal cells by creating an inflammatory BM environment, thereby achieving clonal expansion despite their compromised proliferative capacity.
Essential tumor suppressor and developmental regulator genes remain transcriptionally repressed even in EZH2-insufficient cells [10,65]. HSPCs are maintained in mice with only one Ezh1 allele (that lack Ezh2), while deletion of both Ezh1 and Ezh2 leads to the depletion of HSPCs. Residual PRC2 in mice with only one Ezh1 allele preferentially targeted genes with high levels of H3K27me3 and H2AK119 monoubiquitination (H2AK119ub1) in HSPCs. We designated these genes as Ezh1 core target genes. Most Ezh1 core target genes were bivalent developmental regulators. These results indicate that Ezh1 targets bivalent genes to maintain self-renewing stem cells in EZH2-insufficient MDS [11].
Ezh2 insufficiency in mouse HSPCs leads to a switch from H3K27me3 to acetylation (H3K27ac) at promoter regions of many PRC2 target genes, which is closely associated with increased transcription of a subset of direct polycomb targets, including those with bivalent promoters [56,65,66,68,69,70,71]. The absence of Ezh2 induces activation of a cohort of fetal-specific genes, including let-7 target genes [71]. Many of the fetal-specific let-7 target genes, including Lin28, Hmga2, and Igfbp3, are known as oncofetal genes, and are targeted by Ezh2-mediated H3K27me3 in adult BM HSPCs. Ezh2 insufficiency also activates the production of inflammatory cytokines and proteins, such as interleukin-6 (IL-6), S100a8, and S100a9. In mouse models, a loss of Ezh2 induces a feto-oncogenic program that includes genes such as Plag1, whose overexpression phenocopies a loss of Ezh2 to accelerate induction of AML [56].
Loss-of-function mutations in EZH2 and SUZ12 also have been found in early T-cell precursor (ETP) acute lymphoblastic leukemia (ETP-ALL) and non-ETP T-ALL in children and T-ALL in adults (Table 1). Experiments in several mouse models have demonstrated that Ezh2-deficient mice develop T-ALL [10,72]. Of note, oncogenic NOTCH1 mutations are frequently found in T-ALL and induce the loss of H3K27me3 modifications by antagonizing the functions of PRC2, thus activating the transcription of NOTCH1 targets [39]. We also generated an ETP-ALL mouse model by deleting Ezh2 in p53-null hematopoietic cells. In this ETP-ALL model, a large portion of the PRC2 target genes acquired DNA hypermethylation at their promoters following the loss of Ezh2, which included pivotal T cell differentiation–regulating genes. Thus, PRC2 protects key T cell developmental regulators from DNA hypermethylation to keep them primed for activation during subsequent differentiation phases, while PRC2 insufficiency predisposes ETPs to leukemic transformation [73]. In another mouse model of ETP-ALL, inactivation of PRC2 also caused derepression of Il6ra, resulting in activation of JAK/STAT signaling [74]. Mutations in EZH2 frequently co-occur with inactivating mutations in RUNX1 in ETP-ALL. Combined deletion of Ezh2 and Runx1 in ETPs induced their expansion. The addition of a RAS-signaling pathway mutation (Flt3-ITD) resulted in an aggressive ETP-like leukemia co-expressing myeloid and lymphoid genes [75].
In some cases, mutations in genes encoding histone H3K27 reduce levels of H3K27me3, even in the absence of loss-of-function EZH2 mutations, leading to tumor development. These mutations, initially found in pediatric brain tumors, cause amino acid substitution such as K to I/M and induce a global reduction in H3K27me3 levels by binding to EZH2 and strongly trapping PRC2, despite the fact that only one copy of the 30-copy histone H3 gene has the mutation [76]. Recently, it was reported that there are cases with these mutations in AML as well, although the frequency of H3K27I/M mutations has been found very low, at 4 out of 434 patients (0.9%) [77]. Furthermore, in AML mouse models, H3K27I/M mutations have been reported to be a potent disease promoter in the RUNX1-RUNX1T1 AML mouse model [78].
3.2. BCOR and BCORL1
Two of the noncanonical PRC1.1 genes, BCOR and its homolog BCORL1, are both X-linked and are frequently mutated in patients with various hematological malignancies (Table 1). These are inactivating mutations, and they often co-occur with mutations in DNMT3A, TET2, RUNX1, and STAG2 [34,36,79]. We demonstrated that a Bcor mutant that lacks the PCGF1-binding domain combines with Tet2 loss to induce lethal MDS in mice. These MDS cells reproduced MDS or evolved into lethal MDS/myeloproliferative neoplasms in secondary recipients. These findings indicate that BCOR mutations promote the initiation and progression of MDS in concert with concurrent driver mutations [19]. Transcriptional profiling revealed the de-repression of myeloid regulator genes such as Cebp and HoxA family genes, in HSPCs. The loss of BCOR induced reductions in H2AK119ub1 levels at many promoter regions of PRC1 targets, including Hoxa7, Hoxa9, and Cebpa [19]. This Bcor mutant also combines with an oncogenic Kras mutation, KrasG12D, to initiate a fully transplantable acute leukemia [80]. These findings indicate that BCOR mutations promote the initiation and progression of MDS and acute leukemia in concert with concurrent driver mutations, indicating its function as a tumor suppressor. Of interest, Bcor-insufficient MDS cells do not show activation of Ezh2 target genes, such as let-7 target genes and inflammatory genes described above [19]. These results suggest that PRC1.1 insufficiency is involved in the pathogenesis of MDS in a manner that differs from PRC2 insufficiency (Figure 2).
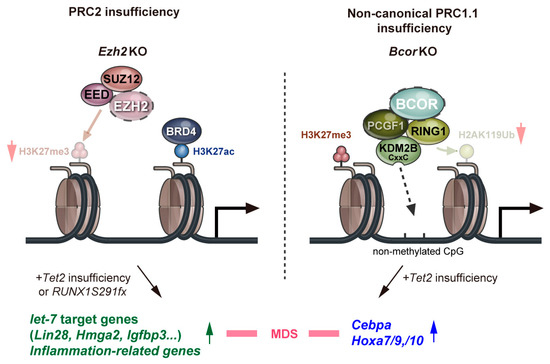
Figure 2.
A model for roles of polycomb repressive complexes in the pathogenesis of MDS. Red arrows indicate a decrease in the levels of H3K27 trimethylation and H2K119 ubiquitination. Green and blue arrows indicate increased expression levels of the respective genes.
Although inactivating mutations in BCOR and BCORL1 are not frequent in T-ALL, mice with both Bcor insufficiency and Kdm2b insufficiency show a strong propensity to develop T-ALL, mostly in a Notch-dependent manner [16,19,81]. BCOR targets many NOTCH1 targets, including Myc, and antagonizes their transcriptional activation [16,81]. A Bcor mutant lacking the PCGF1-binding domain also promoted B-cell lymphomagenesis in Eμ-Myc mice [82].
As described above, BCOR also functions as a corepressor of BCL6 and contributes to the formation of a noncanonical PRC1/BCOR complex containing CBX8 in GC B cells, thereby linking BCL6 to EZH2-containing PRC2. The interaction between BCL6-noncanonical PRC1 and BCOR-PRC2 led to the silencing of B-cell differentiation and cell cycle checkpoint genes to permit immunoglobulin affinity maturation. Thus, excessive gene silencing by this machinery is involved in lymphomagenesis [59]. PRC1.1 component genes are significantly overexpressed in primary AML CD34+ cells, and downmodulation of these components strongly reduced cell proliferation in vitro and attenuated leukemogenesis in humanized xenograft mouse models [83]. Transgenic mice overexpressing Kdm2b developed myeloid or B-lymphoid leukemia [84]. These findings suggest that noncanonical PRC1.1 also has a context-specific oncogenic function.
3.3. ASXL1
ASXL1 is one of the homologs of the additional sex comb gene, which functions as a chromatin-binding scaffold protein for epigenetic regulators. ASXL1 regulates polycomb functions and trithorax in Drosophila [85]. The loss of ASXL1 results in global reductions in H3K27me3 levels, suggesting that ASXL1 regulates the recruitment of PRC2 to their target loci [86,87]. ASXL1 also forms a complex with the deubiquitination enzyme BAP1, and removes monoubiquitin from H2AK119Ub, to de-repress genes targeted by PRC1 [88]. ASXL1 is targeted by deletions or somatic mutations in patients with hematologic malignancies, including MDS (14–23%), AML (5–17%), CMML (40–49%), and primary myelofibrosis (13–32%) [89]. ASXL1 mutations are associated with poor prognosis in many hematologic tumor patients [90,91,92,93]. Recent studies using mice expressing an ASXL1 mutant, which generated a carboxy-terminal truncated ASXL1 protein, demonstrated that an ASXL1 mutation alone was not sufficient for inducing the development of hematologic malignancies, but increased the susceptibility to leukemogenesis in concert with a RUNX1 mutant or in viral insertional mutagenesis, indicating that mice expressing an ASXL1 mutant represent a premalignant condition [94].
4. PRC as a Therapeutic Target
As mentioned above, both loss-of-function and gain-of-function mutations in EZH2 are found in hematological malignancies. Although these mutations are thought to be involved in tumor development in various ways, a comprehensive understanding is lacking, and treatment challenges remain. Several PRC2 inhibitors targeting the oncogenic function of EZH2 have been developed and are currently in preclinical and clinical studies.
4.1. EZH2 Inhibitors
Several EZH2-specific inhibitors have been developed [95] and their safety and efficacy have been evaluated in clinical trials (Table 2) [96,97,98,99]. Except for two clinical trials (NCT03456726, NCT03213665), most of them included patients without EZH2 mutations. Tazemetostat is the most widely studied EZH2-specific inhibitor, and it was the first to be approved by the FDA (2020) for advanced epithelioid sarcoma. The efficacy of tazemetostat for the treatment of malignant lymphoma has been validated in phase I and II studies [96,97], and a phase II study of tazemetostat showed high efficacy for follicular lymphoma with both wild-type and mutant EZH2 mutations [97]. In this phase II study, the objective response rate was 69% (31 of 45 patients) in the cohort with EZH2 mutation and 35% (19 of 54 patients) in those with wild type EZH2. Serious treatment-related adverse events were reported only in 4 out of 99 patients (4%). As tazemetostat was well tolerated and the frequency of serious side effects was low, it is a promising treatment for relapsed and refractory follicular lymphoma.

Table 2.
Clinical trials of PRC2 inhibitors for hematologic malignancies.
The efficacy of tazemetostat has also been tested in combination with other drugs. For instance, attempts have been made to add tazemetostat to RCHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone), which is the standard of care for diffuse large B-cell lymphoma (DLBCL) (NCT02889523) [98]. EZH2 inhibitors and prednisolone were shown to have synergistic anti-tumor activity in a DLBCL mouse model [100]. In addition, the combination of EZH2 inhibitors with immune checkpoint inhibitors appeared to be effective for the treatment of solid tumor-bearing mice. Mechanistically, it has been proposed that EZH2-mediated gene silencing of chemokines, such as CXCL9 and CXCL10, prevents effector T-cell trafficking to the tumor microenvironment, thereby allowing tumor cells to evade immunity [101]. In light of these basic research results, attempts have been made to combine EZH2 inhibitors with immune checkpoint inhibitors, such as antibodies against PD-L1 or CTLA-4, in the treatment of solid tumors (NCT04407741, NCT03525795). This type of combination therapy may be applicable to the treatment of hematological malignancies.
Inhibitors targeting both EZH1 and EZH2 (EZH1/2 dual inhibitors) and EED (EED inhibitor) have also been developed [102,103,104], and these inhibitors show better tumor-eradicating effects than EZH2-specific inhibitors in the treatment of AML, multiple myeloma, and EZH2 inhibitor-resistant DLBCL cells [105,106,107,108,109,110]. EED stabilizes and activates PRC2 to maintain H3K27 methylation for gene silencing. EED inhibitors are also attracting attention as a potentially promising agent as well as EZH2 inhibitors. One of the EED inhibitors, MAK-683, is currently under clinical trial for the patients with advanced malignancies including DLBCL (NCT02900651).
4.2. Hypomethylating Agents
Hypomethylating agents such as azacitidine and decitabine are clinically used in the treatment of high-risk MDS. One of the indications of the hypomethylating agent for MDS treatment is that MDS cells have higher levels of DNA hyper-methylation than de novo AML cells [111]. Azacitidine has been shown to improve the overall survival of patients with high-risk MDS [112]. However, it is not always the case that the mutations of epigenetic modifiers that affect the levels of DNA methylation such as TET2, IDH1/2, and DNMT3A predict the response to the hypomethylating agents [113]. Although MDS harboring TP53 mutations have been reported to temporarily respond better to hypomethylating agent [113], there are few reports that it leads to improved survival. The prediction of response to hypomethylating agents and which cases will obtain the benefits most remain to be elucidated.
The efficacy of the hypomethylating agents for hematologic malignancies harboring EZH2 mutations also remains unclear due to the relatively small number of cases. However, it has been well documented that PRC2 target genes, particularly bivalent genes marked by H3K4me3 and H3K27me3, defined in embryonic stem cells as well as hematopoietic stem and progenitor cells preferentially display DNA hyper-methylation in tumor cells, such as colon cancer, AML, and MDS, regardless of genetic mutations in epigenetic modifiers [114,115]. In addition, a functional link between DNA methylation and PRC2 has been implicated in the regulation of the biological processes of development, differentiation, and cancer [116,117]. We showed that an epigenetic switch from H3K27me3 to DNA methylation occurs at a significant portion of PRC2 targets following the loss of Ezh2 in Tet2 hypomorph (Tet2KD/KDEzh2Δ/Δ) mice, thereby maintaining their transcriptional repression to promote the progression of MDS [118]. Although Ezh2 and Ezh1 targets both underwent DNA hypermethylation in a similar manner, Ezh2 targets gained higher levels of DNA methylation than those of Ezh1 targets, indicating that Ezh2 targets are the major targets of the epigenetic switch in Tet2KD/KDEzh2Δ/Δ MDS cells. The gene silencing of Ezh1 targets was regulated mainly by sustained H3K27me3 and only partly by DNA methylation. An epigenetic switch from H3K27me3 to DNA methylation was also evident in the ETP-ALL mouse model established by Ezh2 loss in p53-null hematopoietic cells [73]. In these mice, a large portion of PRC2 target genes acquired DNA hypermethylation of their promoters following reductions in H3K27me3 levels upon the loss of Ezh2. The epigenetic switch was accentuated at the promoter regions of pivotal transcriptional regulator genes in Ezh2/p53-deficient ETP-ALL cells, the silencing of which impedes the proper ensemble of transcription factors for T cell development. Of note, the treatment of Ezh2/p53-deficient ETP-ALL cells with decitabine resulted in the terminal differentiation of ETP-ALL cells following the reactivation of a set of genes encoding transcriptional regulators of T cell development. These results indicate that the DNA methylation–mediated inhibition of the gene networks of T cell development was largely responsible for the differentiation block of ETPs during the development of ETP-ALL. Thus, PRC2 protects key T cell developmental regulators from DNA hypermethylation in order to keep them primed for activation upon subsequent differentiation phases, while its insufficiency predisposes ETPs to leukemic transformation. These results revealed a previously unrecognized epigenetic switch in response to PRC2 dysfunction and provide the basis for specific rational epigenetic therapy for ETP-ALL with PRC2 insufficiency.
4.3. Novel Strategies for Loss-of-Function Mutations in PRC Genes—Synthetic Lethality
Loss of EZH2 has been shown to lead to chemotherapy resistance in both AML [119] and T-ALL [120]. A number of clinical studies have also shown that loss-of-function mutations in EZH2 are correlated with poor patient outcomes and require specific treatments [121,122,123]. Although drug discovery against loss-of-function mutations has generally been more difficult than drug discovery against gain-of-function mutations, the concept of synthetic lethality, in which cell death results from inhibiting a more dependent pathway, for loss-of-function mutations has recently received much attention. The most well-known genes involved in synthetic lethality are BRCA and PARP [124], and the usefulness of PARP inhibitors for BRCA-deficient breast and ovarian cancers has been demonstrated in clinical trials [125,126]. Vigorous attempts have been made to find synthetic lethal partners for loss-of-function mutations in EZH2.
As described above, PRC2 insufficiency induces leads to a switch from H3K27me3 to acetylation (H3K27ac) at promoter regions of many PRC2 target genes, which is closely associated with increased transcription of a subset of direct polycomb targets, including potential oncogenes in cancer cells. This epigenetic switch may bring about an oncogenic addiction to the H3K27ac modification in EZH2 insufficient tumor cells. NF1 encodes a RasGTPase-activating protein (RasGAP) and its loss drives cancer by activating Ras. SUZ12 loss potentiates the effects of NF1 mutations in NF1-associated cancers by amplifying Ras-driven transcription through enhancing acetylation at H3K27 [127]. Bromodomain inhibitors inhibit the function of enhancers by competitively interfering with the binding of BRD4 to H3K27ac and abrogate the progression of tumors [128]. NF1-associated cancers appeared to be sensitive to bromodomain inhibition, indicating that PRC2 insufficiency might trigger an epigenetic switch that sensitizes these cancers to bromodomain inhibitors. We also found that loss of Ezh2 in JAK2V617F mice promotes an epigenetic switch at promoter regions of PRC2 target genes. This results in the activation of potential oncogenes such as Hmga2, thereby promoting the development of JAK2V617F-induced myelofibrosis in mice [68]. Loss of PRC2 increases sensitivity to bromodomain inhibition of JAK2V617F myelofibrosis initiating cells in vitro and in vivo [68]. Furthermore, a combination of bromodomain and JAK kinase inhibition reduces NF-kB-induced inflammation, which completely reverses fibrosis in JAK2V617F model mice [129]. These studies suggest that bromodomain inhibitors alone and the combination of bromodomain inhibitor and JAK2 inhibitor can be a novel therapeutic strategy for eradicating hematologic malignancies harboring EZH2 mutation [127,130].
There are some other promising synthetic lethal partner candidates for EZH2 that were recently discovered. Gu et al. showed that BCAT1, which catalyzes the reversible transamination of branched-chain amino acids, is aberrantly activated in EZH2-deficient myeloid neoplasms, and inhibition of BCAT1 selectively impairs EZH2-deficient cells. These findings indicate that hematologic malignancies harboring EZH2 loss-of-function mutations have an increased dependence on BCAT1, making BCAT1 a potential therapeutic target [131]. Leon et al. demonstrated that EZH2 loss-of-function mutations have increased sensitivity to CHK1 inhibitors caused by enhanced replication stress due to increased expression of MYC-N in T-ALL cells [132].
5. Conclusions
With the recent development of next-generation sequencing, it has become clear that many hematological tumors harbor mutations in PRC genes. However, in many cases, the mechanisms by which these mutations contribute to tumorigenesis remain unclear. Tumors with mutations in PRC genes often have poor treatment outcomes, and treatment challenges remain. EZH2 inhibitors targeting PRC2 gain-of-function have been developed, and new therapeutic targets, such as loss-of-function mutations in PRC2 genes, are being actively explored by employing the concept of synthetic lethality. These developments should improve the treatment outcomes of patients with hematological malignancies harboring mutations in PRC genes.
Author Contributions
S.K. and A.I. wrote the manuscript. A.I. revised the manuscript. All authors have read and agreed to the final version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Iwama, A. Polycomb repressive complexes in hematological malignancies. Blood 2017, 130, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, Y.; Iwama, A. Emerging role of noncanonical polycomb repressive complexes in normal and malignant hematopoiesis. Exp. Hematol. 2018, 68, 10–14. [Google Scholar] [CrossRef]
- Oguro, H.; Iwama, A.; Morita, Y.; Kamijo, T.; van Lohuizen, M.; Nakauchi, H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J. Exp. Med. 2006, 203, 2247–2253. [Google Scholar] [CrossRef]
- Park, I.K.; Qian, D.; Kiel, M.; Becker, M.W.; Pihalja, M.; Weissman, I.L.; Morrison, S.J.; Clarke, M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 2003, 423, 302–305. [Google Scholar] [CrossRef]
- Iwama, A.; Oguro, H.; Negishi, M.; Kato, Y.; Morita, Y.; Tsukui, H.; Ema, H.; Kamijo, T.; Katoh-Fukui, Y.; Koseki, H.; et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 2004, 21, 843–851. [Google Scholar] [CrossRef]
- Oguro, H.; Yuan, J.; Ichikawa, H.; Ikawa, T.; Yamazaki, S.; Kawamoto, H.; Nakauchi, H.; Iwama, A. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 2010, 6, 279–286. [Google Scholar] [CrossRef]
- Lessard, J.; Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 2003, 423, 255–260. [Google Scholar] [CrossRef]
- Mochizuki-Kashio, M.; Mishima, Y.; Miyagi, S.; Negishi, M.; Saraya, A.; Konuma, T.; Shinga, J.; Koseki, H.; Iwama, A. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood 2011, 118, 6553–6561. [Google Scholar] [CrossRef]
- Mochizuki-Kashio, M.; Aoyama, K.; Sashida, G.; Oshima, M.; Tomioka, T.; Muto, T.; Wang, C.; Iwama, A. Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an Ezh1-dependent manner. Blood 2015, 126, 1172–1183. [Google Scholar] [CrossRef]
- Aoyama, K.; Oshima, M.; Koide, S.; Suzuki, E.; Mochizuki-Kashio, M.; Kato, Y.; Tara, S.; Shinoda, D.; Hiura, N.; Nakajima-Takagi, Y.; et al. Ezh1 Targets Bivalent Genes to Maintain Self-Renewing Stem Cells in Ezh2-Insufficient Myelodysplastic Syndrome. iScience 2018, 9, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, I.; Herrera-Merchan, A.; Ligos, J.M.; Carramolino, L.; Nuñez, J.; Martinez, F.; Dominguez, O.; Torres, M.; Gonzalez, S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell 2012, 11, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Xu, J.; Hsu, J.H.; Nguyen, M.; Fujiwara, Y.; Peng, C.; Orkin, S.H. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 2014, 14, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Miller, S.; Hyland, C.; Kauppi, M.; Lebois, M.; Di Rago, L.; Metcalf, D.; Kinkel, S.A.; Josefsson, E.C.; Blewitt, M.E.; et al. Polycomb repressive complex 2 component Suz12 is required for hematopoietic stem cell function and lymphopoiesis. Blood 2015, 126, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Andricovich, J.; Kai, Y.; Peng, W.; Foudi, A.; Tzatsos, A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J. Clin. Investig. 2016, 126, 905–920. [Google Scholar] [CrossRef]
- Isshiki, Y.; Nakajima-Takagi, Y.; Oshima, M.; Aoyama, K.; Rizk, M.; Kurosawa, S.; Saraya, A.; Kondo, T.; Sakaida, E.; Nakaseko, C.; et al. KDM2B in polycomb repressive complex 1.1 functions as a tumor suppressor in the initiation of T-cell leukemogenesis. Blood Adv. 2019, 3, 2537–2549. [Google Scholar] [CrossRef]
- Konuma, T.; Nakamura, S.; Miyagi, S.; Negishi, M.; Chiba, T.; Oguro, H.; Yuan, J.; Mochizuki-Kashio, M.; Ichikawa, H.; Miyoshi, H.; et al. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 2011, 39, 697–709.e5. [Google Scholar] [CrossRef]
- Cao, Q.; Gearhart, M.D.; Gery, S.; Shojaee, S.; Yang, H.; Sun, H.; Lin, D.C.; Bai, J.W.; Mead, M.; Zhao, Z.; et al. BCOR regulates myeloid cell proliferation and differentiation. Leukemia 2016, 30, 1155–1165. [Google Scholar] [CrossRef]
- Tara, S.; Isshiki, Y.; Nakajima-Takagi, Y.; Oshima, M.; Aoyama, K.; Tanaka, T.; Shinoda, D.; Koide, S.; Saraya, A.; Miyagi, S.; et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood 2018, 132, 2470–2483. [Google Scholar] [CrossRef] [PubMed]
- Van den Boom, V.; Rozenveld-Geugien, M.; Bonardi, F.; Malanga, D.; van Gosliga, D.; Heijink, A.M.; Viglietto, G.; Morrone, G.; Fusetti, F.; Vellenga, E.; et al. Nonredundant and locus-specific gene repression functions of PRC1 paralog family members in human hematopoietic stem/progenitor cells. Blood 2013, 121, 2452–2461. [Google Scholar] [CrossRef]
- Ross, K.; Sedello, A.K.; Todd, G.P.; Paszkowski-Rogacz, M.; Bird, A.W.; Ding, L.; Grinenko, T.; Behrens, K.; Hubner, N.; Mann, M.; et al. Polycomb group ring finger 1 cooperates with Runx1 in regulating differentiation and self-renewal of hematopoietic cells. Blood 2012, 119, 4152–4161. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Helin, K. Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 2009, 9, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Mihara, K.; Chowdhury, M.; Nakaju, N.; Hidani, S.; Ihara, A.; Hyodo, H.; Yasunaga, S.; Takihara, Y.; Kimura, A. Bmi-1 is useful as a novel molecular marker for predicting progression of myelodysplastic syndrome and patient prognosis. Blood 2006, 107, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Saudy, N.S.; Fawzy, I.M.; Azmy, E.; Goda, E.F.; Eneen, A.; Abdul Salam, E.M. BMI1 gene expression in myeloid leukemias and its impact on prognosis. Blood Cells Mol. Dis. 2014, 53, 194–198. [Google Scholar] [CrossRef]
- Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; Baty, J.D.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Yamaguchi, H.; Ueki, T.; Usuki, K.; Kobayashi, Y.; Tajika, K.; Gomi, S.; Kurosawa, S.; Saito, R.; Furuta, Y.; et al. Usefulness of BCOR gene mutation as a prognostic factor in acute myeloid leukemia with intermediate cytogenetic prognosis. Genes Chromosomes Cancer 2018, 57, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Tiacci, E.; Holmes, A.B.; Kohlmann, A.; Martelli, M.P.; Kern, W.; Spanhol-Rosseto, A.; Klein, H.U.; Dugas, M.; Schindela, S.; et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011, 118, 6153–6163. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Collins, R.; Jiao, Y.; Ouillette, P.; Bixby, D.; Erba, H.; Vogelstein, B.; Kinzler, K.W.; Papadopoulos, N.; Malek, S.N. Somatic mutations in the transcriptional corepressor gene BCORL1 in adult acute myelogenous leukemia. Blood 2011, 118, 5914–5917. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Toki, T.; Okuno, Y.; Kanezaki, R.; Shiraishi, Y.; Sato-Otsubo, A.; Sanada, M.; Park, M.J.; Terui, K.; Suzuki, H.; et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat. Genet. 2013, 45, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef]
- Nikoloski, G.; Langemeijer, S.M.; Kuiper, R.P.; Knops, R.; Massop, M.; Tönnissen, E.R.; van der Heijden, A.; Scheele, T.N.; Vandenberghe, P.; de Witte, T.; et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010, 42, 665–667. [Google Scholar] [CrossRef]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef]
- Ernst, T.; Chase, A.J.; Score, J.; Hidalgo-Curtis, C.E.; Bryant, C.; Jones, A.V.; Waghorn, K.; Zoi, K.; Ross, F.M.; Reiter, A.; et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. [Google Scholar] [CrossRef]
- Damm, F.; Chesnais, V.; Nagata, Y.; Yoshida, K.; Scourzic, L.; Okuno, Y.; Itzykson, R.; Sanada, M.; Shiraishi, Y.; Gelsi-Boyer, V.; et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 2013, 122, 3169–3177. [Google Scholar] [CrossRef] [PubMed]
- Brecqueville, M.; Cervera, N.; Adélaïde, J.; Rey, J.; Carbuccia, N.; Chaffanet, M.; Mozziconacci, M.J.; Vey, N.; Birnbaum, D.; Gelsi-Boyer, V.; et al. Mutations and deletions of the SUZ12 polycomb gene in myeloproliferative neoplasms. Blood Cancer J. 2011, 1, e33. [Google Scholar] [CrossRef]
- Score, J.; Hidalgo-Curtis, C.; Jones, A.V.; Winkelmann, N.; Skinner, A.; Ward, D.; Zoi, K.; Ernst, T.; Stegelmann, F.; Döhner, K.; et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood 2012, 119, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Van Vlierberghe, P.; Nedjic, J.; Trimarchi, T.; Flaherty, M.S.; Ferres-Marco, D.; da Ros, V.; Tang, Z.; Siegle, J.; et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012, 18, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Kimura, S.; Isobe, T.; Yoshida, K.; Ueno, H.; Nakajima-Takagi, Y.; Wang, C.; Lin, L.; Kon, A.; Suzuki, H.; et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat. Genet. 2017, 49, 1274–1281. [Google Scholar] [CrossRef] [PubMed]
- Kiel, M.J.; Velusamy, T.; Rolland, D.; Sahasrabuddhe, A.A.; Chung, F.; Bailey, N.G.; Schrader, A.; Li, B.; Li, J.Z.; Ozel, A.B.; et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood 2014, 124, 1460–1472. [Google Scholar] [CrossRef] [PubMed]
- Stengel, A.; Kern, W.; Zenger, M.; Perglerová, K.; Schnittger, S.; Haferlach, T.; Haferlach, C. Genetic characterization of T-PLL reveals two major biologic subgroups and JAK3 mutations as prognostic marker. Genes Chromosomes Cancer 2016, 55, 82–94. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Böttcher, S.; et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef]
- Bödör, C.; Grossmann, V.; Popov, N.; Okosun, J.; O’Riain, C.; Tan, K.; Marzec, J.; Araf, S.; Wang, J.; Lee, A.M.; et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 2013, 122, 3165–3168. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef]
- Lohr, J.G.; Stojanov, P.; Lawrence, M.S.; Auclair, D.; Chapuy, B.; Sougnez, C.; Cruz-Gordillo, P.; Knoechel, B.; Asmann, Y.W.; Slager, S.L.; et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 3879–3884. [Google Scholar] [CrossRef]
- Lee, S.; Park, H.Y.; Kang, S.Y.; Kim, S.J.; Hwang, J.; Lee, S.; Kwak, S.H.; Park, K.S.; Yoo, H.Y.; Kim, W.S.; et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget 2015, 6, 17764–17776. [Google Scholar] [CrossRef] [PubMed]
- Dobashi, A.; Tsuyama, N.; Asaka, R.; Togashi, Y.; Ueda, K.; Sakata, S.; Baba, S.; Sakamoto, K.; Hatake, K.; Takeuchi, K. Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal type. Genes Chromosomes Cancer 2016, 55, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Basheer, F.; Giotopoulos, G.; Meduri, E.; Yun, H.; Mazan, M.; Sasca, D.; Gallipoli, P.; Marando, L.; Gozdecka, M.; Asby, R.; et al. Contrasting requirements during disease evolution identify EZH2 as a therapeutic target in AML. J. Exp. Med. 2019, 216, 966–981. [Google Scholar] [CrossRef]
- Tanaka, S.; Miyagi, S.; Sashida, G.; Chiba, T.; Yuan, J.; Mochizuki-Kashio, M.; Suzuki, Y.; Sugano, S.; Nakaseko, C.; Yokote, K.; et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood 2012, 120, 1107–1117. [Google Scholar] [CrossRef]
- Neff, T.; Sinha, A.U.; Kluk, M.J.; Zhu, N.; Khattab, M.H.; Stein, L.; Xie, H.; Orkin, S.H.; Armstrong, S.A. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. USA 2012, 109, 5028–5033. [Google Scholar] [CrossRef]
- Béguelin, W.; Teater, M.; Gearhart, M.D.; Calvo Fernández, M.T.; Goldstein, R.L.; Cárdenas, M.G.; Hatzi, K.; Rosen, M.; Shen, H.; Corcoran, C.M.; et al. EZH2 and BCL6 Cooperate to Assemble CBX8-BCOR Complex to Repress Bivalent Promoters, Mediate Germinal Center Formation and Lymphomagenesis. Cancer Cell 2016, 30, 197–213. [Google Scholar] [CrossRef]
- Caganova, M.; Carrisi, C.; Varano, G.; Mainoldi, F.; Zanardi, F.; Germain, P.L.; George, L.; Alberghini, F.; Ferrarini, L.; Talukder, A.K.; et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Investig. 2013, 123, 5009–5022. [Google Scholar] [CrossRef]
- Béguelin, W.; Popovic, R.; Teater, M.; Jiang, Y.; Bunting, K.L.; Rosen, M.; Shen, H.; Yang, S.N.; Wang, L.; Ezponda, T.; et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013, 23, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Souroullas, G.P.; Jeck, W.R.; Parker, J.S.; Simon, J.M.; Liu, J.Y.; Paulk, J.; Xiong, J.; Clark, K.S.; Fedoriw, Y.; Qi, J.; et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 2016, 22, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.N.; Jankowska, A.M.; Mahfouz, R.; Dunbar, A.J.; Sugimoto, Y.; Hosono, N.; Hu, Z.; Cheriyath, V.; Vatolin, S.; Przychodzen, B.; et al. Multiple mechanisms deregulate EZH2 and histone H3 lysine 27 epigenetic changes in myeloid malignancies. Leukemia 2013, 27, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Muto, T.; Sashida, G.; Oshima, M.; Wendt, G.R.; Mochizuki-Kashio, M.; Nagata, Y.; Sanada, M.; Miyagi, S.; Saraya, A.; Kamio, A.; et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J. Exp. Med. 2013, 210, 2627–2639. [Google Scholar] [CrossRef] [PubMed]
- Sashida, G.; Harada, H.; Matsui, H.; Oshima, M.; Yui, M.; Harada, Y.; Tanaka, S.; Mochizuki-Kashio, M.; Wang, C.; Saraya, A.; et al. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat. Commun. 2014, 5, 4177. [Google Scholar] [CrossRef]
- Starczynowski, D.T.; Kuchenbauer, F.; Argiropoulos, B.; Sung, S.; Morin, R.; Muranyi, A.; Hirst, M.; Hogge, D.; Marra, M.; Wells, R.A.; et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010, 16, 49–58. [Google Scholar] [CrossRef]
- Sashida, G.; Wang, C.; Tomioka, T.; Oshima, M.; Aoyama, K.; Kanai, A.; Mochizuki-Kashio, M.; Harada, H.; Shimoda, K.; Iwama, A. The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition. J. Exp. Med. 2016, 213, 1459–1477. [Google Scholar] [CrossRef]
- Shimizu, T.; Kubovcakova, L.; Nienhold, R.; Zmajkovic, J.; Meyer, S.C.; Hao-Shen, H.; Geier, F.; Dirnhofer, S.; Guglielmelli, P.; Vannucchi, A.M.; et al. Loss of Ezh2 synergizes with JAK2-V617F in initiating myeloproliferative neoplasms and promoting myelofibrosis. J. Exp. Med. 2016, 213, 1479–1496. [Google Scholar] [CrossRef]
- Yang, Y.; Akada, H.; Nath, D.; Hutchison, R.E.; Mohi, G. Loss of Ezh2 cooperates with Jak2V617F in the development of myelofibrosis in a mouse model of myeloproliferative neoplasm. Blood 2016, 127, 3410–3423. [Google Scholar] [CrossRef]
- Oshima, M.; Hasegawa, N.; Mochizuki-Kashio, M.; Muto, T.; Miyagi, S.; Koide, S.; Yabata, S.; Wendt, G.R.; Saraya, A.; Wang, C.; et al. Ezh2 regulates the Lin28/let-7 pathway to restrict activation of fetal gene signature in adult hematopoietic stem cells. Exp. Hematol. 2016, 44, 282–296.e3. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Chagraoui, J.; Krosl, J.; Gendron, P.; Wilhelm, B.; Lemieux, S.; Boucher, G.; Chagnon, P.; Drouin, S.; Lambert, R.; et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012, 26, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Oshima, M.; Sato, D.; Matsui, H.; Kubota, S.; Aoyama, K.; Nakajima-Takagi, Y.; Koide, S.; Matsubayashi, J.; Mochizuki-Kashio, M.; et al. Ezh2 loss propagates hypermethylation at T cell differentiation-regulating genes to promote leukemic transformation. J. Clin. Investig. 2018, 128, 3872–3886. [Google Scholar] [CrossRef] [PubMed]
- Danis, E.; Yamauchi, T.; Echanique, K.; Zhang, X.; Haladyna, J.N.; Riedel, S.S.; Zhu, N.; Xie, H.; Orkin, S.H.; Armstrong, S.A.; et al. Ezh2 Controls an Early Hematopoietic Program and Growth and Survival Signaling in Early T Cell Precursor Acute Lymphoblastic Leukemia. Cell Rep. 2016, 14, 1953–1965. [Google Scholar] [CrossRef]
- Booth, C.A.G.; Barkas, N.; Neo, W.H.; Boukarabila, H.; Soilleux, E.J.; Giotopoulos, G.; Farnoud, N.; Giustacchini, A.; Ashley, N.; Carrelha, J.; et al. Ezh2 and Runx1 Mutations Collaborate to Initiate Lympho-Myeloid Leukemia in Early Thymic Progenitors. Cancer Cell 2018, 33, 274–291.e8. [Google Scholar] [CrossRef]
- Mohammad, F.; Weissmann, S.; Leblanc, B.; Pandey, D.P.; Højfeldt, J.W.; Comet, I.; Zheng, C.; Johansen, J.V.; Rapin, N.; Porse, B.T.; et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat. Med. 2017, 23, 483–492. [Google Scholar] [CrossRef]
- Boileau, M.; Shirinian, M.; Gayden, T.; Harutyunyan, A.S.; Chen, C.C.L.; Mikael, L.G.; Duncan, H.M.; Neumann, A.L.; Arreba-Tutusaus, P.; De Jay, N.; et al. Mutant H3 histones drive human pre-leukemic hematopoietic stem cell expansion and promote leukemic aggressiveness. Nat. Commun. 2019, 10, 2891. [Google Scholar] [CrossRef]
- Lehnertz, B.; Zhang, Y.W.; Boivin, I.; Mayotte, N.; Tomellini, E.; Chagraoui, J.; Lavallée, V.P.; Hébert, J.; Sauvageau, G. H3(K27M/I) mutations promote context-dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood 2017, 130, 2204–2214. [Google Scholar] [CrossRef]
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M.; et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014, 124, 1513–1521. [Google Scholar] [CrossRef]
- Kelly, M.J.; So, J.; Rogers, A.J.; Gregory, G.; Li, J.; Zethoven, M.; Gearhart, M.D.; Bardwell, V.J.; Johnstone, R.W.; Vervoort, S.J.; et al. Bcor loss perturbs myeloid differentiation and promotes leukaemogenesis. Nat. Commun. 2019, 10, 1347. [Google Scholar] [CrossRef]
- Tanaka, T.; Nakajima-Takagi, Y.; Aoyama, K.; Tara, S.; Oshima, M.; Saraya, A.; Koide, S.; Si, S.; Manabe, I.; Sanada, M.; et al. Internal deletion of BCOR reveals a tumor suppressor function for BCOR in T lymphocyte malignancies. J. Exp. Med. 2017, 214, 2901–2913. [Google Scholar] [CrossRef] [PubMed]
- Lefebure, M.; Tothill, R.W.; Kruse, E.; Hawkins, E.D.; Shortt, J.; Matthews, G.M.; Gregory, G.P.; Martin, B.P.; Kelly, M.J.; Todorovski, I.; et al. Genomic characterisation of Eμ-Myc mouse lymphomas identifies Bcor as a Myc co-operative tumour-suppressor gene. Nat. Commun. 2017, 8, 14581. [Google Scholar] [CrossRef] [PubMed]
- van den Boom, V.; Maat, H.; Geugien, M.; Rodríguez López, A.; Sotoca, A.M.; Jaques, J.; Brouwers-Vos, A.Z.; Fusetti, F.; Groen, R.W.; Yuan, H.; et al. Non-canonical PRC1.1 Targets Active Genes Independent of H3K27me3 and Is Essential for Leukemogenesis. Cell Rep. 2016, 14, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Nagamachi, A.; Takubo, K.; Yamasaki, N.; Matsui, H.; Kanai, A.; Nakata, Y.; Ikeda, K.; Konuma, T.; Oda, H.; et al. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood 2015, 125, 3437–3446. [Google Scholar] [CrossRef]
- Sinclair, D.A.; Milne, T.A.; Hodgson, J.W.; Shellard, J.; Salinas, C.A.; Kyba, M.; Randazzo, F.; Brock, H.W. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development 1998, 125, 1207–1216. [Google Scholar]
- Abdel-Wahab, O.; Adli, M.; LaFave, L.M.; Gao, J.; Hricik, T.; Shih, A.H.; Pandey, S.; Patel, J.P.; Chung, Y.R.; Koche, R.; et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012, 22, 180–193. [Google Scholar] [CrossRef]
- Abdel-Wahab, O.; Gao, J.; Adli, M.; Dey, A.; Trimarchi, T.; Chung, Y.R.; Kuscu, C.; Hricik, T.; Ndiaye-Lobry, D.; Lafave, L.M.; et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 2013, 210, 2641–2659. [Google Scholar] [CrossRef]
- Scheuermann, J.C.; de Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Müller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef]
- Fujino, T.; Kitamura, T. ASXL1 mutation in clonal hematopoiesis. Exp. Hematol. 2020, 83, 74–84. [Google Scholar] [CrossRef]
- Pratcorona, M.; Abbas, S.; Sanders, M.A.; Koenders, J.E.; Kavelaars, F.G.; Erpelinck-Verschueren, C.A.; Zeilemakers, A.; Löwenberg, B.; Valk, P.J. Acquired mutations in ASXL1 in acute myeloid leukemia: Prevalence and prognostic value. Haematologica 2012, 97, 388–392. [Google Scholar] [CrossRef]
- Thol, F.; Friesen, I.; Damm, F.; Yun, H.; Weissinger, E.M.; Krauter, J.; Wagner, K.; Chaturvedi, A.; Sharma, A.; Wichmann, M.; et al. Prognostic significance of ASXL1 mutations in patients with myelodysplastic syndromes. J. Clin. Oncol. 2011, 29, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Adès, L.; Fenaux, P.; Beyne-Rauzy, O.; et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 2013, 31, 2428–2436. [Google Scholar] [CrossRef] [PubMed]
- Guglielmelli, P.; Lasho, T.L.; Rotunno, G.; Score, J.; Mannarelli, C.; Pancrazzi, A.; Biamonte, F.; Pardanani, A.; Zoi, K.; Reiter, A.; et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: An international study of 797 patients. Leukemia 2014, 28, 1804–1810. [Google Scholar] [CrossRef] [PubMed]
- Nagase, R.; Inoue, D.; Pastore, A.; Fujino, T.; Hou, H.A.; Yamasaki, N.; Goyama, S.; Saika, M.; Kanai, A.; Sera, Y.; et al. Expression of mutant Asxl1 perturbs hematopoiesis and promotes susceptibility to leukemic transformation. J. Exp. Med. 2018, 215, 1729–1747. [Google Scholar] [CrossRef] [PubMed]
- Gulati, N.; Béguelin, W.; Giulino-Roth, L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk. Lymphoma 2018, 59, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Soria, J.C.; Toulmonde, M.; Michot, J.M.; Lucchesi, C.; Varga, A.; Coindre, J.M.; Blakemore, S.J.; Clawson, A.; Suttle, B.; et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018, 19, 649–659. [Google Scholar] [CrossRef]
- Morschhauser, F.; Tilly, H.; Chaidos, A.; McKay, P.; Phillips, T.; Assouline, S.; Batlevi, C.L.; Campbell, P.; Ribrag, V.; Damaj, G.L.; et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: An open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 2020, 21, 1433–1442. [Google Scholar] [CrossRef]
- Sarkozy, C.; Morschhauser, F.; Dubois, S.; Molina, T.; Michot, J.M.; Cullières-Dartigues, P.; Suttle, B.; Karlin, L.; Le Gouill, S.; Picquenot, J.M.; et al. A LYSA Phase Ib Study of Tazemetostat (EPZ-6438) plus R-CHOP in Patients with Newly Diagnosed Diffuse Large B-Cell Lymphoma (DLBCL) with Poor Prognosis Features. Clin. Cancer Res. 2020, 26, 3145–3153. [Google Scholar] [CrossRef]
- Yap, T.A.; Winter, J.N.; Giulino-Roth, L.; Longley, J.; Lopez, J.; Michot, J.M.; Leonard, J.P.; Ribrag, V.; McCabe, M.T.; Creasy, C.L.; et al. Phase I Study of the Novel Enhancer of Zeste Homolog 2 (EZH2) Inhibitor GSK2816126 in Patients with Advanced Hematologic and Solid Tumors. Clin. Cancer Res. 2019, 25, 7331–7339. [Google Scholar] [CrossRef]
- Knutson, S.K.; Warholic, N.M.; Johnston, L.D.; Klaus, C.R.; Wigle, T.J.; Iwanowicz, D.; Littlefield, B.A.; Porter-Scott, M.; Smith, J.J.; Moyer, M.P.; et al. Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas. PLoS ONE 2014, 9, e111840. [Google Scholar] [CrossRef]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Konze, K.D.; Ma, A.; Li, F.; Barsyte-Lovejoy, D.; Parton, T.; Macnevin, C.J.; Liu, F.; Gao, C.; Huang, X.P.; Kuznetsova, E.; et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013, 8, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Zhao, K.; Gu, J.; Huang, Y.; Wang, Y.; Zhang, H.; Zhang, M.; Zhang, J.; Yu, Z.; Li, L.; et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017, 13, 381–388. [Google Scholar] [CrossRef]
- Honma, D.; Kanno, O.; Watanabe, J.; Kinoshita, J.; Hirasawa, M.; Nosaka, E.; Shiroishi, M.; Takizawa, T.; Yasumatsu, I.; Horiuchi, T.; et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 2017, 108, 2069–2078. [Google Scholar] [CrossRef]
- Bisserier, M.; Wajapeyee, N. Mechanisms of resistance to EZH2 inhibitors in diffuse large B-cell lymphomas. Blood 2018, 131, 2125–2137. [Google Scholar] [CrossRef]
- Rizk, M.; Rizq, O.; Oshima, M.; Nakajima-Takagi, Y.; Koide, S.; Saraya, A.; Isshiki, Y.; Chiba, T.; Yamazaki, S.; Ma, A.; et al. Akt inhibition synergizes with polycomb repressive complex 2 inhibition in the treatment of multiple myeloma. Cancer Sci. 2019, 110, 3695–3707. [Google Scholar] [CrossRef]
- Rizq, O.; Mimura, N.; Oshima, M.; Saraya, A.; Koide, S.; Kato, Y.; Aoyama, K.; Nakajima-Takagi, Y.; Wang, C.; Chiba, T.; et al. Dual Inhibition of EZH2 and EZH1 Sensitizes PRC2-Dependent Tumors to Proteasome Inhibition. Clin. Cancer Res. 2017, 23, 4817–4830. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; On, D.M.; Ma, A.; Parton, T.; Konze, K.D.; Pattenden, S.G.; Allison, D.F.; Cai, L.; Rockowitz, S.; Liu, S.; et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 2015, 125, 346–357. [Google Scholar] [CrossRef]
- Nakagawa, M.; Fujita, S.; Katsumoto, T.; Yamagata, K.; Ogawara, Y.; Hattori, A.; Kagiyama, Y.; Honma, D.; Araki, K.; Inoue, T.; et al. Dual inhibition of enhancer of zeste homolog 1/2 overactivates WNT signaling to deplete cancer stem cells in multiple myeloma. Cancer Sci. 2019, 110, 194–208. [Google Scholar] [CrossRef]
- Fujita, S.; Honma, D.; Adachi, N.; Araki, K.; Takamatsu, E.; Katsumoto, T.; Yamagata, K.; Akashi, K.; Aoyama, K.; Iwama, A.; et al. Dual inhibition of EZH1/2 breaks the quiescence of leukemia stem cells in acute myeloid leukemia. Leukemia 2018, 32, 855–864. [Google Scholar] [CrossRef]
- Figueroa, M.E.; Skrabanek, L.; Li, Y.; Jiemjit, A.; Fandy, T.E.; Paietta, E.; Fernandez, H.; Tallman, M.S.; Greally, J.M.; Carraway, H.; et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood 2009, 114, 3448–3458. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Welch, J.S.; Petti, A.A.; Miller, C.A.; Fronick, C.C.; O’Laughlin, M.; Fulton, R.S.; Wilson, R.K.; Baty, J.D.; Duncavage, E.J.; Tandon, B.; et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N. Engl. J. Med. 2016, 375, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Ohm, J.E.; McGarvey, K.M.; Yu, X.; Cheng, L.; Schuebel, K.E.; Cope, L.; Mohammad, H.P.; Chen, W.; Daniel, V.C.; Yu, W.; et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007, 39, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, C.; Glatz, D.; Schwarzfischer, L.; Wimmer, J.; Stasik, S.; Nuetzel, M.; Heudobler, D.; Andreesen, R.; Ehninger, G.; Thiede, C.; et al. Profiling of aberrant DNA methylation in acute myeloid leukemia reveals subclasses of CG-rich regions with epigenetic or genetic association. Leukemia 2019, 33, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Manzo, M.; Wirz, J.; Ambrosi, C.; Villaseñor, R.; Roschitzki, B.; Baubec, T. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 2017, 36, 3421–3434. [Google Scholar] [CrossRef]
- Hasegawa, N.; Oshima, M.; Sashida, G.; Matsui, H.; Koide, S.; Saraya, A.; Wang, C.; Muto, T.; Takane, K.; Kaneda, A.; et al. Impact of combinatorial dysfunctions of Tet2 and Ezh2 on the epigenome in the pathogenesis of myelodysplastic syndrome. Leukemia 2017, 31, 861–871. [Google Scholar] [CrossRef]
- Göllner, S.; Oellerich, T.; Agrawal-Singh, S.; Schenk, T.; Klein, H.U.; Rohde, C.; Pabst, C.; Sauer, T.; Lerdrup, M.; Tavor, S.; et al. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 2017, 23, 69–78. [Google Scholar] [CrossRef]
- Ariës, I.M.; Bodaar, K.; Karim, S.A.; Chonghaile, T.N.; Hinze, L.; Burns, M.A.; Pfirrmann, M.; Degar, J.; Landrigan, J.T.; Balbach, S.; et al. PRC2 loss induces chemoresistance by repressing apoptosis in T cell acute lymphoblastic leukemia. J. Exp. Med. 2018, 215, 3094–3114. [Google Scholar] [CrossRef]
- Grossmann, V.; Kohlmann, A.; Eder, C.; Haferlach, C.; Kern, W.; Cross, N.C.; Haferlach, T.; Schnittger, S. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia 2011, 25, 877–879. [Google Scholar] [CrossRef] [PubMed]
- Saygin, C.; Hirsch, C.; Przychodzen, B.; Sekeres, M.A.; Hamilton, B.K.; Kalaycio, M.; Carraway, H.E.; Gerds, A.T.; Mukherjee, S.; Nazha, A.; et al. Mutations in DNMT3A, U2AF1, and EZH2 identify intermediate-risk acute myeloid leukemia patients with poor outcome after CR1. Blood Cancer J. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Bejar, R.; Stevenson, K.E.; Caughey, B.A.; Abdel-Wahab, O.; Steensma, D.P.; Galili, N.; Raza, A.; Kantarjian, H.; Levine, R.L.; Neuberg, D.; et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 2012, 30, 3376–3382. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- De Raedt, T.; Beert, E.; Pasmant, E.; Luscan, A.; Brems, H.; Ortonne, N.; Helin, K.; Hornick, J.L.; Mautner, V.; Kehrer-Sawatzki, H.; et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014, 514, 247–251. [Google Scholar] [CrossRef]
- Lovén, J.; Hoke, H.A.; Lin, C.Y.; Lau, A.; Orlando, D.A.; Vakoc, C.R.; Bradner, J.E.; Lee, T.I.; Young, R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013, 153, 320–334. [Google Scholar] [CrossRef]
- Kleppe, M.; Koche, R.; Zou, L.; van Galen, P.; Hill, C.E.; Dong, L.; De Groote, S.; Papalexi, E.; Hanasoge Somasundara, A.V.; Cordner, K.; et al. Dual Targeting of Oncogenic Activation and Inflammatory Signaling Increases Therapeutic Efficacy in Myeloproliferative Neoplasms. Cancer Cell 2018, 33, 29–43.e7. [Google Scholar] [CrossRef]
- Huang, X.; Yan, J.; Zhang, M.; Wang, Y.; Chen, Y.; Fu, X.; Wei, R.; Zheng, X.L.; Liu, Z.; Zhang, X.; et al. Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors. Cell 2018, 175, 186–199.e19. [Google Scholar] [CrossRef]
- Gu, Z.; Liu, Y.; Cai, F.; Patrick, M.; Zmajkovic, J.; Cao, H.; Zhang, Y.; Tasdogan, A.; Chen, M.; Qi, L.; et al. Loss of EZH2 Reprograms BCAA Metabolism to Drive Leukemic Transformation. Cancer Discov. 2019, 9, 1228–1247. [Google Scholar] [CrossRef] [PubMed]
- León, T.E.; Rapoz-D’Silva, T.; Bertoli, C.; Rahman, S.; Magnussen, M.; Philip, B.; Farah, N.; Richardson, S.E.; Ahrabi, S.; Guerra-Assunção, J.A.; et al. EZH2-Deficient T-cell Acute Lymphoblastic Leukemia Is Sensitized to CHK1 Inhibition through Enhanced Replication Stress. Cancer Discov. 2020, 10, 998–1017. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).