Abstract
The endometrium is an important tissue for pregnancy and plays an important role in reproduction. In this study, high-throughput transcriptome sequencing was performed in endometrium samples of Meishan and Yorkshire pigs on days 18 and 32 of pregnancy. Aldo-keto reductase family 1 member C1 (AKR1C1) was found to be a differentially expressed gene, and was identified by quantitative real-time PCR (qRT-PCR) and Western blot. Immunohistochemistry results revealed the cellular localization of the AKR1C1 protein in the endometrium. Luciferase activity assay demonstrated that the AKR1C1 core promoter region was located in the region from −706 to −564, containing two nuclear factor erythroid 2-related factor 2 (NRF2) binding sites (antioxidant response elements, AREs). XLOC-2222497 was identified as a nuclear long non-coding RNA (lncRNA) highly expressed in the endometrium. XLOC-2222497 overexpression and knockdown have an effect on the expression of AKR1C1. Endocrinologic measurement showed the difference in progesterone levels between Meishan and Yorkshire pigs. Progesterone treatment upregulated AKR1C1 and XLOC-2222497 expression in porcine endometrial epithelial cells. In conclusion, transcriptome analysis revealed differentially expressed transcripts during the early pregnancy process. Further experiments demonstrated the interaction of XLOC-2222497/AKR1C1/progesterone in the endometrium and provided new potential targets for pregnancy maintenance and its control.
1. Introduction
Pregnancy is a key physiological process affecting reproduction [1]. Successful pregnancy requires intricate bidirectional regulation between embryo and the matrix [2]. The pig (Sus scrofa) is one of the most important domesticated animals and is also well-suited as a biomedical model because of similarities in anatomy and physiology between pigs and humans [3,4]. Days 18 and 32 of pig pregnancy are the two peaks of embryonic loss [5,6]. Although the two breeds have a similar number of ovulations, Meishan pigs (MS) have a higher litter size than Yorkshire pigs (YK) due to the high embryo survival rate of Meishan pigs [7,8]. Therefore, it is essential to explore the regulation mechanism of endometrium changes and the genetic difference between the two breeds during the two periods.
Long non-coding RNAs (lncRNAs) are non-protein-coding RNA species longer than 200 nucleotides [9,10]. Many lncRNAs were reported to regulate many biological processes via regulating the expression of target genes at transcriptional and post-transcriptional levels [11]. Some lncRNAs can function as signals, decoys, guides, and scaffolding in regulating nuclear activities [12]. Some lncRNAs have participated in the regulation of the cell cycle [13,14], cell differentiation [15,16], and cell apoptosis [17,18,19]. Some lncRNAs have key roles in cancer progression [20,21], and can also serve as diagnostic or prognostic markers [22,23].
Recent studies have also revealed that several lncRNAs play important roles in reproduction. The studies found that the meiotic recombination hot spot locus (Mrhl, a type of single-axon lncRNA) regulated spermatogenesis through two molecular mechanisms [24]. Yerushalmi et al. found 89 lncRNAs, 12 of which are encoded within introns of genes involved in granulosa cell processes [25]. Nakagawa et al. found that NEAT1-knocked-out mice with normal ovulation were stochastically infertile and NEAT1 is essential for corpus luteum formation and the pregnancy under suboptimal conditions [26]. Rosalia et al. found that 41 lncRNAs could interact with oocyte microRNAs (miRNAs) and may regulate folliculogenesis [27]. lncRNA-TCL6 promotes early abortion and inhibits placenta implantation via the epidermal growth factor receptor (EGFR) pathway [28]. Downregulation of lncRNA-H19 could inhibit ectopic endometrial cell proliferation and invasion by modulating miR-124-3p and ITGB3, offering a novel target for treatment of endometriosis [29]. lncRNA AK124742 may be a biomarker to predict pregnancy [30]. These findings are important in both basic reproductive research and clinical application.
Several hormones such as estrogen [31], progesterone [32,33], prostaglandin E, and prostaglandin F [34,35] are involved in the regulation of the pregnancy process. Aldo-keto reductases (AKRs) are part of the oxidoreductase super family and play an important role in the cellular response to electrophilic, osmotic, and oxidative stress, depending on the presence of the coenzyme nicotinamide adenine dinucleotide phosphate (NADPH) [36,37]. The proteins encoded by Akr genes catalyze a variety of metabolic oxidation–reduction reactions, ranging from the reduction of glucose, glucocorticoids, and small carbonyl metabolites to glutathione conjugates and phospholipid aldehydes. Substrates of the family include glucose, steroids, glycosylation end products, lipid peroxidation products, and environmental pollutants [38]. The aldo-keto reductase type 1C (AKR1C), part of the AKR superfamily, comprises the isoforms AKR1C1-AKR1C4 that catalyze NADPH-dependent reductions and have been implicated in biosynthesis, intermediary metabolism, and detoxification [39]. They serve important roles in the metabolism of steroid hormones, conjugated steroids, neurosteroids, and bile acids [40,41]. AKR1C genotypes were associated with nipple number as well as possible effects on age at puberty and ovulation rate in pigs [42]. Aldo-keto reductase family 1 member C1 (AKR1C1), which possesses 20α-Hydroxysteroid dehydrogenase (20α-HSD) activity, is associated with numerous important biological processes [38,43], and has crucial roles in the biosynthesis and inactivation of all classes of steroid hormones, and also in the biosynthesis of neurosteroids and prostaglandins [44]. Knockout of the gene encoding AKR1C1 in the mice resulted in decreasing the number of pups and prolonging the durations of the estrous cycle, pseudopregnancy, and pregnancy [45].
In this study, high-throughput transcriptome sequencing was performed in endometrium samples of Meishan and Yorkshire pigs on days 18 and 32 of pregnancy. Differentially expressed transcripts (including mRNAs and lncRNAs) were identified. Further experiments demonstrated that XLOC-2222497 regulated AKR1C1 in porcine endometrial epithelial cells, and may play an important role in the pregnancy process.
2. Results
2.1. Analysis of lncRNAs and mRNAs in the Endometrium
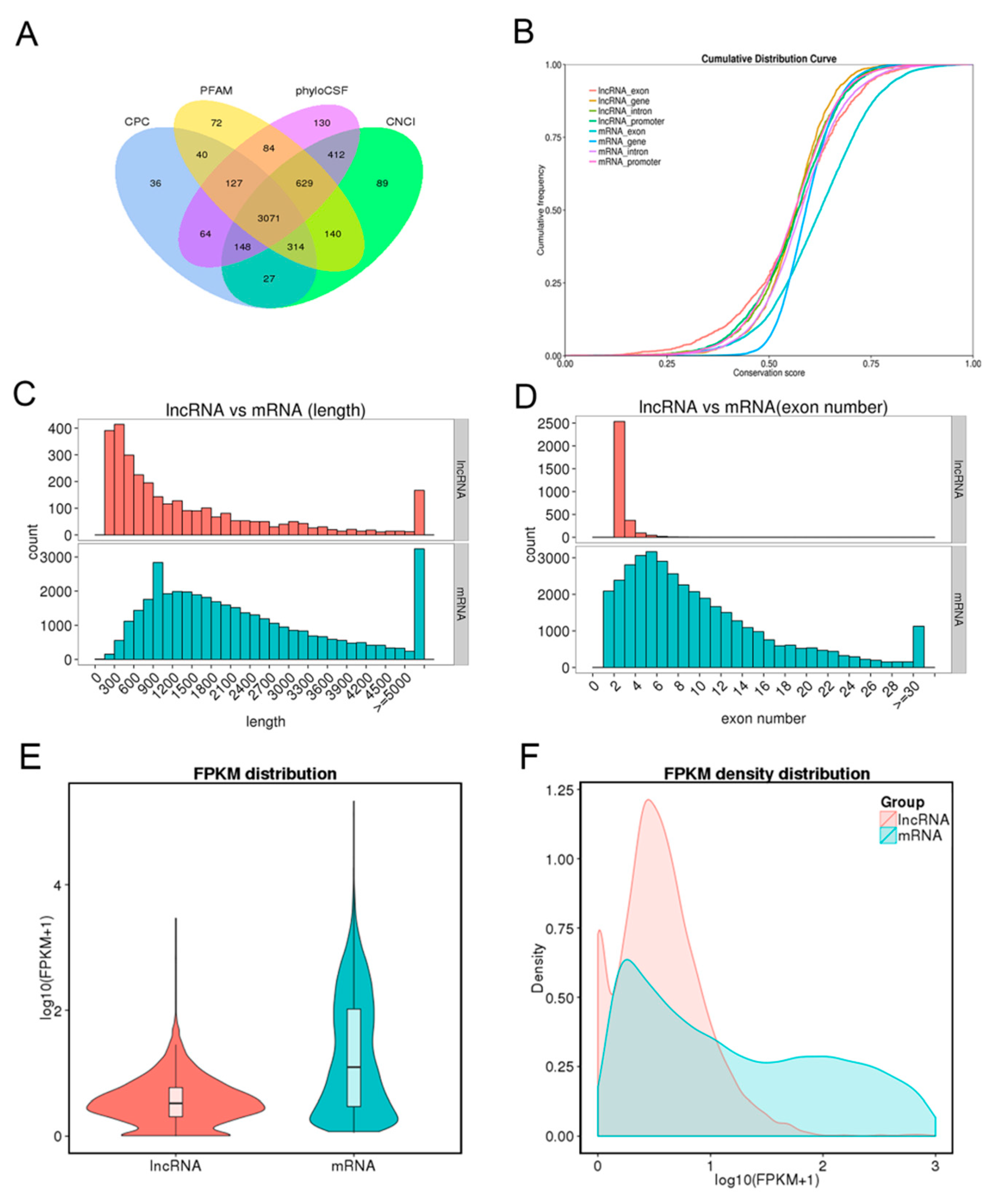
To identify the long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs), eight cDNA libraries were constructed for RNA-seq from endometrium samples of Meishan and Yorkshire pigs on days 18 and 32 of pregnancy. Coding potential calculator (CPC) analysis, coding-non-coding index (CNCI) analysis, protein families (Pfam) protein domain analysis, and phylogenetic codon substitution frequency (PhyloCSF) analysis were performed to identify lncRNAs. As shown in Figure 1A, 3071 lncRNAs were identified in the intersection of the data from four analysis methods. Expression levels of all transcripts were calculated by the HTseq software, and the expression abundance was described by the reads per kilobase per million reads (RPKM). Several highly expressed lncRNAs with RPKM >100 were found in the eight libraries; most of these identified lncRNAs were shared among different libraries (Supplementary Table S1).
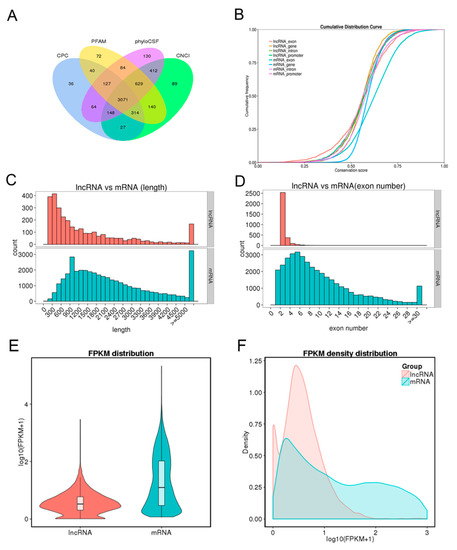
Figure 1.
Long non-coding RNA (lncRNA) identification and comparative analysis. (A) Identification of lncRNAs by coding potential calculator (CPC), protein families (Pfam), phylogenetic codon substitution frequency (PhyloCSF), and coding-non-coding index (CNCI) softwares. (B) Conservation analysis of lncRNAs and mRNAs. (C) Distribution of transcript length. Red for lncRNAs and cyan for mRNAs. (D) Distribution of exon number per transcript. Otherwise, as in C. (E) Fragments per kilobase per million (FPKM) distribution of lncRNAs and mRNAs. (F) FPKM density distribution of lncRNAs and mRNAs.
Conservation analysis between lncRNAs and mRNAs showed the conservation of mRNAs, especially in exon regions, was higher than that of lncRNAs (Figure 1B). Comparison analysis of structure indicated that the transcript length and exon number of lncRNA were less than those of mRNAs (Figure 1C,D). Fragments per kilobase per million reads (FPKM) were calculated to perform expression compare analysis between lncRNAs and mRNAs. As shown in Figure 1E,F, mRNAs had higher expression levels than lncRNAs. At the same time, 1535 differentially expressed mRNAs and 278 differentially expressed lncRNAs were identified in the comparison of Yorkshire pigs on days 32 of pregnancy (YK32) vs. Yorkshire pigs on days 18 of pregnancy (YK18) (Supplementary Table S2 and Table S3).
2.2. Feature Identification of AKR1C1
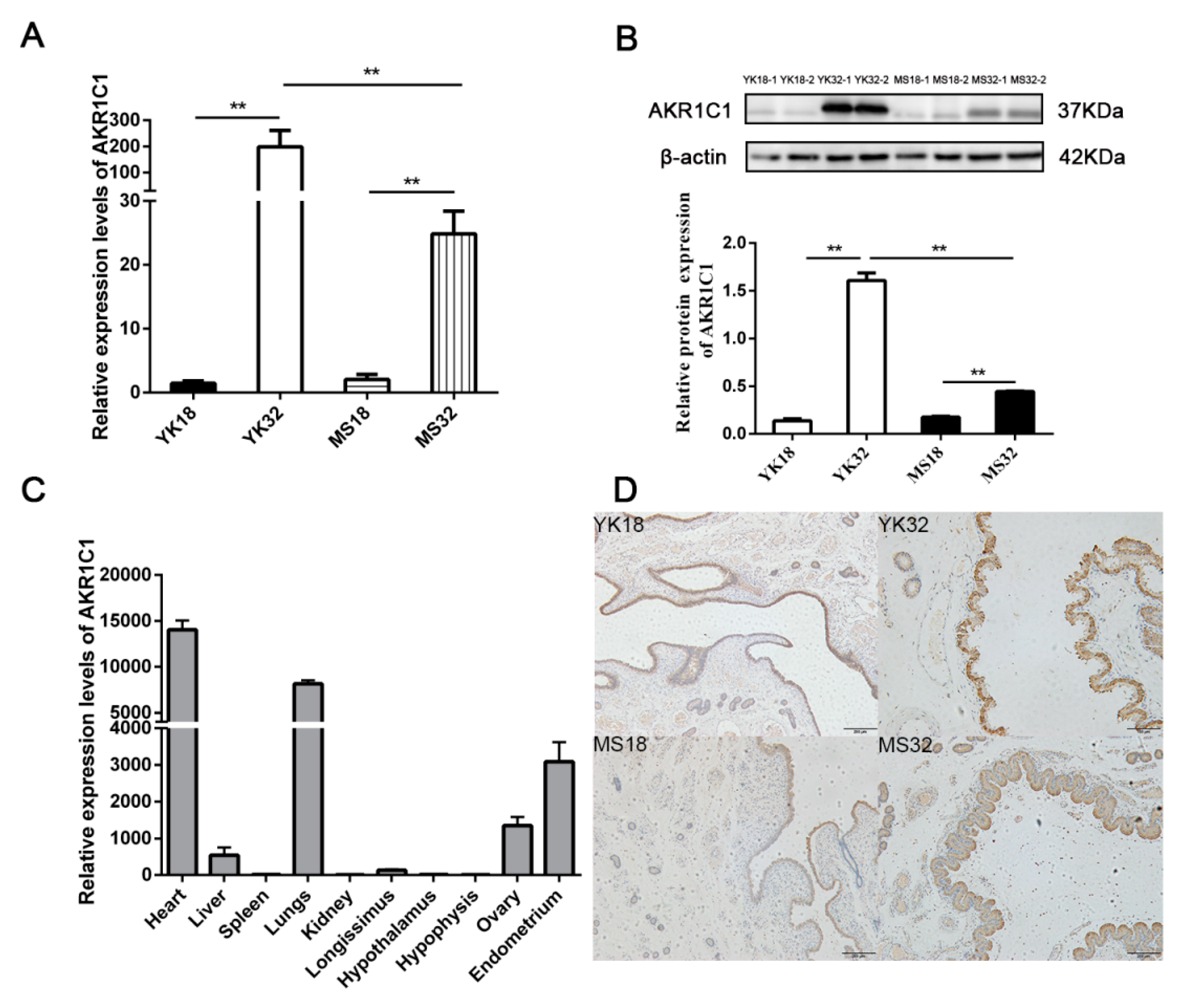
AKR1C1 was found to be a differentially expressed gene in the comparisons of YK32 vs. YK18 and Meishan pigs on days 32 of pregnancy (MS32) vs. Meishan pigs on days 18 of pregnancy (MS18). qRT-PCR and Western blot were used to confirm the expression profile of AKR1C1 (Figure 2A,B). The results revealed that AKR1C1 had high expression levels in 32–day pregnant endometrium, especially in YK32 samples. The tissue expression profile of AKR1C1 was also analyzed by qRT-PCR. As shown in Figure 2C, AKR1C1 had high expression levels in heart, lungs, endometrium, and ovary. Simultaneously, immunohistochemistry results revealed that AKR1C1 protein was located in the endometrium epithelium, including lumen epithelium and glandular epithelium (Figure 2D).
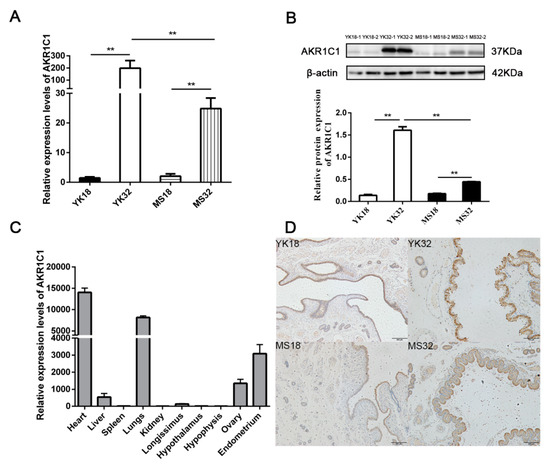
Figure 2.
Feature identification of AKR1C1. (A) RNA expression profile of AKR1C1 in the endometrium from Meishan and Yorkshire pigs on days 18 and 32 of pregnancy by qRT-PCR. ** represents p < 0.01. (B) Protein expression profile of AKR1C1 in the endometrium from Meishan and Yorkshire pigs on days 18 and 32 of pregnancy by Western blot. Down diagram: quantification of Western blot results, ** represents p < 0.01. (C) Expression profile of AKR1C1 by qRT-PCR in different tissues. (D) Immunohistochemistry result of AKR1C1 in the endometrium.
2.3. Identification of AKR1C1 Gene Promoter Region
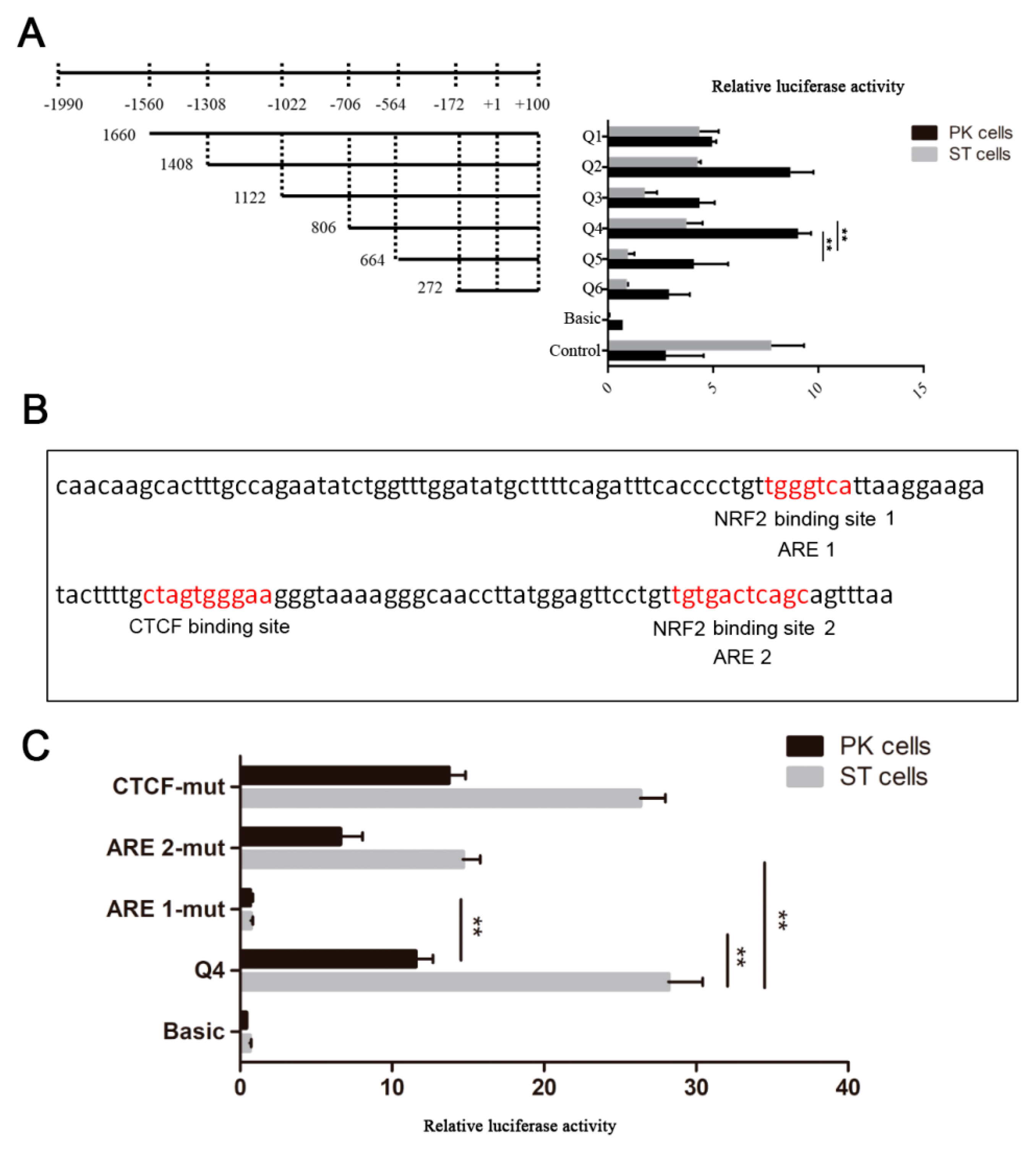
To identify the possible promoter region of AKR1C1 gene, the 1660bp 5’ flanking sequence fragment of AKR1C1 gene was amplified. Then, a series of deletion reporter plasmids (Q1–Q6) were constructed (Figure 3A) and transfected into pig kidney (PK) cells and swine testis cells (ST cells) for 24–48 h to analyze the promoter activity (where + 1 was the transcription start site). As shown in Figure 3A, the luciferase activities of Q5 (−564~ + 100) and basic were extremely significantly lower than that of Q4 in both PK and ST cells (−706~ + 100) (p < 0.01). This result implied that the AKR1C1 core promoter region was in the fragment from −706 to −564. The luciferase activity of Q3 (−1022~ + 100) was extremely significantly lower than that of Q4 (−706~+100) (p < 0.01), which implied that there were negative regulatory elements (NRE) in the region from −1022 to −706.
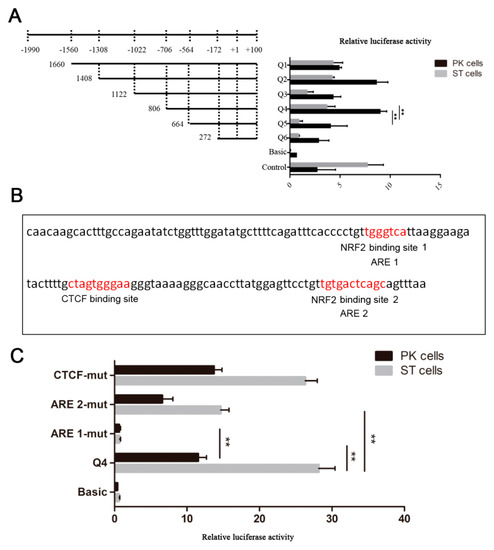
Figure 3.
Identification of AKR1C1 gene promoter region. (A) Luciferase activity assay of AKR1C1 gene promoter. Left: schematic diagram of recombinant plasmids. Right: luciferase activity assay of recombinant plasmids in pig kidney cells (PK cells) and swine testis cells (ST cells). pGL3-basic plasmid was used as negative control. pGL3-control plasmid was used as positive control. ** represents p < 0.01. (B) Transcription factor prediction of AKR1C1 core promoter region. Red means potential binding sites of transcription factor. (C) Luciferase activity assay of site mutant plasmid. pGL3-basic plasmid was used as negative control. Q4 plasmid was used as positive control. ** represents p < 0.01.
The transcription factor binding to the AKR1C1 core promoter region was predicted by JASPAR (http://jaspar.genereg.net/). There were one CCCTC-binding factor (CTCF) binding site and two antioxidant response elements (AREs) in this region (Figure 3B). Nuclear factor erythroid 2-related factor 2 (NRF2, which is also known as NFE2) was predicted to bind to the two AREs. In this study, there were two NRF2 binding sites in the AKR1C1 core promoter region. When ARE 1 and 2 were mutated (ARE 1-mut, ARE 2-mut), the luciferase activity decreased extremely significantly in ST cells (p < 0.01)(Figure 3C). In PK cells, the luciferase activity of ARE 1-mut was also extremely significantly lower than that of Q4 (p < 0.01). These results demonstrated NRF2 may play an important role in the transcriptional regulation of AKR1C1.
2.4. Feature Identification of XLOC-2222497
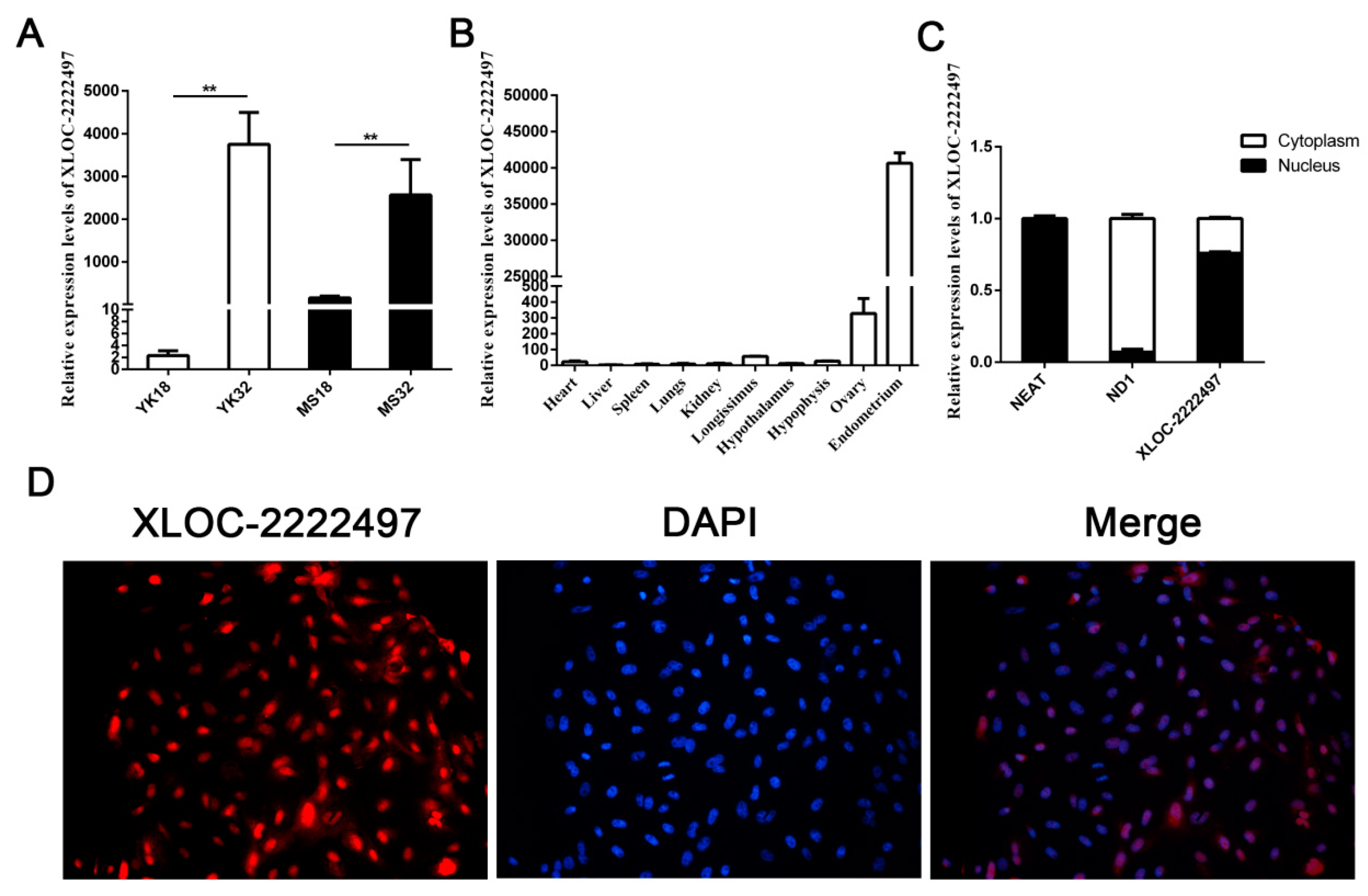
XLOC-2222497 was a differentially expressed lncRNA in our RNA-seq data. qRT-PCR results confirmed its expression profile (Figure 4A). XLOC-2222497 had high expression levels in 32-day pregnant endometrium. The expression level of XLOC-2222497 in Meishan pigs was higher than that in Yorkshire on day 18 of pregnancy. The tissue expression profile revealed that XLOC-2222497 was highly expressed in the endometrium (Figure 4B). To investigate the subcellular localization of XLOC-2222497, the fluorescent probes of XLOC-2219602 were transfected into porcine endometrial epithelial cells for RNA fluorescence in situ hybridization (FISH). As shown in Figure 4D, XLOC-2222497 was mainly located in the nucleus. Cell-fractionation assay obtained the same result (Figure 4C). To predict the coding potential of XLOC-2222497, online software CPC (http://cpc.cbi.pku.edu.cn) was used. As shown in Table 1, XLOC-2222497 had no coding potential.
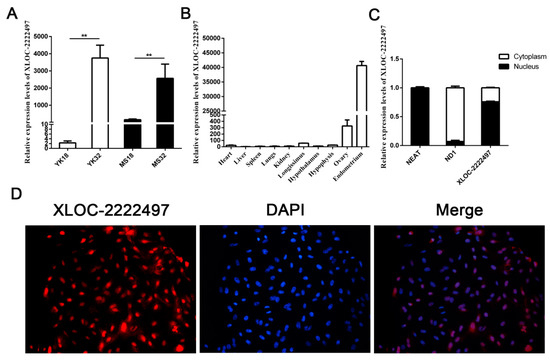
Figure 4.
Feature identification of XLOC-2222497. (A) RNA expression profile of XLOC-2222497 in the endometrium from Meishan and Yorkshire pigs on days 18 and 32 of pregnancy by qRT-PCR. ** represents p < 0.01. (B) Expression profile of XLOC-2222497 in different tissues. (C) qRT-PCR results of XLOC-2222497 cell-fractionation assay. (D) RNA fluorescence in situ hybridization (FISH) results of XLOC-2222497 for subcellular localization. Red fluorescent probe: XLOC-2222497; blue fluorescent probe: 4’, 6-diamidino-2-phenylindole (DAPI).

Table 1.
The predicted coding potential of XLOC-2222497.
2.5. The Regulation of XLOC-2222497 on AKR1C1
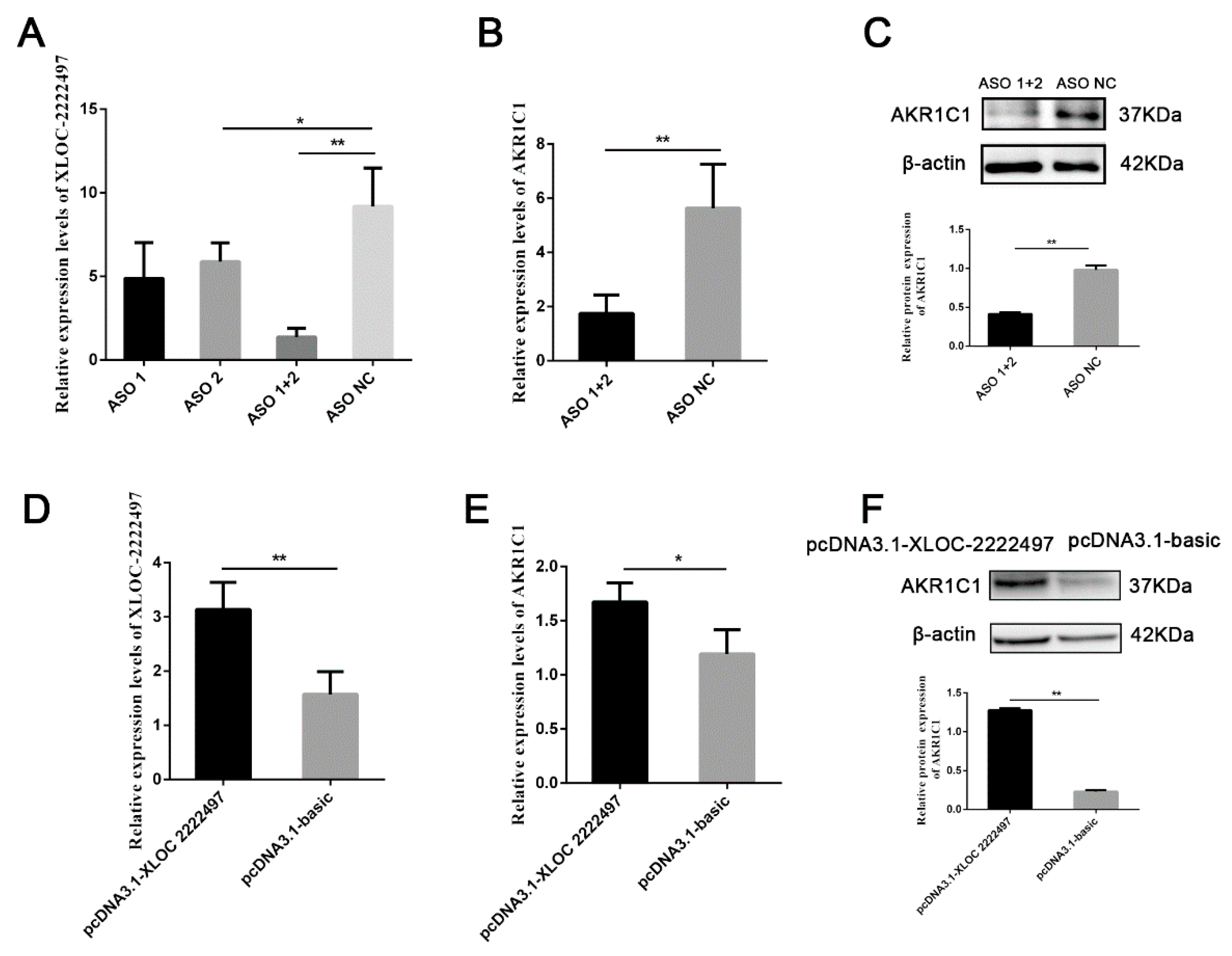
To identify the function of XLOC-2222497, antisense oligonucleotide (ASO) was used to decline the expression of XLOC-2222497. As shown in Figure 5A, XLOC-2222497 expression levels were extremely significantly inhibited after the transfection of ASO1 + 2. ASO1 + 2 combined treatment was used in a further experiment and inhibited AKR1C1 mRNA (Figure 5B) and protein (Figure 5C) expression. To further investigate the effect of XLOC-2222497 on AKR1C1, an overexpression plasmid of XLOC-2222497 was constructed (pcDNA3.1–XLOC-2222497) and transfected into porcine endometrial epithelial cells. As shown in Figure 5D, the overexpression of XLOC-2222497 significantly promoted the expression of AKR1C1 at the mRNA level (Figure 5E) and protein level (Figure 5F). These results demonstrated the positive regulation of XLOC-2222497 on AKR1C1.
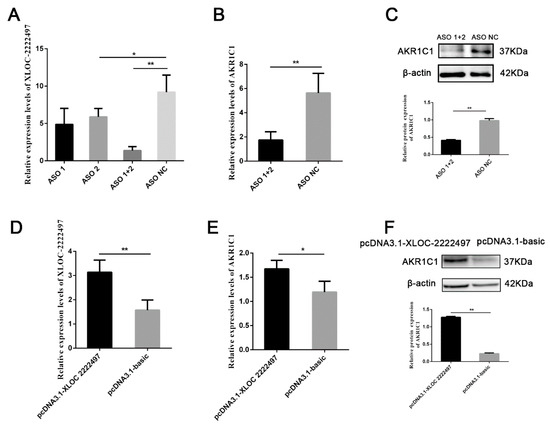
Figure 5.
The regulation of XLOC-2222497 on AKR1C1. (A) qRT-PCR results of antisense oligonucleotide (ASO) knockdown efficiency detection. * represents p < 0.05. ** represents p < 0.01. (B) qRT-PCR results of AKR1C1 after the transfection of ASOs. ** represents p < 0.01. (C) Western blot results of AKR1C1 after the transfection of ASOs. Down diagram: quantification of Western blot results, ** represents p < 0.01. (D) qRT-PCR results of AKR1C1 overexpression efficiency detection. ** represents p < 0.01. (E) qRT-PCR results of AKR1C1 after the transfection of pcDNA3.1–XLOC-2222497. * represents p < 0.05. (F) Western blot results of AKR1C1 after the transfection of pcDNA3.1–XLOC-2222497. Down diagram: quantification of Western blot results, ** represents p < 0.01.
2.6. Progesterone Measurement and Regulation on XLOC-2222497 and AKR1C1
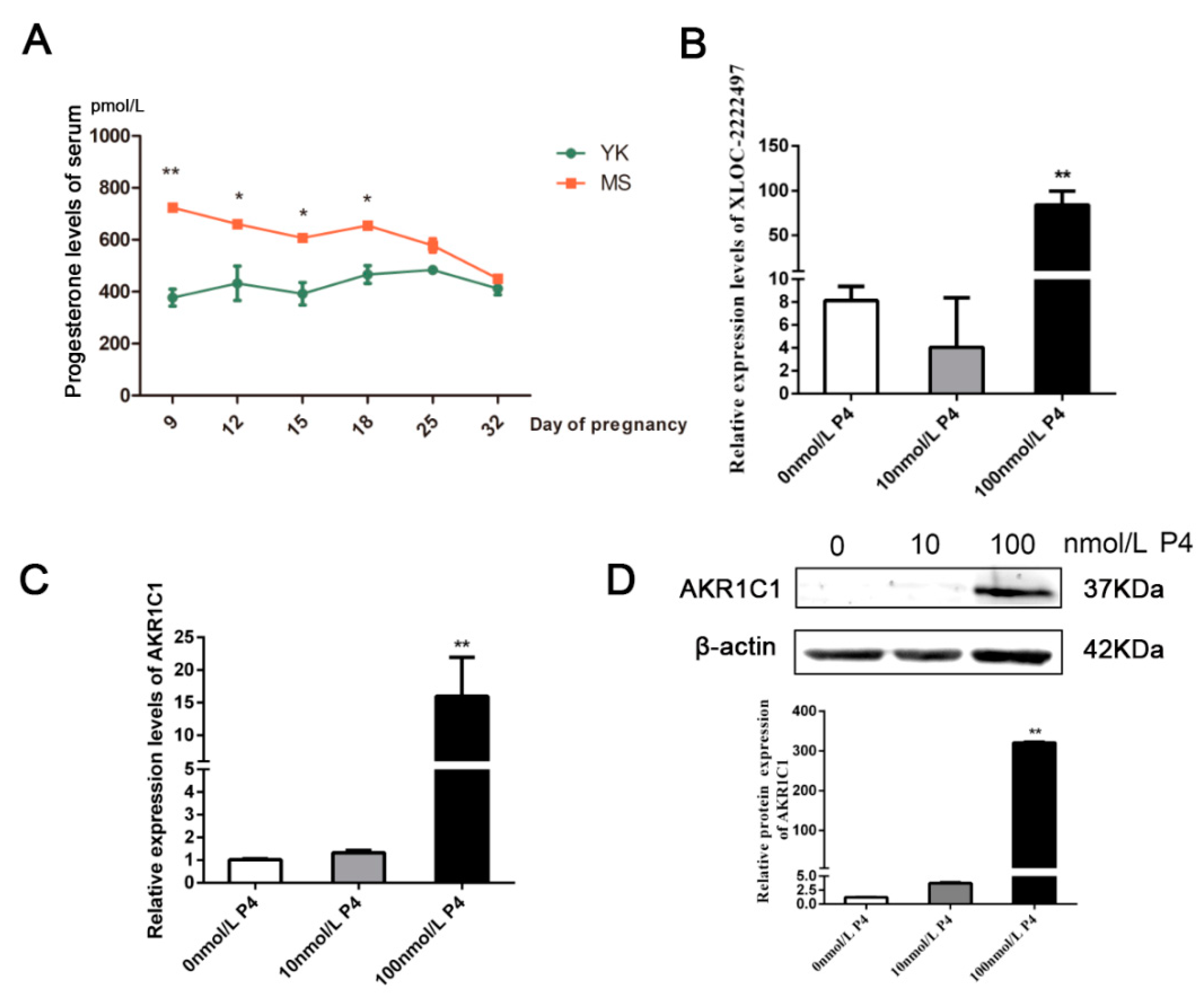
AKR1C1 plays a key role in the progesterone metabolism process [40]. Therefore, progesterone levels were measured in the serum of Meishan and Yorkshire pigs in different early pregnancy stages (Figure 6A). On days 9, 12, 15, 18 of pregnancy, the level of progesterone in Meishan sows was significantly higher than that in Yorkshire sows (p < 0.05). After day 18 of gestation, the level of progesterone in the two breeds tended to be identical. In Meishan pigs, progesterone levels on day 32 of pregnancy were significantly lower than that on day 18 of pregnancy and had an opposite difference with XLOC-2222497 and AKR1C1. Meanwhile, the porcine endometrial epithelial cells were treated with different concentrations of progesterone (P4) for 48 h. As shown in Figure 6B, 100 nmol/L P4 treatment significantly promoted the expression levels of XLOC-2222497. P4 treatment in porcine endometrial epithelial cells also promoted the expression of AKR1C1 (Figure 6C). The Western blot results were consistent with qRT-PCR results (Figure 6D). The above results demonstrated the regulation of progesterone on XLOC-2222497 and AKR1C1.
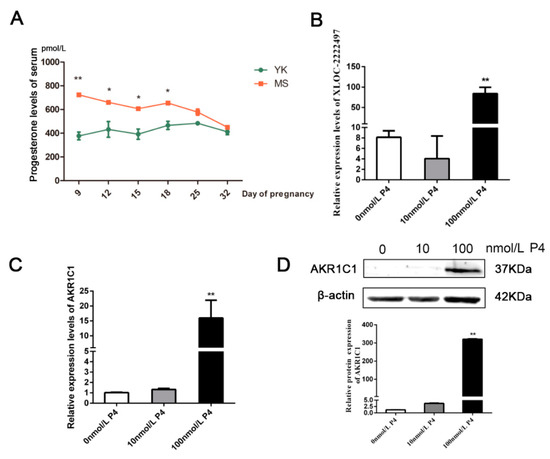
Figure 6.
Progesterone measurement and regulation on XLOC-2222497 and AKR1C1. (A) Progesterone levels in serum of Meishan and Yorkshire pigs at different pregnancy stages. * represents p < 0.05, ** represents p < 0.01,. (B) Expression levels of XLOC-2222497 after the treatment of progesterone in porcine endometrial epithelium cells. ** represents p < 0.01. (C) RNA expression levels of AKR1C1 after the treatment of progesterone in porcine endometrial epithelium cells. ** represents p < 0.01. (D) Protein expression levels of AKR1C1 after the treatment of progesterone in porcine endometrial epithelium cells. Down diagram: quantification of Western blot results, ** represents p < 0.01.
3. Discussion
Progesterone, a natural female hormone, is an essential hormone for pregnancy [46,47]. AKR1C1 belongs to the aldo-keto reductase (AKR) superfamily of nicotinamide adenine dinucleotide phosphate (NADPH)-dependent oxidoreductases [48,49] and has a major role in progesterone metabolism [40,47,50]. Moreover, AKR1C1 can bind to the promoter region of the progesterone receptor and hereby decreases receptor activity [51]. AKR1C1 is expressed ubiquitously [52,53]. Our qRT-PCR result also confirmed the ubiquitous expression of AKR1C1. In steroidogenic tissues, AKR1C1 catalyzes the final steps in progesterone biosynthesis. In peripheral tissues, including steroid hormone target tissues, AKR1C1 converts progesterone to its inactive form of 20-alpha-hydroxy-progesterone and regulates the amount of hormone that can bind to members of the nuclear receptor superfamily, ultimately regulating gene expression [50]. Recent studies showed that AKR1C1 could induce signal transducer and activator of transcription (STAT) activation [54,55], which was involved in pregnancy [56]. In this study, the expression level of progesterone in the serum of pregnant sows decreased from days 18 to 32 of pregnancy, and AKR1C1 was highly expressed on day 32 of pregnancy. These results were consistent with previous studies that AKR1C1 played a critical role in controlling the progesterone concentration [47,57,58,59].
To explore the transcriptional regulation of AKR1C1, luciferase activity assay was performed and demonstrated that the AKR1C1 core promoter region was located in the region from −706 to −564. In this region, there was one CTCF binding site and two AREs. The site-mutation experiment showed that ARE was important for the promoter activity of AKR1C1. NRF2 is a redox-regulated transcription factor that coordinates the basal and inducible expression of a vast array of cytoprotective and antioxidant genes through binding to ARE [60,61,62]. Previous studies also confirmed that NRF2 was a regulator of the AKR1C family via direct binding to the ARE located in the promoter regions of the AKR1Cs [49,63,64]. Wentilactone A (WA) inhibited the expression of AKR1C1 via the insulin like growth factor 1 receptor (IGF-1R)/ insulin receptor substrate 1 (IRS-1)/ phosphatidylinositol 3-kinase (PI3K)/ protein kinase B (AKT)/NRF2 signaling pathway [43]. Accumulation of p62 inhibits Keap1-mediated NRF2 protein degradation by competing with NRF2 for the binding site on Kelch-like enyol-CoA hydratase (ECH)-associated protein 1 (Keap1), a cytosolic repressor protein of NRF2, resulting in transcriptional upregulation of NRF2 downstream genes [65,66]. NRF2 also contains a phosphodegron, phosphorylation of which promotes nuclear export and a return to basal antioxidant signaling. A central regulator of NRF2 in this manner is glycogen synthase kinase 3β (GSK3β) [67]. GSK3β phosphorylates Fyn at the threonine residue. Phosphorylated Fyn accumulates in the nucleus and phosphorylates NRF2, and brings about the nuclear export of NRF2, resulting in NRF2 rebinding Keap1 and being rapidly degraded [68,69]. Wang et al. found the expression levels of AKR1C1 and NRF2 were elevated in progestin-resistant endometrial epithelia. The NRF2/AKR1C1 pathway may represent a new therapeutic strategy for treatment of endometrial hyperplasia/cancer [70].
Due to the powerful and diverse functions of long non-coding RNAs, a large number of lncRNAs have been found and identified. However, few studies focus on lncRNAs related to pig pregnancy. In porcine endometrial tissue, Wang et al. found several differentially expressed lncRNAs that may play a vital role in the process of implantation using RNA sequencing [71,72,73]. In this study, a novel lncRNA XLOC-2222497 was screened by RNA-seq. The online software CPC was used to predict the coding potential of XLOC-2222497 and showed no coding potential. Cell-fractionation assay and RNA FISH results demonstrated that XLOC-2222497 was mainly located in the nucleus. Then the XLOC-2222497 overexpression and knockdown results showed the positive regulation of XLOC-2222497 on AKR1C1. Furthermore, XLOC-2222497 and AKR1C1 were both highly expressed on day 32 of pregnancy, which was consistent with their positive regulation. lncRNAs can regulate the expression of target genes via a variety of mechanisms [74]. Because XLOC-2222497 was mainly located in the nucleus, there were two possible molecular mechanisms underlying the regulation of XLOC-2222497 on AKR1C1: (1) XLOC-2222497 increased the stability of AKR1C1 mRNA. (2) XLOC-2222497 recruited transcription factor to the promoter region of the AKR1C1 gene to promote the transcription of AKR1C1. The specific molecular mechanism needs further studies. In addition, one of the most notable features of lncRNAs is their tissue specificity as compared to protein coding genes [75,76]. The tissue expression profile revealed that XLOC-2222497 was highly expressed in the endometrium, which was consistent with the tissue specificity of lncRNAs. To our knowledge, XLOC-2222497 is the first identified lncRNA regulating AKR1C1.
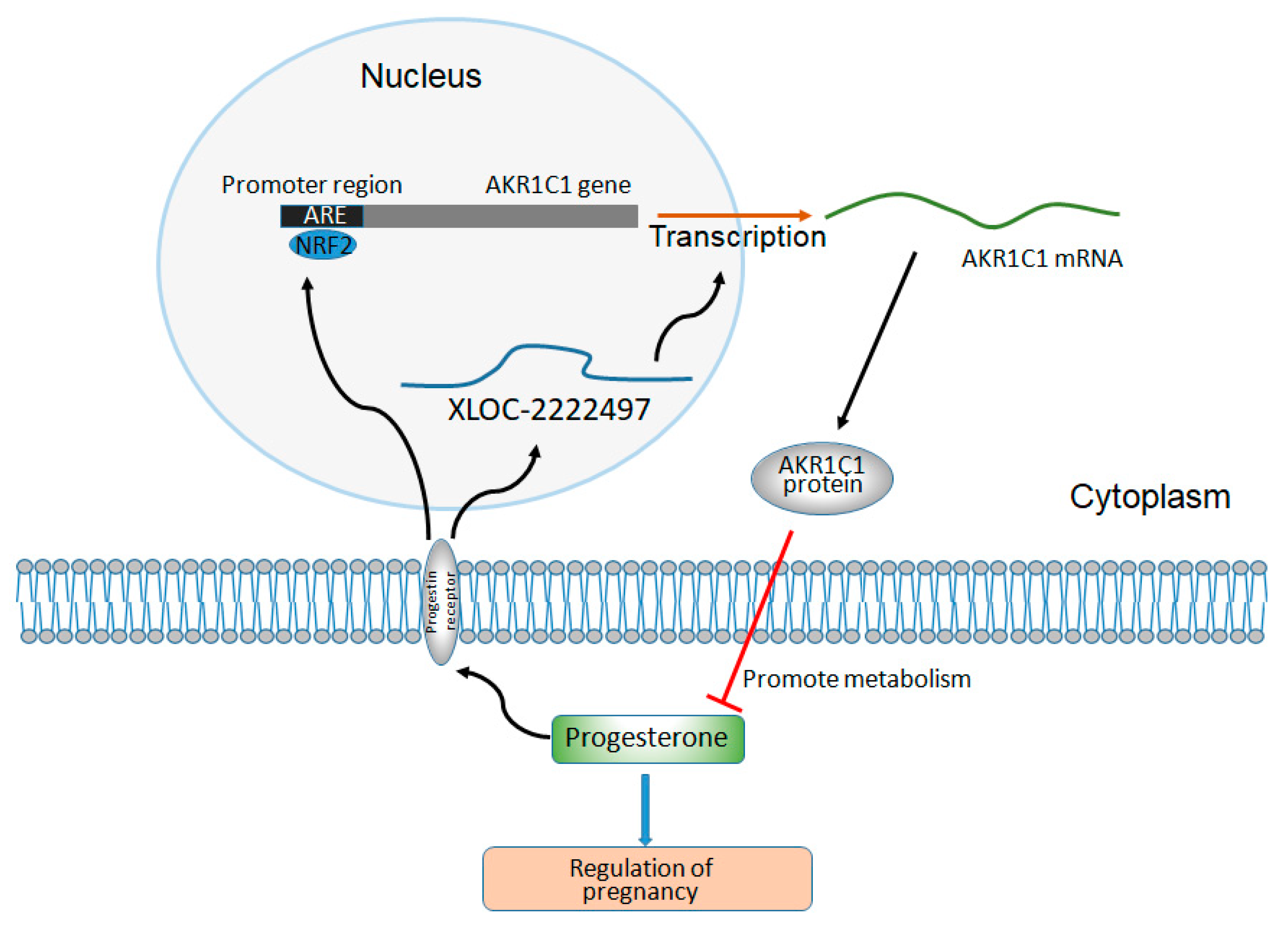
Moreover, the expression level of AKR1C1 and XLOC-2222497 in endometrial cells increased significantly after the treatment of progesterone. This result was consistent with the previous result [77]. The administration of progesterone might activate the NRF2/ARE signal pathway [78]. Ghadiri et al. found that progesterone at both 16 and 32 mg/kg doses induced expression of NRF2 [79]. Byrne et al. revealed norgestrel, an FDA-approved synthetic analog of progesterone, inhibited GSK3β and modulated NRF2 expression at the post-translational level, bringing about its phosphorylation, and subsequent translocation into the nucleus where it bound antioxidant response elements (AREs), bringing about the upregulation of various antioxidants, and detoxifying and cytoprotective genes [69,80,81]. Moreover, cytoplasmic NRF2 expression is significantly correlated with the expression of progesterone receptor (PR), which suggested a possible functional interaction between NRF2 and PR [51]. Taken together, it might corroborate a possible feedback regulation mechanism which merits further investigations: the high progesterone level leads to the increase of XLOC-2222497 and NRF2 by PR, and thus activates AKR1C1, metabolizing progesterone to its inactive form and maintaining a mutual balance (Figure 7). P4-mediated XLOC-2222497 and AKR1C1 expression in the endometrium was conducive to maintaining the stability of the intrauterine environment. In addition, these effects should be also taken into account when exogenous progesterone is administered. The above results demonstrated the important roles of the XLOC-2222497/AKR1C1/progesterone signaling pathway in pregnancy and provided new potential targets for pregnancy maintenance and its control.
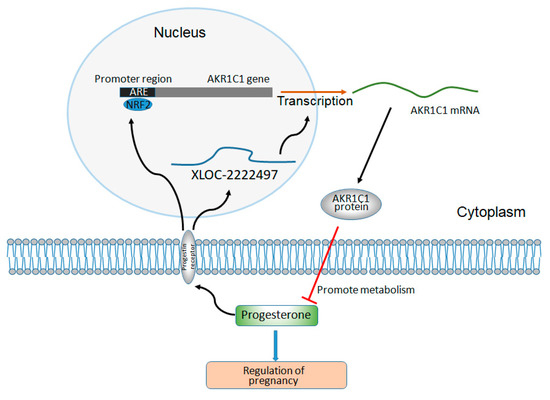
Figure 7.
The regulation diagram of XLOC-2222497, AKR1C1, and progesterone.
4. Materials and Methods
4.1. Ethics Statement
In our research, all animal procedures were approved by The Scientific Ethics Committee of Huazhong Agricultural University, Wuhan, China (ID number: HZAUSW2015-017, permitted at February 2015).
4.2. Animal Sources and Sample Collection
The experimental populations consisted of four Yorkshire pigs and four Meishan pigs of similar age with genetic background from one commercial herd. All pigs were raised under the same conditions and under a standardized feeding regimen with free access to water. Each breed was randomly assigned to two groups: day 18 (n = 2) and day 32 (n = 2), and was artificially inseminated at the same time. Uteri were obtained from animals slaughtered on days 18 and 32 of pregnancy. Each uterine horn was flushed with sterile phosphate buffer saline (PBS) (pH 7.4), and subsequently opened longitudinally on the inner side. Samples from the endometrium of Meishan pigs and Yorkshire pigs on days 18 and 32 of pregnancy were taken. Tissue samples were frozen in liquid nitrogen and stored at −80 °C before RNA isolation. All animal procedures were approved by The Scientific Ethics Committee of Huazhong Agricultural University, Wuhan, China (ID number: HZAUSW2015-017, permitted at February 2015).
4.3. Library Construction and Sequencing
Total RNA was extracted from the endometrium using the TRIzol reagent (Invitrogen, Life Technologies, CA, USA). The RNA samples were quantitated and subjected to quality inspection. Total DNA was extracted using the TIANamp Genomic DNA kit (Tiangen, CA, China) according to the manufacturer’s protocol.
4.4. Identification of Differentially Expressed Genes (DEGs)
First, the expression level of each gene was calculated using the HTseq software (0.6.1) [82], and was normalized based on the reads per RPKM method [83]. Subsequently, DEGs were identified using the R packages DEGseq (1.18.0) [84]. The RNA-seq data was deposited in the Gene Expression Omnibus (accession GSE141564).
4.5. Quantitative Real-Time PCR for Gene Expression
MiRNAs and mRNAs were reverse transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo, Wuhan, China) in accordance with the manufacturer’s instructions. qPCR was performed using a standard UltraSYBR Mixture (CWBIO, Beijing, China) in the Roche LightCyler 480 system (Roche, Mannheinm, Germany) according to the manufacturer’s instructions. Porcine β-actin gene was used as the endogenous control gene for mRNA RT-PCR. The RT-PCR data were analyzed using the 2-∆∆CT method, as previously described [85]. The relative fold changes of mRNA expression were quantified by normalizing the cycle threshold (CT) value of the experimental gene to the mean CT value of the control β-actin gene.
4.6. Western Blot
Cells were split by radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Jiangsu, China) and supplemented with 0.01% of phenylmethanesulfonyl fluoride (PMSF) (Beyotime, Jiangsu, China). The protein was separated by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis, and then was transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Boston, USA). Next, the protein was incubated with the corresponding primary antibody and secondary antibody. Antibodies included AKR1C1 (1:1000, ABclonal, Wuhan, China), and β-actin (1:1000, Boster, Wuhan, China).
4.7. Cell Culture and Transfection
Primary endometrial cells were isolated from the uteri of Yorkshire pigs. For in vitro culture of endometrial cells, PBS (pH = 7.4) was used to wash the external surface of the uteri. When opened longitudinally, the endometrium was separated from the myometrium by using a sterilized blade. The endometrium tissues were collected and flushed with PBS (pH = 7.4). Minced tissues mixed with Dulbecco’s modified eagle medium/Nutrient mixture F-12 (DMEM/F-12) (Hyclone, Logan, USA) culture medium containing 0.1% collagenase I were placed in a 5% CO2 incubator at 37 °C for 2.5 h, and the mixture was shaken every half hour. The DMEM/F-12 culture medium containing 10% fetal bovine serum (FBS) (CLARK, Richmond, USA) supplemented with 1% penicillin–streptomycin was used to terminate the digestion. Afterwards, the mixture was filtered by using the sterilized cell screen and the solution was transferred to the tube. The precipitates were collected to release the endometrial cells after twice centrifugation. The porcine endometrium cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2 in Dulbecco’s modified Eagle medium (Hyclone, Logan, USA) supplemented with 10% fetal bovine serum (FBS) (CLARK, Richmond, USA). Epithelial and stromal cells were isolated by the differential adhesion method. The purified epithelial cells were plated and grown until they reached 70–80% confluent. Then, overexpression plasmid and small interfering RNAs (siRNAs) were transfected using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
4.8. Immunohistochemistry
Partial samples of the endometrium from Meishan pigs and Yorkshire pigs were fixed in 4% formaldehyde solution to make a paraffin section. The immunohistochemistry analysis was performed following the procedures: xylene dewaxing, xylene replaced by gradient alcohol, treatment of peroxide, antigen repair, serum blocking, anti-AKR1C1 incubation, secondary antibody incubation, diaminobenzidine (DAB) coloration, hematoxylin staining, 0.5% hydrochloric acid differentiation, wash with water, gradient alcohol dehydration, xylene transparentization, and mounting. Stained slices were observed under a 400-fold microscope.
4.9. Endocrinologic Measurement
Progesterone levels of serum from Yorkshire pigs and Meishan pigs on days 9, 12, 15, 18, and 32 of pregnancy were measured by an enzyme linked immunosorbent assay (ELISA) kit (Fusheng, CA, China) according to the manufacturer’s protocol. The brief steps are described as follows: standard preparation, sample dilution and incubation, incubation of conjugate reagent, coloration, and spectrophotometry. There were three Yorkshire pigs and two Meishan pigs in each stage.
5. Conclusions
This study investigated long non-coding RNA regulation in the endometrium of Yorkshire and Meishan pigs at different days of pregnancy using high-throughput sequencing. We first identified an AKR1C1-related lncRNA in porcine endometrium. Further experimental results showed that XLOC-2222497 regulated AKR1C1 in porcine endometrial epithelial cells, thus participating in the regulation of progesterone metabolism. This study provides valuable information for future transcriptome studies of porcine endometrium, and the molecular regulation of pregnancy.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/9/3232/s1. Supplementary Table S1: List of highly expressed lncRNAs with RPKM >100. Supplementary Table S2: Differentially expressed mRNA between YK32 and YK18. Supplementary Table S3: Differentially expressed lncRNA between YK32 and YK18.
Author Contributions
D.X. and M.L. conceived and designed this study. T.S., H.Y., G.L., M.W., C.Z., B.H., L.Z., and C.Z. performed the experiments. T.S., H.Y., G.L., M.L., D.X., C.Z., and B.H. conducted the data analysis and prepared figures and tables. M.L., D.X., and T.S. wrote the manuscript. All authors reviewed and approved the manuscript.
Funding
National Science and Technology Major Project of China (2016ZX08006002), State Key Development Program for Basic Research of China (2014CB138504), Hubei Agricultural Sciences And Technology Innovation Center (2019-620-000-001-30), Agricultural Science and Technology Innovation Action of Hubei Province (2018skjcx01), National Natural Science Foundation of China (31201764, 31172179), Natural Science Foundation of Hubei Province (2015CFA101), Animal Husbandry Major Technology Collaborative Extension Project of Hubei Province, Research Project of Huazhong Agricultural University, China.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Norwitz, E.R.; Schust, D.J.; Fisher, S.J. Implantation and the survival of early pregnancy. N Engl. J. Med. 2001, 345, 1400–1408. [Google Scholar] [CrossRef]
- Carson, D.D.; Bagchi, I.; Dey, S.K.; Enders, A.C.; Fazleabas, A.T.; Lessey, B.A.; Yoshinaga, K. Embryo implantation. Dev. Biol. 2000, 223, 217–237. [Google Scholar] [CrossRef]
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabria, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom. Clin. Appl. 2014, 8, 715–731. [Google Scholar] [CrossRef]
- Zhao, P.; Zheng, X.; Feng, W.; Wang, H.; Kang, H.; Ning, C.; Du, H.; Yu, Y.; Li, B.; Zhao, Y.; et al. Profiling long noncoding rna of multi-tissue transcriptome enhances porcine noncoding genome annotation. Epigenomics 2018, 10, 301–320. [Google Scholar] [CrossRef]
- Kyriazakis, I.; Whittemore, C.T.; Whittemore, C.T. Whittemore’s Science and Practice of Pig Production, 3rd ed.; Blackwell Pub.: Oxford, UK; Ames, Iowa, 2006; p xvi; p. 685. [Google Scholar]
- Wessels, J.M.; Linton, N.F.; Croy, B.A.; Tayade, C. A review of molecular contrasts between arresting and viable porcine attachment sites. Am. J. Reprod. Immunol. 2007, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Lambert, E.; Williams, D.H.; Lynch, P.B.; Hanrahan, T.J.; McGeady, T.A.; Austin, F.H.; Boland, M.P.; Roche, J.F. The extent and timing of prenatal loss in gilts. Theriogenology 1991, 36, 655–665. [Google Scholar] [CrossRef]
- Christenson, R.K.; Vallet, J.L.; Leymaster, K.A.; Young, L.D. Uterine function in meishan pigs. J. Reprod. Fertil. Suppl. 1993, 48, 279–289. [Google Scholar] [PubMed]
- Perkel, J.M. Visiting "noncodarnia". BioTechniques 2013, 54, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Cabianca, D.S.; Casa, V.; Bodega, B.; Xynos, A.; Ginelli, E.; Tanaka, Y.; Gabellini, D. A long ncrna links copy number variation to a polycomb/trithorax epigenetic switch in fshd muscular dystrophy. Cell 2012, 149, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.S.; Li, T.P.; Ton, H.; Mao, X.D.; Chen, Y.J. Advances of long noncoding rnas-mediated regulation in reproduction. Chin. Med. J. 2018, 131, 226–234. [Google Scholar] [CrossRef]
- Ma, H.; Hao, Y.; Dong, X.; Gong, Q.; Chen, J.; Zhang, J.; Tian, W. Molecular mechanisms and function prediction of long noncoding rna. Sci. World J. 2012, 2012, 541786. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xiao, Y.; Ma, L.; Wang, J. Regulating of cell cycle progression by the lncrna cdkn2b-as1/mir-324-5p/rock1 axis in laryngeal squamous cell cancer. Int. J. Biol. Markers 2020, 35, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Sahu, M.; Mallick, B. Modulation of specific cell cycle phases in human embryonic stem cells by lncrna rna decoys. J. Mol. Recognit. JMR 2019, 32, e2763. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wan, Q.; Zhang, L.; Li, Y.; Zhang, P.; Li, D.; Feng, C.; Yi, F.; Zhang, L.; Ding, X.; et al. Analysis of lncrna expression in cell differentiation. RNA Biol. 2018, 15, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Shui, X.; Chen, S.; Lin, J.; Kong, J.; Zhou, C.; Wu, J. Knockdown of lncrna neat1 inhibits th17/cd4(+) t cell differentiation through reducing the stat3 protein level. J. Cell. Physiol. 2019, 234, 22477–22484. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zeng, C.; Gan, S.; Li, H.; Cheng, Y.; Chen, D.; Li, R.; Zhu, W. Lncrna hottip-mediated hoxa11 expression promotes cell growth, migration and inhibits cell apoptosis in breast cancer. Int. J. Mol. Sci. 2018, 19, 472. [Google Scholar] [CrossRef]
- Li, L.; Zhu, J.; Ye, F.; Duan, Z.; Zhou, J.; Huang, Z.; Wang, L. Upregulation of the lncRNA SRLR in polycystic ovary syndrome regulates cell apoptosis and IL-6 expression. Cell Biochem. Funct. 2020. Epub ahead of print. [Google Scholar] [CrossRef]
- Yong, H.; Wu, G.; Chen, J.; Liu, X.; Bai, Y.; Tang, N.; Liu, L.; Wei, J. Lncrna malat1 accelerates skeletal muscle cell apoptosis and inflammatory response in sepsis by decreasing brca1 expression by recruiting ezh2. Mol. Ther. Nucleic Acids 2019, 19, 97–108. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long noncoding rnas in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Qi, P.; Zhou, X.Y.; Du, X. Circulating long non-coding rnas in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding rna hotair reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding rna activated by tgf-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, G.; Rao, S.M. A novel noncoding rna processed by drosha is restricted to nucleus in mouse. RNA 2008, 14, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, G.M.; Salmon-Divon, M.; Yung, Y.; Maman, E.; Kedem, A.; Ophir, L.; Elemento, O.; Coticchio, G.; Dal Canto, M.; Mignini Renzinu, M.; et al. Characterization of the human cumulus cell transcriptome during final follicular maturation and ovulation. Mol. Hum. Reprod. 2014, 20, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncrna neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Vento, M.E.; Borzi, P.; Ragusa, M.; Barbagallo, D.; Arena, D.; Purrello, M.; Di Pietro, C. Non-coding rnas in the ovarian follicle. Front. Genet. 2017, 8, 57. [Google Scholar] [CrossRef]
- Liu, L.P.; Gong, Y.B. Lncrna-tcl6 promotes early abortion and inhibits placenta implantation via the egfr pathway. Eur. Rev. Med Pharmacol. Sci. 2018, 22, 7105–7112. [Google Scholar]
- Liu, S.; Qiu, J.; Tang, X.; Cui, H.; Zhang, Q.; Yang, Q. Lncrna-h19 regulates cell proliferation and invasion of ectopic endometrium by targeting itgb3 via modulating mir-124-3p. Exp. Cell Res. 2019, 381, 215–222. [Google Scholar] [CrossRef]
- Xu, X.F.; Li, J.; Cao, Y.X.; Chen, D.W.; Zhang, Z.G.; He, X.J.; Ji, D.M.; Chen, B.L. Differential expression of long noncoding rnas in human cumulus cells related to embryo developmental potential: A microarray analysis. Reprod. Sci. 2015, 22, 672–678. [Google Scholar] [CrossRef]
- Rothschild, M.; Jacobson, C.; Vaske, D.; Tuggle, C.; Wang, L.; Short, T.; Eckardt, G.; Sasaki, S.; Vincent, A.; McLaren, D.; et al. The estrogen receptor locus is associated with a major gene influencing litter size in pigs. Proc. Natl. Acad. Sci. USA 1996, 93, 201–205. [Google Scholar] [CrossRef]
- MacLaughlin, D.T.; Harding, G.B.; Westphal, U. Steroid-protein interactions. Xxv. Binding of progesterone and cortisol in pregnancy sera; progesterone-binding globulin and uterine cytosol receptor in the pregnant guinea pig. Am. J. Anat. 1972, 135, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Magness, R.R.; Reynolds, L.P.; Ford, S.P. Evidence for uterine metabolism of progesterone during early pregnancy in the pig. Theriogenology 1986, 25, 551–558. [Google Scholar] [CrossRef]
- Bazer, F.W.; Thatcher, W.W.; Martinat-Botte, F.; Terqui, M. Conceptus development in large white and prolific chinese meishan pigs. J. Reprod. Fertil. 1988, 84, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Waclawik, A. Novel insights into the mechanisms of pregnancy establishment: Regulation of prostaglandin synthesis and signaling in the pig. Reproduction 2011, 142, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Penning, T.M. Aldo-keto reductases and bioactivation/detoxication. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 263–292. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Sun, H.; Zhang, J.; Hou, Y.; Zhang, T.; Kang, J.; Wang, Z.; Yang, Q.; Long, R. Analysis of aldo-keto reductase gene family and their responses to salt, drought, and abscisic acid stresses in medicago truncatula. Int. J. Mol. Sci. 2020, 21, 754. [Google Scholar] [CrossRef]
- Barski, O.A.; Tipparaju, S.M.; Bhatnagar, A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab. Rev. 2008, 40, 553–624. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Yu, G. Avasimibe inhibits tumor growth by targeting foxm1-akr1c1 in osteosarcoma. Oncotargets Ther. 2019, 12, 815–823. [Google Scholar] [CrossRef]
- Hevir, N.; Vouk, K.; Sinkovec, J.; Ribic-Pucelj, M.; Rizner, T.L. Aldo-keto reductases akr1c1, akr1c2 and akr1c3 may enhance progesterone metabolism in ovarian endometriosis. Chem. Biol. Interact. 2011, 191, 217–226. [Google Scholar] [CrossRef]
- Zhao, S.F.; Wang, S.G.; Zhao, Z.Y.; Li, W.L. Akr1c1-3, notably akr1c3, are distinct biomarkers for liver cancer diagnosis and prognosis: Database mining in malignancies. Oncol. Lett. 2019, 18, 4515–4522. [Google Scholar] [CrossRef]
- Nonneman, D.J.; Wise, T.H.; Ford, J.J.; Kuehn, L.A.; Rohrer, G.A. Characterization of the aldo-keto reductase 1c gene cluster on pig chromosome 10: Possible associations with reproductive traits. Bmc Vet. Res. 2006, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Meng, L.; Xu, G.; Lv, C.; Wang, H.; Tian, H.; Chen, R.; Jiao, B.; Wang, B.; Huang, C. Wentilactone a induces cell apoptosis by targeting akr1c1 gene via the igf-1r/irs1/pi3k/akt/nrf2/flip/caspase-3 signaling pathway in small cell lung cancer. Oncol. Lett. 2018, 16, 6445–6457. [Google Scholar] [CrossRef] [PubMed]
- Rizner, T.L.; Penning, T.M. Role of aldo-keto reductase family 1 (akr1) enzymes in human steroid metabolism. Steroids 2014, 79, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Choi, J.H.; Hirabayashi, K.; Matsuwaki, T.; Suzuki, M.; Yamanouchi, K.; Horai, R.; Sudo, K.; Iwakura, Y.; Nishihara, M. Reproductive phenotypes in mice with targeted disruption of the 20alpha-hydroxysteroid dehydrogenase gene. J. Reprod. Dev. 2007, 53, 499–508. [Google Scholar] [CrossRef]
- Romero, R.; Yeo, L.; Chaemsaithong, P.; Chaiworapongsa, T.; Hassan, S.S. Progesterone to prevent spontaneous preterm birth. Semin. Fetal Neonatal Med. 2014, 19, 15–26. [Google Scholar] [CrossRef]
- Zeng, C.; Zhu, D.; You, J.; Dong, X.; Yang, B.; Zhu, H.; He, Q. Liquiritin, as a natural inhibitor of akr1c1, could interfere with the progesterone metabolism. Front. Physiol. 2019, 10, 833. [Google Scholar] [CrossRef]
- Jez, J.M.; Bennett, M.J.; Schlegel, B.P.; Lewis, M.; Penning, T.M. Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 1997, 326, 625–636. [Google Scholar] [CrossRef]
- Huebbers, C.U.; Verhees, F.; Poluschkin, L.; Olthof, N.C.; Kolligs, J.; Siefer, O.G.; Henfling, M.; Ramaekers, F.C.S.; Preuss, S.F.; Beutner, D.; et al. Upregulation of akr1c1 and akr1c3 expression in opscc with integrated hpv16 and hpv-negative tumors is an indicator of poor prognosis. Int. J. Cancer 2019, 144, 2465–2477. [Google Scholar] [CrossRef]
- Penning, T.M. Molecular endocrinology of hydroxysteroid dehydrogenases. Endocr. Rev. 1997, 18, 281–305. [Google Scholar]
- Czogalla, B.; Kahaly, M.; Mayr, D.; Schmoeckel, E.; Niesler, B.; Hester, A.; Zeder-Goss, C.; Kolben, T.; Burges, A.; Mahner, S.; et al. Correlation of nrf2 and progesterone receptor and its effects on ovarian cancer biology. Cancer Manag. Res. 2019, 11, 7673–7684. [Google Scholar] [CrossRef]
- Penning, T.M.; Burczynski, M.E.; Jez, J.M.; Hung, C.F.; Lin, H.K.; Ma, H.; Moore, M.; Palackal, N.; Ratnam, K. Human 3alpha-hydroxysteroid dehydrogenase isoforms (akr1c1-akr1c4) of the aldo-keto reductase superfamily: Functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones. Biochem. J. 2000, 351, 67–77. [Google Scholar] [PubMed]
- Nishizawa, M.; Nakajima, T.; Yasuda, K.; Kanzaki, H.; Sasaguri, Y.; Watanabe, K.; Ito, S. Close kinship of human 20alpha-hydroxysteroid dehydrogenase gene with three aldo-keto reductase genes. Genes Cells Devoted Mol. Cell. Mech. 2000, 5, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.M.; Chang, Y.C.; Yang, Y.C.; Lin, S.K.; Chang, P.M.; Hsiao, M. Akr1c1 controls cisplatin-resistance in head and neck squamous cell carcinoma through cross-talk with the stat1/3 signaling pathway. J. Exp. Clin. Cancer Res. Cr 2019, 38, 245. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Chang, L.L.; Yan, F.J.; Hu, Y.; Zeng, C.M.; Zhou, T.Y.; Yuan, T.; Ying, M.D.; Cao, J.; He, Q.J.; et al. Akr1c1 activates stat3 to promote the metastasis of non-small cell lung cancer. Theranostics 2018, 8, 676–692. [Google Scholar] [CrossRef]
- Maj, T.; Chelmonska-Soyta, A. Pleiotropy and redundancy of stat proteins in early pregnancy. Reprod. Domest. Anim. Zuchthyg. 2007, 42, 343–353. [Google Scholar] [CrossRef]
- Ji, Q.; Aoyama, C.; Nien, Y.D.; Liu, P.I.; Chen, P.K.; Chang, L.; Stanczyk, F.Z.; Stolz, A. Selective loss of akr1c1 and akr1c2 in breast cancer and their potential effect on progesterone signaling. Cancer Res. 2004, 64, 7610–7617. [Google Scholar] [CrossRef]
- Lewis, M.J.; Wiebe, J.P.; Heathcote, J.G. Expression of progesterone metabolizing enzyme genes (akr1c1, akr1c2, akr1c3, srd5a1, srd5a2) is altered in human breast carcinoma. BMC Cancer 2004, 4, 27. [Google Scholar] [CrossRef]
- Piekorz, R.P.; Gingras, S.; Hoffmeyer, A.; Ihle, J.N.; Weinstein, Y. Regulation of progesterone levels during pregnancy and parturition by signal transducer and activator of transcription 5 and 20alpha-hydroxysteroid dehydrogenase. Mol. Endocrinol. 2005, 19, 431–440. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Ryoo, I.G.; Kwak, M.K. Regulatory crosstalk between the oxidative stress-related transcription factor nfe2l2/nrf2 and mitochondria. Toxicol. Appl. Pharmacol. 2018, 359, 24–33. [Google Scholar] [CrossRef]
- Fao, L.; Mota, S.I.; Rego, A.C. Shaping the nrf2-are-related pathways in alzheimer’s and parkinson’s diseases. Ageing Res. Rev. 2019, 54, 100942. [Google Scholar] [CrossRef] [PubMed]
- Zenkov, N.K.; Menshchikova, E.B.; Tkachev, V.O. Keap1/nrf2/are redox-sensitive signaling system as a pharmacological target. Biochem. Biokhimiia 2013, 78, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Wise, R.A.; Holbrook, J.T.; Criner, G.; Sethi, S.; Rayapudi, S.; Sudini, K.R.; Sugar, E.A.; Burke, A.; Thimmulappa, R.; Singh, A.; et al. Lack of effect of oral sulforaphane administration on nrf2 expression in copd: A randomized, double-blind, placebo controlled trial. PLoS ONE 2016, 11, e0163716. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor nrf2 through inactivation of keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Wu, X.; Sun, R.; Wang, H.; Yang, B.; Wang, F.; Xu, H.; Chen, S.; Zhao, R.; Pi, J.; Xu, Y. Enhanced p62-nrf2 feedback loop due to impaired autophagic flux contributes to arsenic-induced malignant transformation of human keratinocytes. Oxidative Med. Cell. Longev. 2019, 2019, 1038932. [Google Scholar] [CrossRef]
- Rojo, A.I.; Medina-Campos, O.N.; Rada, P.; Zuniga-Toala, A.; Lopez-Gazcon, A.; Espada, S.; Pedraza-Chaverri, J.; Cuadrado, A. Signaling pathways activated by the phytochemical nordihydroguaiaretic acid contribute to a keap1-independent regulation of nrf2 stability: Role of glycogen synthase kinase-3. Free Radic. Biol. Med. 2012, 52, 473–487. [Google Scholar] [CrossRef]
- Jain, A.K.; Jaiswal, A.K. Gsk-3beta acts upstream of fyn kinase in regulation of nuclear export and degradation of nf-e2 related factor 2. J. Biol. Chem. 2007, 282, 16502–16510. [Google Scholar] [CrossRef]
- Byrne, A.M.; Ruiz-Lopez, A.M.; Roche, S.L.; Moloney, J.N.; Wyse-Jackson, A.C.; Cotter, T.G. The synthetic progestin norgestrel modulates nrf2 signaling and acts as an antioxidant in a model of retinal degeneration. Redox Biol. 2016, 10, 128–139. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhang, Z.; Park, J.Y.; Guo, D.; Liao, H.; Yi, X.; Zheng, Y.; Zhang, D.; Chambers, S.K.; et al. Mechanism of progestin resistance in endometrial precancer/cancer through nrf2-akr1c1 pathway. Oncotarget 2016, 7, 10363–10372. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, S.; Liu, X.; Liu, H.; Hu, T.; Qiu, X.; Zhang, J.; Lei, M. Analyses of long non-coding rna and mrna profiling using rna sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 2016, 6, 20238. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, T.; Wu, L.; Liu, X.; Xue, S.; Lei, M. Identification of non-coding and coding rnas in porcine endometrium. Genomics 2017, 109, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hua, R.; Xue, S.; Li, W.; Wu, L.; Kang, T.; Lei, M. Mrna/lncrna expression patterns and the function of fibrinogen-like protein 2 in meishan pig endometrium during the preimplantation phases. Mol. Reprod. Dev. 2019, 86, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.J.; Kim, H.J.; Park, K.K. Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis. Int. J. Mol. Sci. 2020, 21, 2698. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding rnas. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; Srikanth, K.; Park, W.; Lee, S.H.; Choi, B.H.; Kim, H.; Kim, Y.M.; Cho, E.S.; Kim, J.H.; Lee, J.H.; et al. Transcriptome analysis to identify long non coding rna (lncrna) and characterize their functional role in back fat tissue of pig. Gene 2019, 703, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Beranic, N.; Lanisnik Rizner, T. Progestin effects on expression of akr1c1-akr1c3, srd5a1 and pgr in the z-12 endometriotic epithelial cell line. Chem. -Biol. Interact. 2013, 202, 218–225. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, J.; Ding, H.; Wu, W.; Xiao, G. Progesterone provides the pleiotropic neuroprotective effect on traumatic brain injury through the nrf2/are signaling pathway. Neurocritical Care 2017, 26, 292–300. [Google Scholar] [CrossRef]
- Ghadiri, T.; Vakilzadeh, G.; Hajali, V.; Khodagholi, F. Progesterone modulates post-traumatic epileptogenesis through regulation of bdnf-trkb signaling and cell survival-related pathways in the rat hippocampus. Neurosci. Lett. 2019, 709, 134384. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the keap1-nrf2-are pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Wyse Jackson, A.C.; Cotter, T.G. The synthetic progesterone norgestrel is neuroprotective in stressed photoreceptor-like cells and retinal explants, mediating its effects via basic fibroblast growth factor, protein kinase a and glycogen synthase kinase 3beta signalling. Eur. J. Neurosci. 2016, 43, 899–911. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq-A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by rna-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. Degseq: An r package for identifying differentially expressed genes from rna-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Xu, Z.L.; Wang, X.W.; Zhao, X.F.; Wang, J.X. Clip domain serine protease and its homolog respond to vibrio challenge in chinese white shrimp, fenneropenaeus chinensis. Fish. Shellfish Immunol. 2009, 26, 787–798. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).