Abstract
The 15q11.2 BP1-BP2 microdeletion (Burnside–Butler) syndrome is emerging as the most frequent pathogenic copy number variation (CNV) in humans associated with neurodevelopmental disorders with changes in brain morphology, behavior, and cognition. In this study, we explored functions and interactions of the four protein-coding genes in this region, namely NIPA1, NIPA2, CYFIP1, and TUBGCP5, and elucidate their role, in solo and in concert, in the causation of neurodevelopmental disorders. First, we investigated the STRING protein-protein interactions encompassing all four genes and ascertained their predicted Gene Ontology (GO) functions, such as biological processes involved in their interactions, pathways and molecular functions. These include magnesium ion transport molecular function, regulation of axonogenesis and axon extension, regulation and production of bone morphogenetic protein and regulation of cellular growth and development. We gathered a list of significantly associated cardinal maladies for each gene from searchable genomic disease websites, namely MalaCards.org: HGMD, OMIM, ClinVar, GTR, Orphanet, DISEASES, Novoseek, and GeneCards.org. Through tabulations of such disease data, we ascertained the cardinal disease association of each gene, as well as their expanded putative disease associations. This enabled further tabulation of disease data to ascertain the role of each gene in the top ten overlapping significant neurodevelopmental disorders among the disease association data sets: (1) Prader–Willi Syndrome (PWS); (2) Angelman Syndrome (AS); (3) 15q11.2 Deletion Syndrome with Attention Deficit Hyperactive Disorder & Learning Disability; (4) Autism Spectrum Disorder (ASD); (5) Schizophrenia; (6) Epilepsy; (7) Down Syndrome; (8) Microcephaly; (9) Developmental Disorder, and (10) Peripheral Nervous System Disease. The cardinal disease associations for each of the four contiguous 15q11.2 BP1-BP2 genes are NIPA1- Spastic Paraplegia 6; NIPA2—Angelman Syndrome and Prader–Willi Syndrome; CYFIP1—Fragile X Syndrome and Autism; TUBGCP5—Prader–Willi Syndrome. The four genes are individually associated with PWS, ASD, schizophrenia, epilepsy, and Down syndrome. Except for TUBGCP5, the other three genes are associated with AS. Unlike the other genes, TUBGCP5 is also not associated with attention deficit hyperactivity disorder and learning disability, developmental disorder, or peripheral nervous system disease. CYFIP1 was the only gene not associated with microcephaly but was the only gene associated with developmental disorders. Collectively, all four genes were associated with up to three-fourths of the ten overlapping neurodevelopmental disorders and are deleted in this most prevalent known pathogenic copy number variation now recognized among humans with these clinical findings.
1. Introduction
The 15q11.2 BP1-BP2 deletion (Burnside–Butler) syndrome is an emerging condition that encompasses four protein-coding genes (NIPA1, NIPA2, CYFIP1, and TUBGCP5) within this chromosome region. When disturbed, these four genes lead to cognitive impairment with speech and/or motor delay along with dyslexia and psychiatric/behavior problems (attention deficit hyperactivity, autism, schizophrenia or psychosis), ataxia or poor coordination, seizures, congenital anomalies and structural brain defects [1]. These genes are associated with neurological, cognitive, or behavior problems as well as playing a role in both Prader–Willi and Angelman syndromes, first examples in humans of genomic imprinting [2].
These imprinting disorders typically are caused by a deletion in the majority of cases involving the 15q11-q13 chromosome region of different parental origin [i.e., paternal in Prader–Willi syndrome (PWS) and maternal in Angelman syndrome (AS)] [1,2]. The typical 15q11-q13 deletion involves proximal chromosome 15 breakpoints BP1 or BP2 and the more distally placed BP3 containing repetitive DNA segments allowing malalignment in meiosis of the homologous chromosome 15s leading to deletions or duplications in the region. The individuals with the larger typical type I deletion (involving BP1 and BP3) are found in both Prader–Willi and Angelman syndromes. They are reported with more severe developmental symptoms and clinical severity than individuals with the smaller typical type II deletion (involving BP2 and BP3) [3]. The 15q11.2 BP1-BP2 microdeletion encompasses a 500kb region located between breakpoints BP1 and BP2 proximal to the centromere of chromosome 15 including the four protein-coding genes (i.e., NIPA1, NIPA2, CYFIP1, TUBGCP5) that become the major focus of this review.
Hundreds of patients have now been reported with the 15q11.2 BP1-BP2 microdeletion often associated with the above listed neurodevelopmental disorders. A summary of the clinical features reported in over 200 individuals were described by Cox and Butler [4] and further reviewed by Butler [1] by grouping into five categories. These categories are (1) growth and development; (2) dysmorphic features; (3) intelligence and academic achievement; (4) behavioral and psychiatric problems, and (5) other related medical concerns. Developmental problems were reported in 73% of cases and speech delay in 67%; dysmorphic ears (46%) and palatal anomalies (46%); writing (60%) and reading (57%) difficulties, memory problems (60%) and verbal IQ scores ≤75 (50%); behavior problems, unspecified (55%); and abnormal brain imaging findings (43%). Additional clinical features were not as common and included motor delay (42%), ADD/attention deficit hyperactivity disorder (35%), autism spectrum disorder (27%), seizures or epilepsy (26%), and schizophrenia/paranoid psychosis (20%).
A recent report on structural brain anomalies using detailed imaging in hundreds of patients with the chromosome 15q11.2 BP1-BP2 microdeletion and controls showed a smaller brain surface area with a thicker cortex. These findings were more common in the frontal, cingulate, and parietal lobes in those with the deletions, supporting clinical observations and evidence as an emerging syndrome. Aberrations, leading to copy number variation (CNV) in these genes are estimated to be present in 0.5% to 1.0% of the population, making this specific site as the most prevalent known pathogenic copy number variation in humans [5]. However, not all individuals with this microdeletion are clinically affected, yet the collection of findings appears to share biological pathways that are hitherto unexplored. Their presumed genetic mechanisms require further analysis, as illustrated and discussed in our report.
Summarized results from chromosomal microarray analysis by a certified commercial clinical testing laboratory of patients presenting with genetic services were reported by Ho et al. [6]. The microarray analysis included 2.8 million probes optimized for the detection of CNVs associated with neurodevelopmental disorders. They reported an overall CNV detection rate of 28.1% in 10,351 consecutive patients which rose to nearly 33% of cases without ASD but with developmental delay/intellectual disability and/or multiple congenital anomalies. The overall rate of detection for those with ASD was also significant at 24.4%. The 15q11.2 BP1-BP2 deletion (Burnside–Butler) syndrome was found to be the most common disturbance (9%) seen in 85 genetic defects associated with neurodevelopmental disorders in this large cohort of consecutive patients and the most common finding in those presenting with ASD.
The larger 15q11-q13 type I deletion is approximately 6.6Mb in size seen in both PWS and AS and includes four genes (NIPA1, NIPA2, CYFIP1, and TUBGCP5) located within the proximal 15q11.2 BP1-BP2 microdeletion region. The smaller typical 15q11-q13 type II deletion has these four genes intact (see Figure 1). These genes are highly conserved and expressed in the brain (GeneCards.org & UniProtKB/Swiss-Prot). The NIPA1 gene causes autosomal dominant hereditary spastic paraplegia and postural disturbances when disturbed (https://www.malacards.org/card/spastic_paraplegia_6_autosomal_dominant) and functions as a magnesium transporter (https://www.genecards.org/cgibin/carddisp.pl?gene=NIPA1&keywords=NIPA1).
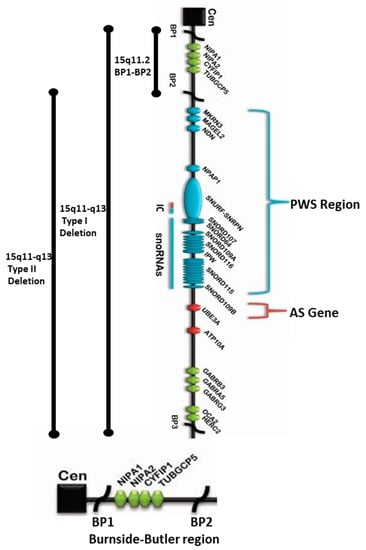
Figure 1.
15q11.2 BP1-BP2 microdeletion (Burnside–Butler) syndrome region found at the proximal end of Prader–Willi syndrome (PWS) / Angelman syndrome (AS) regions within the 15q11-q13 Type I deletion depicting the location and order of the four protein-coding genes therein: NIPA1, NIPA2, CYFIP1, and TUBGCP5 within the 15q11.2 region distal to the centromere and proximal to the imprinted PWS/AS genes. The bottom enlarged horizontal chromatin figure exclusively depicts NIPA1, NIPA2, CYFIP1, and TUBGCP5 genes in the BP1-BP2 region.
Mutations of the NIPA2 gene are reported in patients with childhood absence epilepsy with decreased intracellular magnesium concentration in neurons [7,8,9,10,11]. The CYFIP1 gene encodes a protein product that interacts with FMRP, the protein coded by the FMR1 gene causing fragile X syndrome (Entrez Gene: 23191; https://www.genecards.org/cgi-bin/carddisp.pl?gene=CYFIP1&keywords=CYFIP1). The fourth gene is TUBGCP5 associated with the chromosome 15q11.2 deletion syndrome and obsessive-compulsive disorder when disturbed. It also plays a role in microtubule nucleation at the centrosome in cells (UniProtKB: Q96RT8; https://www.genecards.org/cgi-bin/carddisp.pl?gene=TUBGCP5&keywords=TUBGCP5) [8,9,10,11,12,13,14,15,16,17,18].
Overview of 15q11-q13 BP1-BP3 Region Depicting the Proximal Location of BP1-BP2 Microdeletion (Burnside–Butler) Syndrome Region Within the Broader Type I Deletion Adjacent to Prader–Willi Syndrome (PWS)/Angelman Syndrome (AS) Regions.
The location, as well as the order of genes and transcripts (e.g., snoRNAs), are also shown in Figure 1 above that causes PWS/AS. Those genes that are imprinted and paternally expressed causing PWS (in blue) and those causing AS (in red) are also imprinted and maternally expressed. The location and the relative size of the 15q11.2 BP1–BP2 microdeletion region, the typical larger 15q11–q13 Type I deletion involving breakpoints BP1 and BP3, and the typical smaller 15q11–q13 Type II deletion involving breakpoints BP2 and BP3 in both PWS and AS are illustrated. IC: imprinting center controlling the activity of imprinted genes in the 15q11–q13 region.
2. Results
2.1. Overview of the Four Genes in the 15q11.2 BP1-BP2 Region
All four syntenic and bi-allelically conserved expressed genes in the 15q11.2 region between breakpoints BP1 and BP2 are functionally predicted to interact with each other along with seven other genes. The predicted STRING functional interactions network encompasses these four genes as illustrated in Figure 2.
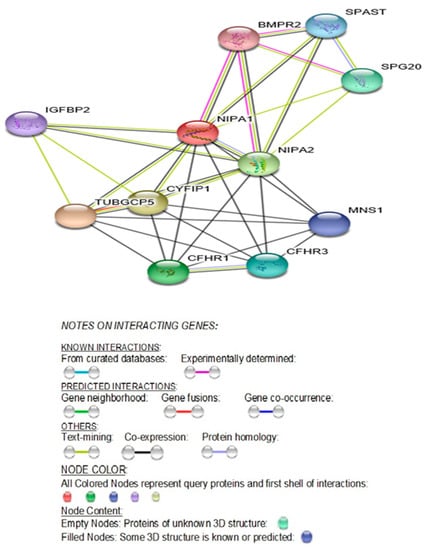
Figure 2.
STRING Protein-Protein Interaction network involving NIPA1, NIPA2, CYFIP1, and TUBGCP5 genes with functional interactions showing 11 nodes (Table 1) and 34 edges and predicted functional interactions, such as, biological process (GO) and molecular function (GO) (Table 2), as designated in STRING: 9606.ENSP00000337452 (STRING Consortium 2019). Network nodes represent proteins (Table 1) with splice isoforms or post-translational modifications collapsed into each node for all proteins produced by a single protein-coding gene. Edges represent protein-protein associations that are considered specific and meaningful, i.e., proteins jointly contribute to a shared function, such as, Biological Process (GO) and Molecular Function (GO) (Table 2).
The STRING diagram and protein interactions involving all four genes of this microdeletion syndrome are depicted in Figure 2 and their predicted functions are presented in Table 1. The identified biological processes (GO) and molecular functions (GO) are presented in Table 2 (https://version11.stringdb.org/cgi/network.pl?taskId=XJOUKPc8icHM).

Table 1.
STRING: Predicted Functions for NIPA1, NIPA2, CYFIP1, and TUBGCP5 Genes.

Table 2.
Predicted STRING Protein–Protein Functional Interactions for NIPA1, NIPA2, CYFIP1, and TUBGCP5 Genes.
Protein Network (STRING) Interactions Encompassing the Four Genes: NIPA1, NIPA2, CYFIP1 and TUBGCP5
2.2. NIPA1 (Non-Imprinted in Prader–Willi/Angelman Syndrome Region Protein 1) Gene
Attributes, Location, Description, Function and Associated Disorders for NIPA1 Gene.
The chromosomal band location for the NIPA1 gene is 15q11.2 with the genomic location (GRCh38/hg38) at 22,773,063–22,829,789 with a size of 56,727 bases in a plus-strand orientation. This gene has the following attributes: Size: 329 amino acids; Molecular mass: 34,562 Da; Quaternary structure:Homodimer (https://www.genecards.org/cgi-bin/carddisp.pl?gene=NIPA1&keywords=NIPA1). NIPA1 has two alternative splice isoforms; isoform 2 differs from the canonical sequence by missing the first 75 amino acids (GeneCards & UniProtKB/Swiss-Prot). It is a multi-pass membrane protein recruited to the cell membrane in response to low extracellular magnesium (GeneCards & UniProtKB/Swiss-Prot). This gene is widely expressed with highest levels in neuronal tissues and overexpressed in the spinal cord (20.8), frontal cortex (17.0), and fetal liver (7.3) (Protein differential expression in normal tissues from Human Integrated Protein Expression Database (HIPED) for NIPA1 Gene: https://www.genecards.org/Guide/GeneCard#protein-differential-expression; https://www.genecards.org/cgi-bin/carddisp.pl?gene=NIPA1&keywords=NIPA1).
NIPA1 protein plays a potential role in nervous system development and maintenance and is most ubiquitously expressed in the brain with a mean RPKM (Reads Per Kilobase per Million reads placed) of 16.56 + 3.055 (https://www.ncbi.nlm.nih.gov/gene/123606/?report=expression; https://www.gtexportal.org/home/gene/NIPA1) [14]. The multiple transmembrane domains of this protein localize to endosomes and plasma membrane. This protein is recruited in response to low extracellular magnesium and functions in Mg2 transport (Q7RTP0-NIPA1_HUMAN) [15]. Subcellular localization with immunofluorescence shows that endogenous NIPA1 protein associates with early endosomes and the cell surface in a variety of neuronal and epithelial cells. As expected of a magnesium-responsive gene, altered magnesium concentration leads to redistribution between the endosomal compartment and the plasma membrane; high magnesium results in diminished cell surface NIPA1 whereas low magnesium leads to accumulation in early endosomes and recruitment to the plasma membrane [15].
An important paralog of this gene, NIPAL1 is also a magnesium ion transmembrane transporter. In addition, there are three other protein-coding transcript variants (paralogs): NIPAL2, NIPAL3, and NIPAL4, which are also magnesium ion transmembrane transporters. Other genes also perform a similar magnesium transport function, namely, MAGT1, MMGT1, and MRS2 (GeneCards.Org). Although NIPA1 protein acts as a Mg (2+) transporter, it can also transport other divalent cations such as Fe(2+), Sr(2+), Ba(2+), Mn(2+), and Co(2+), but to a much less extent than Mg(2+) (UniProtKB/Swiss-Prot Summary). Among its other related pathways are the transport of glucose and other sugars, bile salts and organic acids, metal ions, and amine compounds.
Only mutations (not haploinsufficiency due to 15q11.2 BP1-BP2 microdeletions including the NIPA1 gene) have thus far been associated with autosomal dominant spastic paraplegia 6 / SPG6- linked hereditary spastic paraplegia: HSP (https://www.genecards.org/cgi-bin/carddisp.pl?gene=NIPA1&keywords=NIPA1). Spastic paraplegia 6 is a neurodegenerative disorder characterized by slow, gradual, progressive weakness and spasticity of the lower limbs. Rate of progression and the severity of symptoms are quite variable with initial symptoms of difficulty with balance, weakness, and stiffness in the legs, muscle spasms, and dragging the toes when walking. In some forms of the disorder, bladder symptoms (such as incontinence) may appear, or weakness and stiffness may be present to other parts of the body (UniProtKB/Swiss-Prot) [16,17,18]. Two variants in the NIPA1 gene, namely, p.Thr45Arg (VAR_023440; SNP ID: rs104894496) and p.Gly106Arg (VAR_023441; SNP ID: rs104894490) are disease-causing variations for spastic paraplegia type 6 (SPG6), autosomal dominant form. A rare NIPA1 deletion was also found in a patient with pervasive developmental disorder not otherwise specified and with mild intellectual disability [19]; this deletion is also linked to autism (SFARI.org).
The mouse NIPA1 mutants, p.Thr39Arg and p.Gly100Arg, corresponding to the respective human mutants are associated with hereditary spastic paraplegia (HSP) showing a loss-of-function when expressed in oocytes and altered trafficking in transfected COS7 cells [15]. The NIPA1 gene normally encodes an Mg2+ transporter protein and the loss-of-function of NIPA1, due to abnormal trafficking of the mutated protein provides the basis for the HSP phenotype [15,16,20]. Only abnormal trafficking of the mutated protein is causative for HSP and not from its deficiency as evident by Prader–Willi syndrome (PWS) or Angelman syndrome (AS) with only one copy of the NIPA1 gene in the typical 15q11-q13 type I deletion in both PWS or AS involving breakpoints BP1 and BP3 including the four genes in the 15q11.2 BP1-BP2 region. Other genes or transcripts, both imprinted and biallelic, do not have HSP [16,21]. Key findings in PWS and AS develop from errors of imprinting dependent on the parent of origin [22,23]. Therefore, SPG6 linked HSP is more likely caused by a dominant-negative effect or a toxic gain-of-function mechanism rather than a loss-of-function [16,17,21,24,25].
A perusal of genes involved in numerous other forms of spastic paraplegia indicates causation by genetic defects in various other metabolic pathways unrelated to magnesium transporter protein deficiency (see GeneCards.org). The syntenic NIPA2 gene, which also codes a magnesium ion transmembrane transporter protein and along with NIPA1 in the same region are both deleted in the Type I deletion but not deleted in the smaller typical 15q11-q13 Type II deletion in PWS or AS. However, patients with the larger Type I deletion do not have spastic paraplegia (GeneCards.org). Mutations in the NIPA2 gene have been reported in generalized epilepsy and childhood absence epilepsy [10,11,26]. In addition to the magnesium ion transmembrane transporter activity, NIPA1 protein inhibits Bone Morphogenetic Protein signaling by regulating the endosomal trafficking and degradation of type 2 BMP receptors (BMPR2) in Drosophila and HeLa cells [27,28].
Bone Morphogenetic Proteins (BMPs) are a group of signaling molecules that belong to the Transforming Growth Factor-β (TGFβ) superfamily of proteins. Initially discovered for their ability to induce bone formation, BMPs are now known to play crucial roles in all organ systems. BMPs are important in embryogenesis and development, and also in the maintenance of adult tissue homeostasis. Another relevant role of BMP in the neurological system is neurogenesis. Neural defects are associated with loss of BMP function in mouse models [29]. For instance, BMP11 is involved in spinal cord neurogenesis and secreted from neurons themselves serving as an inhibitory signal in the generation of new neurons from progenitors in the olfactory epithelium [30].
In rodents, BMP signaling is upregulated following lesions of the corticospinal track and suppressing upregulation that promotes regrowth of axons [31]. Targeting specific BMP receptor subunits for therapeutic purposes may provide an approach for manipulating gliosis and enhancing functional outcomes after spinal cord injury [32]. Therefore, it is no surprise that BMP signaling cuts across all hereditary spastic paraplegia (HSP) categories and are widely implicated in neurodegenerative diseases [33]. Among the HSP proteins, NIPA1 is best characterized mechanistically to inhibit BMP signaling [15]. In summary, these studies support abnormal BMP signaling in many cases probably resulting from abnormal BMP receptor trafficking and could be a unifying pathogenic mechanism for some forms of hereditary spastic paraplegia [34].
Further highlighting the importance of BMP regulation in neural development is the role of BMP7 in corticogenesis; BMP7 deletions result in reduced cortical thickening and impaired neurogenesis [35]. Interestingly, BMPR type 1A (BMPR1A) is important in the establishment of neurons involved in regulating feeding behavior [32]. Similarly, NIPA1 was shown to inhibit BMP signaling by regulating the endosomal trafficking and degradation of type 2 BMP receptors (BMPR2) in Drosophila and HeLa cells [27,28]. Perhaps, haploinsufficiency of NIPA1 protein in cases of 15q11-q13 BP1 or BP2/BP3 deletions might partially affect its inhibition of BMP signaling by regulating endosomal trafficking and degradation of type 2 BMP receptors, thereby affecting neurons. This might be the causal factor for the reported developmental and language delay, neurobehavioral disturbances, and psychiatric problems, such as autism, seizures, and schizophrenia with occasional mild dysmorphic features seen to a varying degree in patients with the 15q11.2 BP1-BP2 deletion (Burnside–Butler) syndrome [1,36]. Biological processes (GO), molecular functions (GO), cellular components with KEGG, and Reactome pathways are summarized in Table 3. In Table 4 examples of diseases or disorders which have been recognized and associated with the NIPA1 gene when disturbed are collated and ranked by order using MalaCards (MalaCards.org) from GeneCards (GeneCards.org). STRING functional interactions with nodes and edges (see Figure 3). Further illustrations of interactions and functions of NIPA1 with relationship to other proteins can be found in Table Table 5.

Table 3.
STRING: Biological Processes (GO), Molecular Function (GO), Cellular Components with KEGG, and Reactome Pathway for the NIPA1 Gene.

Table 4.
Putative Diseases or Disorders Identifiable Using GeneCards.org, HGMD, and DISEASES as Referenced Sources for the NIPA1 Gene.
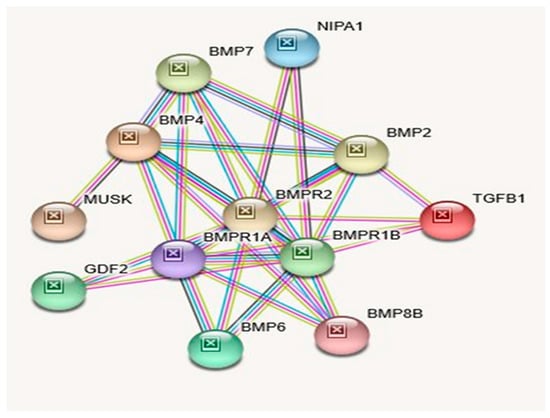
Figure 3.
STRING Protein–Protein Interaction Network for NIPA1 gene with functional interactions with 12 nodes (Source: STRING.org). Network nodes represent proteins with splice isoforms or post-translational modifications collapsed into each node for all proteins produced by a single protein-coding gene. Edges represent protein-protein associations that are considered specific and meaningful (i.e., proteins jointly contribute to a shared function, such as, Biological Process (GO) and Molecular Function (GO) (Table 3). STRING interactants and their functions related to other proteins associated with NIPA1 gene are listed in Table 5. (See Figure 2 for legend with a description of symbols and notes on interpreting the interaction of genes and their encoded proteins).

Table 5.
STRING Interactants and Their Functions Related to Other Proteins Associated with NIPA1 Gene.
Interacting Proteins for NIPA1 Gene: STRING Interaction Network.
2.3. NIPA2 (Non-Imprinted in Prader–Willi/Angelman Syndrome Region Protein 2) Gene
Attributes, Location, Description, Function, and Associated Disorders for NIPA2 Gene.
The chromosomal band location for the NIPA2 gene is 15q11.2 with genomic location (GRCh38/hg38) at 22,838,641- 22,868,384 with a size of 29,744 bases in a plus-strand orientation. This gene is protein-coding specifically for NIPA magnesium transporter 2 (https://www.genecards.org/cgi-bin/carddisp.pl?gene=NIPA2&keywords=nipa2). When disturbed (deleted), this gene is associated with both Angelman and Prader–Willi syndromes due to specific parent of origin deletions of the 15q11-q13 region. The typical 15q11-q13 type I deletion also includes the other three protein-coding genes in the 15q11.2 BP1-BP2 region, namely, NIPA1, CYFIP1, and TUBGCP5. In the case of the smaller typical 15q11-q13 type II deletion, these four genes remain intact in the region (Figure 1) [1,2,3].
The NIPA2 protein has the following attributes: Size: 360 amino acids; Molecular mass:39185 Da. Quaternary structure: No Data Available. This protein has two isoforms; isoform 2 differs from the canonical sequence as follows: 47–66: GQGGHAYLKEWLWWAGLLSM → V. Similar to NIPA1, NIPA2 is also a multi-pass membrane protein found in early endosomes recruited to the cell membrane in response to low extracellular magnesium (UniProtKB/Swiss-Prot for NIPA2 Gene) (Table 6).

Table 6.
Biological Process (GO), Molecular Function (GO), Cellular Component (GO) with KEGG, and Reactome Pathways for the NIPA2 Gene.
Unlike NIPA1, this gene is overexpressed in B-lymphocytes and the placenta ((UniProtKB/Swiss-Prot). In addition to PWS and AS involvement for this gene, it is also associated with childhood absence epilepsy [26], childhood electroclinical syndrome [10,11], and possibly autosomal recessive congenital ichthyosis (MalaCards/GeneCards) and important biological processes and functions (see Table 7).

Table 7.
Putative Associated Diseases for the NIPA2 Gene.
Among related pathways of this gene are miscellaneous transport and binding events and transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds. Gene Ontology (GO) annotations related to this gene also include magnesium ion transmembrane transporter activity. An important paralog of this gene is NIPAL1 (GeneCards). Chai et al. [9] determined that the NIPA2 gene contains 7 exons and spans 29 kb. The coding region extends between exons 3 and 7, and alternative splicing utilizes alternate exons 2 and 2b which results in multiple transcript variants. Pseudogenes of this gene are found on chromosomes 3, 7, and 21 (Entrez Gene: 81614).
Similar to NIPA1, this multi-pass cell membrane protein is localized in the plasma membrane and early endosomes (Gene Ontology (GO)—GO:0005769; GO:0005886) and recruited to the cell membrane in response to low extracellular magnesium (GeneCards; ECO:0000250). Paralogs for the NIPA2 gene are NIPA1, NIPAL1, NIPAL4, NIPAL2, and NIPAL3 (PMID: 28514442; [35]). Although NIPA2 is highly conserved (PMID: 14508708; [9]) and expressed in parts of the brain as well as in most other organ systems, it is significantly overexpressed in B-lymphocytes and the placenta. In addition, five variants have been reported for this gene with either loss or gain of function or both (GeneCards). Table 8 describes the STRING protein interactions and their functions with this gene and description while Table 6 shows biological processes, molecular functions, cellular components, and KEGG and Reactome pathways attributed to the NIPA2 gene. To examine NIPA2 protein interactions and connectivity with other genes and their interrelated proteins, see the STRING interaction network displayed in Figure 4.

Table 8.
STRING Protein Interactants and Their Functions for NIPA2.
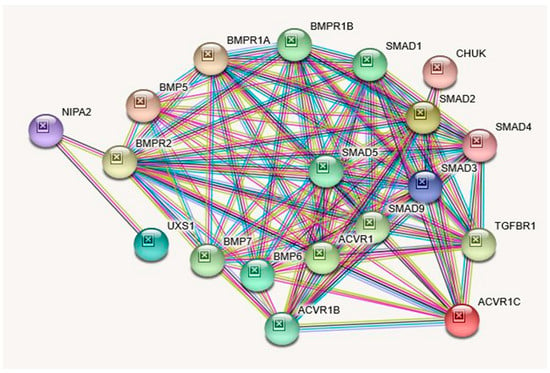
Figure 4.
STRING Protein–Protein Interaction Network for NIPA2 gene with functional interactions with 17 nodes. (Source: STRING.org). Network nodes represent proteins with splice isoforms or post-translational modifications collapsed into each node for all proteins produced by a single protein-coding gene. Edges represent protein-protein associations that are considered specific and meaningful (i.e., proteins jointly contribute to a shared function, such as, Biological Process (GO) and Molecular Function (GO) (Table 6). NIPA2 gene protein Interactants and their functions are listed in Table 8. (See Figure 2 for legend with a description of symbols and notes on interpreting the interaction of genes and their encoded proteins). STRING Protein Interactants and Their Functions for NIPA2 gene are listed in Table 8.
Protein–Protein Inter-Relationships Involving the NIPA2 Gene.
2.4. CYFIP1 (Cytoplasmic FMR1 Interacting Protein 1) Gene
Attributes, Location, Description, Function, and Associated Disorders for CYFIP1 Gene.
The chromosomal band location for the CYFIP1 gene is 15q11.2 with the genomic location (GRCh38/hg38) at 22,867,052–22,981,063 with a size of 114,012 bases in a minus strand orientation (Genecards.org). The CYFIP1 protein has the following attributes: Size: 1253 amino acids; Molecular mass: 145,182 Da. Quaternary structure: Component of the WAVE1 complex composed of ABI2, CYFIP1 or CYFIP2, BRK1, NCKAP1, and WASF1/WAVE1. CYFIP1 and CYFIP2 are part of the Wiskott–Aldrich syndrome protein-family verprolin-homologous protein (WAVE) complex that regulates actin polymerization at synapses and CYFIP1 protein has 3 described isoforms produced by alternative splicing (https://www.uniprot.org/uniprot/Q7L576) (see Table 9). This protein is required for neuronal and bristle development in Drosophila [37]. Through the CYFIP1 protein, the fragile X syndrome protein represses activity-dependent translation, and thus implicated in fragile X syndrome [38].

Table 9.
STRING: Biological Process (GO), Molecular Function (GO), Cellular Component (GO) with KEGG, and Reactome Pathways for the CYFIP1 Gene.
The CYFIP1 gene is highly expressed in the perinuclear region and enriched in synaptosomes. It is also enriched in membrane ruffles and at the tips of lamellipodia with the following subcellular localizations (GeneCards & UniProtKB/Swiss-Prot). The CYFIP1 gene is widely expressed in all tissues (GenecCards). In addition to this gene’s association with fragile X syndrome, it is also associated with autism (see Table 10). A large chromosomal deletion including this gene is associated with increased risk of schizophrenia and epilepsy in human patients and reduced expression of this gene has also been observed in various human cancers, the encoded protein may inhibit tumor invasion (Entrez Gene summary: RefSeq, May 2017). Among this gene’s related pathways are the regulation of actin dynamics for phagocytic cup formation and signaling by Rho GTPases (Table 9). Gene Ontology (GO) annotations related to this gene include Rac GTPase binding. An important paralog of this gene is CYFIP2 (GeneCards). Table 10 lists putative associated diseases for CYFIP1 Gene. STRING protein interactions with their functions and descriptions related to CYFIP1 are presented in Table 11. An illustration of the identified interactions can also be visualized in Figure 5.

Table 10.
Putative Associated Diseases for CYFIP1 Gene.

Table 11.
STRING Protein–Protein Interactants and Their Functions for CYFIP1.
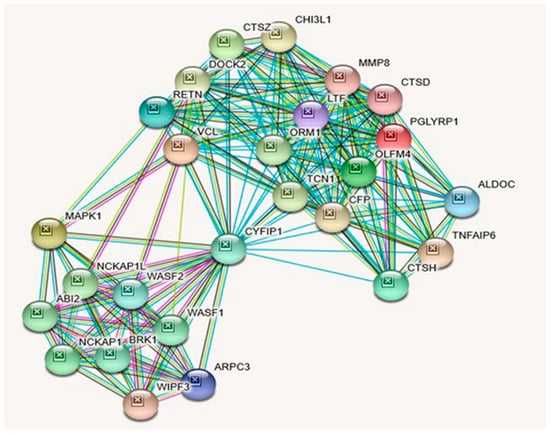
Figure 5.
STRING Protein–Protein Interaction Network for CYFIP1 gene with functional interactions with 26 nodes. (Source: STRING.org). Network nodes represent proteins with splice isoforms or post-translational modifications collapsed into each node for all proteins produced by a single protein-coding gene. Edges represent protein-protein associations that are considered specific and meaningful, i.e., proteins jointly contribute to a shared function, such as, Biological Process (GO) and Molecular Function (GO) (Table 9). CYFIP1 gene protein–protein interactants and their functions are listed in Table 11. (See Figure 2 for legend with a description of symbols and notes on interpreting the interaction of genes and their encoded proteins).
Protein–Protein Inter-Relationships Involving CYFIP1 Gene.
2.5. TUBGCP5 (Tubulin Gamma Complex Associated Protein 5) Gene
Attributes, Location, Description, Function, and Associated Disorders for TUBGCP5 Gene.
The chromosomal band location for TUBGCP5 gene is 15q11.2 with the genomic location (GRCh38/hg38) at 22,983,192–23,039,673 with a size of 56,482 bases in a minus strand orientation (Genecards.org). TUBGCP5 protein has the following attributes: Size: 1024 amino acids; Molecular mass:118321 Da; Quaternary structure: Gamma-tubulin complex which is composed of gamma-tubulin, TUBGCP2, TUBGCP3, TUBGCP4, TUBGCP5 and TUBGCP6 proteins (GeneCards & UniProtKB/Swiss-Prot). This protein has 2 isoforms produced by alternative splicing; the sequence of this isoform differs from the canonical sequence as follows: 1010–1024: LESLALSLMAGMEQS → CEYIMLKYFYLCISL. TUBGCP5 gene is widely expressed with the highest levels in the heart and skeletal muscle with moderate levels in the brain (GeneCards & UniProtKB/Swiss-Prot). An important paralog of this gene is TUBGCP6.
Table 12 shows biological processes, molecular functions, cellular components with KEGG and Reactome pathways related to TUBGCP5. Among its related pathways are Nanog in Mammalian ESC Pluripotency and G-Beta Gamma Signaling. Gene Ontology (GO) annotations related to this gene include microtubule binding, and components of the cytoskeleton (Table 12). Table 13 shows diseases associated with TUBGCP5 gene disturbances based on MalaCards and arranged in descending order including Prader–Willi syndrome as the top disorder followed by schizophrenia and then autism. Table 13 shows proteins and their functions that interact with TUBGCP5, as depicted in Figure 6, illustrating the interactions of the TUBGCP5 gene with other related genes.

Table 12.
STRING: Biological Process (GO), Molecular Functions (GO), Cellular Components (GO) with KEGG, and Reactome Pathways for the TUBGCP5 Gene.

Table 13.
Putative Associated Diseases for the TUBGCP5 Gene.
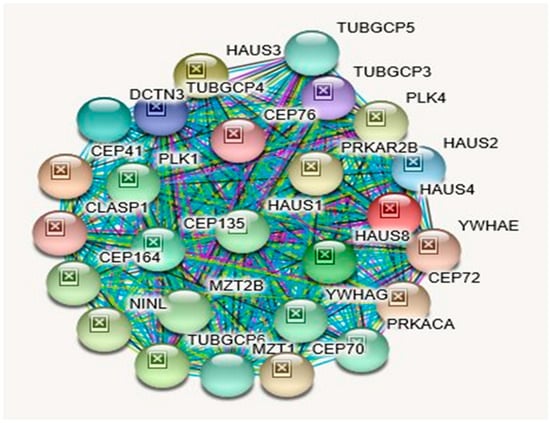
Figure 6.
STRING Protein–Protein Interaction Network for TUBGCP5 gene with functional interactions with 26 nodes (Source: STRING.org). Network nodes represent proteins with splice isoforms or post-translational modifications collapsed into each node for all proteins produced by a single protein-coding gene. Edges represent protein-protein associations that are considered specific and meaningful i.e., proteins jointly contribute to a shared function, such as, Biological Process (GO) and Molecular Function (GO) (Table 12). TUBGCP5 gene interactants and their functions are listed in Table 14. (See Figure 2 for legend with a description of symbols and notes on interpreting the interaction of genes and their encoded proteins).
Protein–Protein Inter-Relationships Involving the TUBGCP5 Gene.
3. Discussion
Features seen in patients with the 15q11.2 BP1-BP2 deletion (Burnside–Butler) syndrome can be quite variable. Different clinical phenotypes are seen in children with this disorder and can depend on the source of the parental deletion [36]. Recent evidence of partial expression or bias of these four genes depending on the parent of origin leads to possible clinical differences in the offspring. For example, when the deletion is paternal, there is a greater risk of having congenital heart defects [36]. If the mother transmits the deletion, then there is a greater risk of intellectual disability and autism in their affected children [36]. A recent review further showed a phenotype with global and regional measures of surface area and cortical thickness in the brain with subcortical volumes and cognition differences compared to controls. This investigation consisted of 203 individuals compared with 4500 controls without the deletion collected from years 2015–2019 and the average age was 56 years [5]. This international study analyzed the largest CNV and neuroimaging results to date in those with the 15q11.2 BP1-BP2 deletions and non-deletion controls. They reported reduced brain surface area, a thicker cortex, and a smaller nucleus accumbens in the 15q11.2 BP1-BP2 deletion subjects. The significant difference in cortical thickness was more evident in the frontal, cingulate and parietal lobes. Furthermore, cognitive ability was lower for those with the deletion compared with non-deletion individuals suggesting involvement in neural plasticity and development leading to functional brain differences identified clinically [5].
As noted in Table 15, all four genes are protein-coding genes and neither were maternally nor paternally imprinted. Their encoded proteins interact with each other in crucial biological processes and molecular pathways (Figure 2, Table 1 and Table 2). The predicted functional interactions encompassing all four genes included 11 nodes with 34 edges that pertain to biological processes and molecular functions utilizing searchable genomic databases involving the four genes in this 15q11.2 BP1-BP2 microdeletion syndrome (see Table 1). Among them, in addition to magnesium ion transport molecular function, significant other biological processes, such as regulation of axonogenesis and axon extension, regulation and production of bone morphogenetic protein (BMP), regulation of cellular growth and development were observed. These biological processes are relevant to variable clinical phenotypes, such as, autism, seizures, and schizophrenia which are also seen in this microdeletion syndrome. These clinical phenotypes often affect neurological development and are occasionally accompanied by mild dysmorphic features [1,9,10,11,12,13].

Table 15.
Summary of Functions, Nature, Compartmentalization, Related Pathways, and Cardinal Diseases Associated with NIPA1, NIPA2, CYFIP1, and TUBGCP5 Genes in the 15q11.2 BP1-BP2 Region.
Although NIPA1 and NIPA2 share related KEGG and Reactome pathways (Table 3 and Table 6), their cardinal disease associations are different (Table 15): Spastic Paraplegia 6, Autosomal Dominant and Spastic Paraplegia 6 for NIPA1 and Angelman Syndrome and Prader–Willi Syndrome for NIPA2. However, as shown in Table 16, they both are also associated with PWS and AS. Unlike NIPA1 and NIPA2, CYFIP1, and TUBGCP5 genes do not share any KEGG or Reactome pathways (Table 9 and Table 12). As shown in Table 15, cardinal diseases that are associated with the CYFIP1 gene are Fragile X Syndrome and Autism. The cardinal disease that is associated with the TUBGCP5 gene is Prader–Willi syndrome. However, as depicted in Table 16, they both are additionally associated with PWS, while the CYFIP1 gene alone is additionally associated with AS. Most notably, all four syntenic genes in this region are associated with Autism Spectrum Disorder; Schizophrenia; Epilepsy and Down Syndrome (Table 16). Except for TUBGC5, all three genes are associated with the 15q11.2 BP1-BP2 Deletion Syndrome with Attention Deficit Hyperactive Disorder & Learning Disability (Table 16). It should be noted that interacting proteins for CYFIP1 and TUBGCP5 proteins are quite different (Figure 5, Table 11; Figure 6, Table 14, respectively), which is unlike the case of NIPA1 and NIPA2 (Figure 3, Table 5; Figure 4, Table 8, respectively).

Table 16.
Summary of Associated Neurodevelopmental Maladies Across All Four Genes.

Table 14.
STRING Interactants and Their Functions for the TUBGCP5 Gene.
It is interesting to note that none of the protein-protein interacting genes for the four genes, which total 70, are the same (Table 5, Table 8, Table 11, and Table 14). However, NIPA1 and NIPA2 proteins, given their involvement with common biological processes (GO) and molecular functions (GO) along with BMP6, BMP7, BMPR1A, BMPR1B, and BMPR2-proteins interact with both. In contrast, none of the protein-protein interactants are similar between CYFIP1 and TUBGCP5 proteins (Table 11 and Table 14), thus affirming their functional dissimilarities.
It should be noted, as illustrated in STRING Figure 2 and Table 1, NIPA1, NIPA2, CYFIP1, and TUBGCP5 proteins show protein–protein interactions among themselves with the 11 nodes and 34 edges addressed earlier. As shown in Table 1, NIPA1 protein interacts with TUBGCP5, CYFIP1, and NIPA2 proteins (scores: 0.995, 0.967, and 0.941, respectively) along with seven other protein interactants, and thus predicted to cause significant biological processes (Table 2), and a significant molecular function is magnesium ion transmembrane transporter activity with a 0.0042 false discovery rate (Table 2).
The varied biological processes and predicted functions of these four genes, as noted above, through their protein–protein interactions with seven other proteins (see Figure 2) collectively could play a role in the neurodevelopmental disorders. They complement regulation associated with lipoproteins and lipid metabolism and adipogenesis, encompassing CFHR1, CFHR3, and BMPR2 protein–protein interactions, given marked obesity is a common finding in PWS.
Examining the interaction of NIPA1 protein with its 11 other interacting proteins indicates that three-fourths of the interactions are important for developmental bone morphogenesis or multifunctional proteins that control proliferation, differentiation, and other functions in many cell types (see Figure 3; Table 5). The interactions of NIPA2 protein with 19 other proteins indicate their collective role as intracellular signal transducers and transcriptional modulators that are activated by TGFbeta, and thereby impacting bone morphogenesis (see Figure 4; Table 8). CYFIP1 protein interacts with 25 other proteins. These proteins have a wide range of activity with functions including cytoskeleton organization and actin filament binding with cell-matrix adhesion, MAP kinase signal transduction impacts cell growth, survival and differentiation, stimulation of glucose uptake in cells, intracellular protein breakdown and tissue remodeling (see Figure 5; Table 11). TUBGCP5 protein interacts with 25 other proteins. These proteins are focused on mitotic spindle formation and assembly, microtubule organization, and production of centrosomal proteins involved in the regulation of centriole duplication during cell division (see Figure 6; Table 14).
3.1. Overlapping Association of PWS and AS Specific Genes with Both Syndromes at Varying Degrees
Although UBE3A is a classical AS gene (9.31 score), it is also associated with PWS to a lesser degree (4.106 score) (https://www.malacards.org/search/results/UBE3A). Further, it should be noted that the 15q11-q13 region includes the imprinted gene cluster with two maternally expressed protein-coding genes UBE3A and ATP10A (https://www.malacards.org/search/results?query=ATP10A). Similarly, all of the classical PWS genes overlap with both PWS and AS, albeit to varying degrees of significance: MAGEL2: PWS/AS: SCORE: 7.035/3.937; NDN: PWS/AS: SCORE: 7.435/4.528; NDAP1: PWS/AS: SCORE: 6.321/4.816; SNRPN: PWS/AS: SCORE: 6.563/5.584; SNORD116: PWS/AS: SCORE: 0.426/0.121; and even the IPW region: PWS/AS: SCORE: 7.761/0.100 (https://www.malacards.org). Therefore, it may not be appropriate to assume that PWS/AS are exclusively caused only by the designated PWS and AS regional genes per se, as shown in Figure 1, since most of these genes are also overlappingly associated with both PWS and AS, although to varying degrees. Moreover, PWS/AS phenotype is also overlappingly associated with three proximally located contiguous genes within the 15q11.2 BP1-BP2 region: NIPA1, NIPA2, and CYFIP1, as depicted in Table 16.
PWS is caused by loss of expression of the paternally inherited genes on chromosome 15q11.2-q13, and the cardinal features of PWS are attributable to the critical interval within the 15q11.2-q13 imprinted gene cluster, containing the small nucleolar RNA (snoRNA) SNORD116 and non-coding RNA IPW (Imprinted in Prader–Willi) exons [39,40,41]. Similarly, the cardinal features of AS are attributable to the loss of expression of maternally inherited genes, and UBE3A in particular, which causes a distinct AS neurodevelopmental disorder [42].
Although the PWS core features are attributable to the critical interval within the 15q11.2-q13 imprinted gene cluster and the AS core features are attributable to the loss of expression of maternally inherited gene UBE3A, these core features might be influenced by several other genes that lie within the broader BP1-BP3 region. The functions of all of these genes are in-turn likely modulated by hundreds of their interacting genes that lie throughout the genome, as shown in the STRING protein interactions for NAPI1, NAPI2, CYFIP1, and TUBGCP5: Figure 2, Table 1; Figure 3, Table 5; Figure 4, Table 8; Figure 5, Table 11; Figure 6, Table 14; Table 15 and Table 16. One could speculate that PWS/AS, similar to that of ASD, depends on either the involvement of the PWS critical region in the imprinted segment within the 15q11.2-q13 BP2-BP3 region or the involvement of a critical gene, namely, UBE3A in the maternally imprinted segment [43] and in concert with other genes that lie within chromosome 15, not excluding the 15q11.2 BP1-BP2 region genes.
Every one of these genes along with their hundreds of interacting genes within the genome as a whole, as depicted and tabulated in this study pertaining to NIPA1, NIPA2, CYFIP1, and TUBGCP5 genes all in concert could impact on the PWS/AS signature phenotype in any given proband. Thus, each PWS/AS proband’s clinical presentation is likely to be a unique genomic signature, within the broader spectrum of PWS/AS clinical presentations, which is akin to Autism Spectrum Disorder.
3.2. 15q11.2 B-P1-BP2 Microdeletion (Burnside–Butler) Syndrome: Characterization of Genes Within the BP1-BP2 Region, Meta-Analysis, and Parent-of-origin Effects Reveal Neurodevelopmental Associated Phenotypes
Each of the four evolutionarily conserved genes that escape imprinting and contiguously lie within the BP1-BP2 region are associated with distinct cardinal diseases: NIPA1: Spastic Paraplegia 6, Autosomal Dominant and Spastic Paraplegia 6; NIPA2: Angelman Syndrome and Prader–Willi Syndrome; CYFIP1: Fragile X Syndrome and Autism; and TUBGCP5: Prader–Willi Syndrome (see Table 15). Based on reports of their mutations or disturbed expression patterns, as reported in authentic web resources, such as, GeneCards.org, SAFARI.org, Gene Ontology (GO), OMIM.org, Entrez and UniProtKB/Swiss-Prot, their “collective loss or dosage duplication” is due to the BP1-BP2 microdeletion or microduplication that appears to impact only behavioral and neurological functions. These disorders include speech and motor delays, behavioral problems, seizures, and autism in affected individuals [44]. A recent review of the 15q11.2 BP1–BP2 microdeletion syndrome [1] found common phenotypic features, which included autism, developmental delay, motor and language delays, and behavioral problems.
Parental studies among these subjects demonstrated phenotypically normal carriers in several instances, and mildly affected carriers in others, complicating phenotypic association and/or causality. This could be due to either reduced penetrance or altered gene dosage on a particular genetic background [1,4]. The four non-imprinted and biallelically expressed genes, NIPA1, NIPA2, CFYIP1 and TUBGCP5, in this microdeletion were initially noted to impact the severity of clinical presentation and neurological impairment in these two classical genomic imprinting disorders, Prader–Willi and Angelman syndromes with typical 15q11–q13 deletions depending on the absence or presence of the genomic area between breakpoints BP1 and BP2 containing these four genes, which led to the recognition of this microdeletion (Burnside–Butler) syndrome [4].
Therefore, it is intriguing to note that the review [4] of over 200 15q11.2 BP1–BP2 microdeletion syndrome individuals, as meta-analysis, did not find either the NIPA1 associated cardinal disease: Spastic Paraplegia, the NIPA2 associated cardinal diseases: Angelman Syndrome and Prader–Willi Syndrome, the CYFIP1 associated cardinal disease: Fragile X Syndrome, or the TUBGCP5 associated cardinal disease: Prader–Willi Syndrome, except for the CYFIP1 associated cardinal disease: Autism, as presented in Table 15.
The clinical features as noted in the review [4] included developmental (73%) and speech (67%) delays; dysmorphic ears (46%) and palatal anomalies (46%); writing (60%) and reading (57%) difficulties, memory problems (60%) and verbal IQ scores ≤75 (50%); general behavioral, unspecified (55%) and abnormal brain imaging (43%). Other clinical features noted in this review [4], but not considered as common were seizures/epilepsy (26%), autism spectrum disorder (27%), attention deficit disorder (ADD)/attention deficit hyperactivity disorder (ADHD) (35%), schizophrenia/paranoid psychosis (20%), and motor delay (42%).
The review [4] also found that not all individuals with this deletion were clinically affected, but neuropsychiatric and behavior disturbances and mild dysmorphic features were associated with genomic imbalances of the 15q11.2 BP1–BP2 region, including microdeletions, but with an apparent incomplete penetrance and variable expressivity.
3.3. All Four BP1-BP2 Region Genes Are Significantly Associated with Autism Spectrum Disorder
From reported patient cohorts presenting for genetic services and microarray analysis, this microdeletion syndrome can now be recognized as the most common cytogenetic abnormality found in ASD instead [6], which is not only a cardinal CYFIP1 associated disease (see Table 15), but ASD is also consistently associated with TUBGCP5, NIPA1, and NIPA2 genes (see Table 4, Table 7, Table 10, Table 13, and Table 16).
Thus, all four genes in this narrow segment between BP1 and BP2 are significantly associated with autism spectrum disorder, with an average of >68 MalaCards InFormaTion Score (MIFTS) with annotation strength (max 100) and with Solr (an open-source enterprise-search platform) relevance scores 0.171, 0.088, 4.578/0.377, and 0.21/0.094 for NIPA1, NIPA2, CYFIP1, and TUBGCP5, respectively (Table 4, Table 7, Table 10, Table 13, and Table 16). Moreover, even PWS and AS specific genes that lie in the BP2-BP3 PWS/AS region, such as, MAGEL2, SNRPN, UBE3A, and ATP10A are also significantly co-associated with autism (Malacards.org; SFARI.org) [45]. Therefore, it seems fair to conclude that except for PWS specific genes or transcripts, such as, snoRNAs, most other genes that flank on either side in broadly designated PWS and AS regions are all co-associated with autism. Most of the genes within the broader BP1-BP3 region are also co-associated with not only PWS and AS, but also with autism (Malacards.org; SFARI.org) [45]. Even the GABRB3 gene that is distal to the ATP10A gene is a SFARI (SFARI.org) recognized autism gene (Malacards.org; SFARI.org) [45].
3.4. 15q11.2 B–P1–BP2 Microdeletion (Burnside–Butler) Syndrome: Frequency
The 15q11.2 BP1–BP2 microdeletion syndrome has a reported de novo frequency between 5%–22%, with 51% having inherited the microdeletion from an apparently unaffected parent and 35% having inherited the microdeletion from an affected parent [4,6]. These low penetrance estimates may relate to subclinical manifestations of neuropsychiatric/behavioral problems, incomplete information, or lack of detailed clinical or psychiatric studies in the parents of individuals with 15q11.2 BP1–BP2 microdeletion or members of control cohorts.
3.5. Summary
The four genes within the narrower proximal BP1-BP2 region, NIPA1, NIPA2, CYFIP1, and TUBGCP5, as well as those that lie within the broader BP2-BP3 region, such as MAGEL2, SNRPN, UBE3A, ATP10A, as well as GABRB3 gene that are farther away from the distal ATP10A gene- are all recognized ASD genes. Furthermore, the 15q11.2 region collectively represents PWS, AS, and autism phenotypes (Malacards.org; SFARI.org) [45].
In addition to the above consistent correlation of each of the four genes that lie within the narrow proximal BP1-BP2 region with autism, in a more recent study [36] parent-of-origin effects (POE) of the 15q11.2 BP1-BP2 microdeletion were found to be associated with differences in clinical features in individuals inheriting the deletion. Among all probands studied, maternal deletions were found to be associated with epilepsy, autism spectrum disorder (p = 0.02) and macrocephaly (p = 0.016), while paternal deletions were associated with congenital heart disease (CHD) (p = 0.004) and abnormal muscular phenotypes (p < 0.05), while CHD and abnormal muscular phenotypes were seen in paternal deletions. This study not only supported POEs of this deletion, but notably included ASD, macrocephaly, and epilepsy.
The above key processes and summary of the four genes within the narrower proximal 15q11.2 BP1-BP2 region are collectively critical for normal neuronal development, plasticity, and function [1,5]. Their loss or gain (CNV) are associated with neurodevelopmental disorders, spastic paraplegia, seizures, learning, and gait disturbances with motor delay and neuro-behavioral psychiatric problems including autism, dyslexia, and schizophrenia/paranoid psychosis [1].
The most common maladies found to be individually associated with each of the four genes are: spastic paraplegia found in the top 12 of 15 disorders associated with NIPA1; PWS/AS, epilepsy and psychiatric/behavioral problems (autism or schizophrenia) in the top 10 disorders for NIPA2; fragile X syndrome, autism, schizophrenia, PWS and pervasive developmental disorder in the top 10 disorders for CYFIP1; and PWS, schizophrenia, autism, microcephaly, essential hypertension, body mass index (quantitative trait), epilepsy, and Down syndrome in the top 10 disorders associated with TUBGCP5 (see Table 16). As noted in Table 15, all four genes are neither maternally nor paternally imprinted, but their proteins interact with each other as described and illustrated (see Figure 2, Table 1 and Table 2) in crucial biological processes (GO) and molecular pathways (GO). Collectively, either due to the microdeletion or microduplication, these genes predominantly affect brain morphology [1,4].
Our detailed exploration of the various aspects of each of these four genes, in solo and in concert is meant to enable an in-depth understanding of their individual and collective contribution in the causation of different neurodevelopmental phenotypes that have been variably reported in the literature. All four genes are individually associated with Prader–Willi syndrome, autism spectrum disorder, schizophrenia, epilepsy, and Down syndrome (Maladies.org; GeneCards.org). Except for the TUBGCP5 gene, all three remaining genes are associated with Angelman syndrome, although the TUBGCP5 gene is located most proximal to the Angelman syndrome gene, UBE3A. TUBGCP5 gene is also not associated with attention deficit hyperactivity disorder and learning disability, developmental disorder, and peripheral nervous system disease. CYFIP1 is the only gene that is not associated with microcephaly, but it is the only gene that is associated with a developmental disorder (see Table 16).
Thus, collectively, all four genes are associated with ten overlapping neurodevelopmental maladies up to 77.5% of the time.
4. Materials and Methods
We characterized the four genes (NIPA1, NIPA2, CYFIP1, and TUBGCP5) found within the 15q11.2 BP1-BP2 chromosome region of about 500 bp in size. When this region is deleted, an emerging disorder (i.e., Burnside–Butler syndrome) is being characterized leading to the importance of our description of these four genes. We undertook a literature review of hundreds of patients reported and summarized clinically with this microdeletion reported as the most common genetic defect found in consecutive patients presenting for genetic testing with high-resolution microarray analysis and autism spectrum disorder (e.g., [6]). Genome databases, search platforms for gene interaction, gene ontology (GO) biological process, molecular function, cellular components, along with KEGG and Reactome pathways using STRING network data along with sources, GeneCards.org, SAFARI.org, KEGG, PubMed, Gene Ontology (GO), OMIM.org, Entrez and UniProtKB/Swiss-Prot were summarized in tabular form and STRING Network results illustrated in figures and discussed in the text involving all four genes. An overview of each gene was also described throughout the summary process utilizing an approach reported previously [46].
The Gene Ontology Resource (GO; http://geneontology.org) provides structured, computable knowledge regarding the functions of genes and gene products. Founded in 1998, GO has become widely adopted in the life sciences, and its contents are under continual improvement, both in quantity and in quality. The Gene Ontology resource (GO; http://geneontology.org) is the most comprehensive and widely used knowledge base concerning the functions of genes. In GO, all functional knowledge is structured and represented in a form amenable to computational analysis, which is essential to support modern biological research. The GO knowledge base is structured using a formal ontology, by defining classes of gene functions (GO terms) developed over the past 20 years that have specified relations to each other [47]. GO terms include definitions, or equivalence axioms, that define the term relative to other terms in the GO or other ontologies. Their relationships can be computationally inferred using logical reasoning. The GO structure is constantly evolving in response to new scientific discoveries and are continuously refined using current biological information. The GO knowledgebase consists of ontology and annotations. As of the 5 September 2018 release (doi:10.5281/zenodo.1410625), there were ∼45000 terms in GO: 29698 biological processes, 11147 molecular functions, and 4201 cellular components, linked by almost 134000 relationships [47].
MalaCards Relevance Scores provide information used for the construction of our tables of Putative Associated Diseases (see Table 4, Table 7, Table 10 and Table 13) for the readership. MalaCards is an integrated database of human maladies and their annotations modeled from the architecture and information from the popular GeneCards database of human genes. The search platform used by MalaCards to obtain these data [48] is SOLR, an open-source full-text search platform widely used for analytics based on Apache’s Lucene text search API. When a term is searched by Lucene then it returns a set of scored hits. A “hit” represents a document (a MalaCard), whose fields (actual annotations) were previously indexed by Lucene.
The scoring is calculated by a Lucene defined algorithm:
score(q,d) = coord(q,d) · queryNorm(q) · ∑ tf(t in d) · idf(t) · t.getBoost() · norm(t,d)
t in q
t in q
The factors in this formula are:
- tf stands for term frequency—the more times a search term appears in a document, the higher the score
- idf stands for inverse document frequency—matches on rarer terms count more than matches on common terms
- coord is the coordination factor—if there are multiple terms in a query, the more terms that match, the higher the score
- lengthNorm—matches on a smaller field score higher than matches on a larger field
- index-time boost—if a boost was specified for a document at index time, scores for searches that match that document will be boosted.
- query clause boost—a user may explicitly boost the contribution of one part of a query over another.
To ensure showing the best precision, MalaCards displays the score as (base 2 log of the score) + 10
For more about Lucene’s scoring mechanism see Apache Lucene—Scoring [48].
GeneCards.org—the human gene database [49] is from the GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analysis.
STRING protein network was utilized to generate Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6 and their accompanying data on the protein–protein interactions as well as their Biological Processes (GO), Molecular Functions (GO), Cellular Component (GO) and KEGG and Reactome Pathways. The data are presented in Table 1, Table 2 and Table 3, Table 5, Table 6, Table 8, Table 9, Table 11, Table 12, and Table 14 and are adopted from STRING CONSORTIUM 2019. Data in Table 15 were derived from the GeneCards website (GeneCards.org) for respective genes, while the data for Table 16 were derived from MalaCards diseases (Malacards.org) as seen in Table 4, Table 7, Table 10, and Table 13 in this study.
Author Contributions
Based on M.G.B.’s suggestion of this research topic, S.K.R. independently researched and originally drafted the entire content of this manuscript for which M.G.B. contributed to Figure 1, contributed towards drafting the introduction and critically reviewed and edited the entire manuscript. Title, abstract, all figure legends, Figure 2, Figure 3, Figure 4, Figure 5 and Figure 6, Tabulations 1 through 16, results, interpretations, discussions, and conclusions were exclusively by S.K.R. Table 15 and Table 16 were originally synthesized by S.K.R. from various reputable databases, such as GeneCards.org, MalaCards.org, SAFARI.Org, KEGG, PubMed, Gene Ontology (GO), OMIM.org, Entrez, UniProtKB/Swiss-Prot and from https://compartments.jensenlab.org. All authors have read and agree to the published version of the manuscript.
Funding
We acknowledge the National Institute of Child Health and Human Development (NICHD) grants number HD02528 and KUMC Research Institute and the KUMC Research Institute, Clinical Pilot Research Grant Program.
Acknowledgments
We thank the reviewers for their constructive and valid suggestions and comments. We thank Waheeda Hossain at KUMC for her help in formatting this manuscript and amending Figure 1. We also thank Grace Graham at KUMC for her technical help. Syed Rafi obtained permissions via emails from the following organizations for the reproduction of their website figures and data pertaining to the four genes studied and we are grateful: 1. STRING Corporation 2019: On Nov. 11, 2019 under STRING’s CC BY 4.0′ license. 2. GeneCards.org, on Nov. 11, 2019, Marilyn Safran, Head of GeneCard’s Development. 3. MalaCards.org, on Dec. 23, 2019, Marilyn Safran, Head of GeneCard’s Development, and 4. UniProtKB/Swiss-Prot; Uniprot.Org, on Dec. 23, 2019.
Conflicts of Interest
There are no conflicts of interest noted by either author.
References
- Butler, M.G. Clinical and genetic aspects of the 15q11.2 BP1-BP2 microdeletion disorder. J. Intellect. Disabil. Res. 2017, 61, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Bittel, D.C.; Butler, M.G. Prader-Willi syndrome: Clinical genetics, cytogenetics and molecular biology. Expert Rev. Mol. Med. 2005, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Lee, P.D.K.; Whitman, B.Y. Management of Prader-Willi Syndrome, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Cox, D.M.; Butler, M.G. The 15q11.2 BP1-BP2 microdeletion syndrome: A review. Int. J. Mol. Sci. 2015, 16, 4068–4082. [Google Scholar] [PubMed]
- Writing Committee for the ENIGMA-CNV Working Group; Van Der Meer, D.; Sønderby, I.E.; Kaufmann, T.; Walters, G.B.; Abdellaoui, A.; Ames, D.; Amunts, K.; Andersson, M.; Armstrong, N.J.; et al. Association of copy number variation of the 15q11.2 BP1-BP2 region with cortical and subcortical morphology and cognition. JAMA Psychiatry 2019, 30, 1–11. [Google Scholar]
- Ho, K.S.; Wassman, E.R.; Baxter, A.L.; Hensel, C.H.; Martin, M.M.; Prasad, A.; Twede, H.; Vanzo, R.J.; Butler, M.G. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders using an ultra-high-resolution chromosomal microarray optimized for neurodevelopmental disorders. Int. J. Mol. Sci. 2016, 17, 2070. [Google Scholar] [CrossRef]
- Butler, M.G. Magnesium supplement and the 15q11.2 BP1-BP2 microdeletion (Burnside-Butler) syndrome: A potential treatment? Int. J. Mol. Sci. 2019, 20, 2914. [Google Scholar] [CrossRef]
- Das, D.K.; Tapias, V.; D’’Aiuto, L.; Chowdari, K.V.; Francis, L.; Zhi, Y.; Ghosh, A.; Surti, U.; Tischfield, J.; Sheldon, M.; et al. Genetic and morphological features of human iPSC-derived neurons with chromosome 15q11.2 (BP1-BP2) deletions. Mol. Neuropsychiatry 2015, 1, 116–123. [Google Scholar] [CrossRef]
- Chai, J.-H.; Locke, D.; Greally, J.M.; Knoll, J.H.M.; Ohta, T.; Dunai, J.; Yavor, A.; Eichler, E.E.; Nicholls, R.D. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am. J. Hum. Genet. 2003, 73, 898–925. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, Y.; Zhang, P.; Sang, T.; Zhang, F.; Ji, T.; Huang, Q.; Xie, H.; Du, R.; Cai, B.; et al. NIPA2 located in 15q11.2 is mutated in patients with childhood absence epilepsy. Hum. Genet. 2012, 131, 1217–1224. [Google Scholar]
- Jiang, Y.; Zhang, Y.; Zhang, P.; Zhang, F.; Xie, H.; Chan, P.; Wu, X. NIPA2 mutations are correlative with childhood absence epilepsy in the Han Chinese population. Hum. Genet. 2014, 133, 675–676. [Google Scholar] [CrossRef]
- Jerkovich, A.M.; Butler, M.G. Further phenotypic expansion of 15q11.2 BP1-BP2 microdeletion (Burnside-Butler) syndrome. J. Pediatric Genet. 2014, 3, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Jerkovich, A.M.; Butler, M.G. 15q11.2 Microdeletion (BP1-BP2) and developmental delay, behaviour issues, epilepsy and congenital heart disease: A series of 52 patients. Eur. J. Med. Genet. 2015, 58, 140–147. [Google Scholar]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteomics 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Goytain, A.; Hines, R.; El-Husseini, A.; Quamme, G.A. NIPA1(SPG6), the basis for autosomal dominant form of hereditary spastic paraplegia, encodes a functional Mg2+ transporter. J. Biol. Chem. 2007, 282, 8060–8068. [Google Scholar] [CrossRef]
- Rainier, S.; Chai, J.-H.; Tokarz, D.; Nicholls, R.D.; Fink, J.K. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6). Am. J. Hum. Genet. 2003, 73, 967–971. [Google Scholar] [CrossRef]
- Zhao, J.; Matthies, D.S.; Botzolakis, E.J.; Macdonald, R.L.; Blakely, R.D.; Hedera, P. Hereditary spastic paraplegia-associated mutations in the NIPA1 gene and its Caenorhabditis elegans homolog trigger neural degeneration in vitro and in vivo through a gain-of-function mechanism. J. Neurosci. 2008, 28, 13938–13951. [Google Scholar] [CrossRef]
- Chen, C.-P.; Lin, S.-P.; Lee, C.; Chern, S.-R.; Wu, P.-S.; Chen, Y.-N.; Chen, S.-W.; Wang, W. Familial transmission of recurrent 15q11.2 (BP1-BP2) microdeletion encompassing NIPA1, NIPA2, CYFIP1, and TUBGCP5 associated with phenotypic variability in developmental, speech, and motor delay. Taiwan J. Obstet. Gynecol. 2017, 56, 93–97. [Google Scholar] [CrossRef]
- Leblond, C.S.; Heinrich, J.; Delorme, R.; Proepper, C.; Betancur, C.; Huguet, G.; Konyukh, M.; Chaste, P.; Ey, E.; Råstam, M.; et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012, 8, E1002521. [Google Scholar] [CrossRef]
- Klebe, S.; Durr, A.; Bouslam, N.; Grid, D.; Paternotte, C.; Depienne, C.; Hanein, S.; Bouhouche, A.; Elleuch, N.; Azzedine, H.; et al. Spastic Paraplegia 5: Locus refinement, candidate gene analysis and clinical description. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144B, 854–861. [Google Scholar] [CrossRef]
- Botzolakis, E.J.; Zhao, J.; Gurba, K.N.; Macdonald, R.L.; Hedera, P. The effect of HSP-causing mutations in SPG3A and NIPA1 on the assembly, trafficking, and interaction between atlastin-1 and NIPA1. Mol. Cell Neurosci. 2011, 46, 122–135. [Google Scholar] [CrossRef]
- Butler, M.G. Single gene and syndromic causes of obesity: Illustrative examples. In Progress in Molecular Biology and Translational Science; Tao, Y., Ed.; Elsevier Inc: Chennai, India, 2016; Volume 140, pp. 1–45. [Google Scholar]
- Butler, M.G.; Hartin, S.N.; Hossain, W.A.; Manzardo, A.M.; Kimonis, V.E.; Dykens, E.; Gold, J.A.; Kim, S.-J.; Weisensel, N.; Tamura, R.; et al. Molecular genetic classification in Prader-Willi syndrome: A multisite cohort study. J. Med. Genet. 2019, 56, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Song, C.; Guo, H.; Xu, P.; Huang, W.; Zhou, Y.; Sun, J.; Li, C.-X.; Du, Y.; Li, X.; et al. Distinct novel mutations affecting the same base in the NIPA1 gene cause autosomal dominant hereditary spastic paraplegia in two Chinese families. Hum. Mutat. 2005, 25, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Chen, Y.-J.; Wang, M.-W.; Lin, X.-H.; Dong, E.-L.; Chen, W.-J.; Wang, N.; Lin, X. Genetic and clinical profile of Chinese patients with autosomal dominant spastic paraplegia. Mol. Diagn. Ther. 2019, 23, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, M.S.; Damiano, J.A.; Mullen, S.A.; Bellows, S.; Scheffer, I.E.; Berkovic, S.F. Does variation in NIPA2 contribute to genetic generalized epilepsy? Hum. Genet. 2014, 133, 673–674. [Google Scholar] [CrossRef]
- Cowan, C.M.; Jiang, X.; Hsu, T.; Soo, C.; Zhang, B.; Wang, J.Z.; Kuroda, S.; Wu, B.; Zhang, Z.; Zhang, X.; et al. Synergistic effects of Nell-1 and BMP-2 on the osteogenic differentiation of myoblasts. J. Bone Miner. Res. 2007, 22, 918–930. [Google Scholar] [CrossRef]
- Wang, X.; Shaw, W.R.; Tsang, H.T.H.; Reid, E.; O’Kane, C.J. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neursci. 2007, 10, 177–185. [Google Scholar] [CrossRef]
- Lee, K.J.; Mendelsohn, M.; Jessel, T.M. Neuronal patterning by BMPs: A requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998, 12, 3394–3407. [Google Scholar] [CrossRef]
- Wu, H.-H.; Ivkovic, S.; Murray, R.C.; Jaramillo, S.; Lyons, K.S.; Johnson, J.E.; Calof, A.L. Autoregulation of neurogenesis by GDF11. Neuron 2003, 37, 197–207. [Google Scholar] [CrossRef]
- Matsuura, I.; Taniguchi, J.; Hata, K.; Saeki, N.; Yamashita, T. BMP inhibition enhances axonal growth and functional recovery after spinal cord injury. J. Neurochem. 2008, 105, 1471–1479. [Google Scholar] [CrossRef]
- Sahni, V.; Mukhopadhyay, A.; Tysseling, V.; Hebert, A.; Birch, D.; Mcguire, T.L.; Kessler, J.A. BMPR1a and BMPR1b signaling exert opposing effects on gliosis after spinal cord injury. J. Neurosci. 2010, 30, 1839–1855. [Google Scholar] [CrossRef]
- Bayat, V.; Jaiswal, M.; Bellen, H.J. The BMP signaling pathway at the Drosophila neuromuscular junction and its links to neurodegenerative diseases. Curr. Opin. Neurobiol. 2011, 21, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Blackstone, C. Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 2012, 35, 25–47. [Google Scholar] [CrossRef] [PubMed]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein commmunities and disease networks. Nature 2017, 25, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.W.; Serrano, M.A.; Loddo, S.; Robinson, C.; Alesi, V.; Dallapiccola, B.; Novelli, A.; Butler, M.G. Parent-of-origin effects in 15q11.2 BP1-BP2 microdeletion (Burnside-Butler) syndrome. Int. J. Mol. Sci. 2019, 20, 1459. [Google Scholar] [CrossRef]
- Bogdan, S.; Grewe, O.; Strunk, M.; Mertens, A.; Klämbt, C. Sra-1 interacts with Kette and Wasp and is required for neuronal and bristle development in Drosophila. Development 2004, 131, 3981–3989. [Google Scholar] [CrossRef]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; De Rubeis, S.; Di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef]
- Doe, C.M.; Relkovic, D.; Garfield, A.S.; Dalley, J.W.; Theobald, D.E.; Humby, T.; Wilkinson, L.S.; Isles, A.R. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum. Mol. Genet. 2009, 18, 2140–2148. [Google Scholar] [CrossRef]
- Stelzer, Y.; Sagi, I.; Yanuka, O.; Eiges, R.; Benvenisty, N. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat. Genet. 2014, 46, 551–557. [Google Scholar] [CrossRef]
- Garfield, A.; Davies, J.; Burke, L.; Furby, H.; Wilkinson, L.; Heisler, L.; Isles, A. Increased alternate splicing of Htr2c in a mouse model for Prader-Willi syndrome leads disruption of 5HT2C receptor mediated appetite. Mol. Brain 2016, 9, 95. [Google Scholar] [CrossRef]
- Clayton-Smith, J.; Laan, L. Angelman syndrome: A review of the clinical and genetic aspects. J. Med. Genet. 2003, 40, 87–95. [Google Scholar] [CrossRef]
- Galiveti, C.R.; Raabe, C.A.; Konthur, Z.; Rozhdestvensky, T.S. Differential regulation of non-protein coding RNAs from Prader-Willi syndrome locus. Sci. Rep. 2014, 4, 6445. [Google Scholar] [CrossRef] [PubMed]
- Burnside, R.D.; Pasion, R.; Mikhail, F.M. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: A susceptibility region for neurological dysfunction including developmental and language delay. Hum. Genet. 2011, 130, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.G.; Rafi, S.K.; Manzardo, A.M. High-resolution chromosome ideogram representation of currently recognized genes for autism spectrum disorders. Int. J. Mol. Sci. 2015, 16, 6464–6495. [Google Scholar] [CrossRef] [PubMed]
- Rafi, S.; Fernández-Jaén, A.; Alvarez, S.; Nadeau, O.W.; Butler, M.G. High functioning autism with missense mutations in Synaptotagmin-like protein 4 (SYTL4) and Transmembrane protein 187 (TMEM187) genes: SYTL4- protein modeling, protein-protein interaction, expression profiling and microRNA studies. Int. J. Mol. Sci. 2019, 20, 3358. [Google Scholar] [CrossRef]
- Ontology, G.; Genet, N. The Gene Ontology Consortium. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Kaplan, S. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).