Abstract
Deinococcus radiodurans is a polyextremophilic bacterium well known for its extreme resistance to irradiation, oxidative stress, and other damaging conditions. Many small noncoding RNAs (ncRNAs) in D. radiodurans have been identified by deep sequencing analysis and computational predictions. However, the precise roles of ncRNAs and their target genes in the oxidative stress response have not been investigated. Here, we report the identification and characterization of a novel ncRNA named OsiR (for oxidative stress-induced ncRNA). Oxidative stress tolerance analysis showed that deleting osiR significantly decreased viability, total antioxidant capacity, and catalase activity in D. radiodurans under oxidative stress conditions. Comparative phenotypic and qRT-PCR analyses of an osiR mutant identify a role of OsiR in regulating the expression of the catalase gene katE2. Microscale thermophoresis and genetic complementation showed that a 21-nt sequence in the stem–loop structure of OsiR (204–244 nt) directly base pairs with its counterpart in the coding region of katE2 mRNA (843–866 nt) via a 19 nt region. In addition, deletion of katE2 caused a significant reduction of catalase activity and oxidative stress tolerance similar to that observed in an osiR mutant. Our results show that OsiR positively regulates oxidative stress tolerance in D. radiodurans by increasing the mRNA stability and translation efficiency of katE2. This work provides a new regulatory pathway mediated by ncRNA for the oxidative stress response that most likely contributes to the extreme tolerances of D. radiodurans.
1. Introduction
The polyextremophilic bacterium Deinococcus radiodurans can survive under high-intensity ionizing radiation, ultraviolet radiation, desiccation, and oxidative stresses [1,2,3,4]. Recent studies have shown that the strong resistance of D. radiodurans to various damaging conditions can be explained by its highly efficient antioxidative cell protection [4]. Indeed, strong resistance to oxidative stress plays an important role in the environmental adaptability [4,5]. Oxidative stress results from the formation of reactive oxygen species (ROS), of which the three main types are hydroxyl radicals, superoxide radicals, and hydrogen peroxide [6]. The ROS produced by ionizing radiation, UV radiation, desiccation, and oxidative stresses can damage proteins, lipids, nucleic acids, and carbohydrates and induce potentially lethal double-strand DNA breaks (DSBs) in the bacterial genome [7,8,9]. Therefore, it is not surprising that this bacterium has, over a long period of evolution, developed an effective antioxidant system to cope with oxidative stress. D. radiodurans displays pronounced resistance to all ROS-generating agents [10,11], and its ROS-scavenging enzymes include two major antioxidant enzymes: superoxide dismutase, which eliminates superoxide radicals from the cells, and catalase, which degrades H2O2. It has been reported that the ability of protein extracts of D. radiodurans to scavenge H2O2 was 30 times higher than that of E. coli extracts [12]; moreover, the catalase activity of D. radiodurans was 127 times and 32 times higher than that of E. coli during the exponential and stationary phases, respectively [13]. The D. radiodurans genome encodes two KatE-type catalases, DR1998 (KatE1), and DRA0259 (KatE2), and one eukaryotic-type catalase, DRA0146 [4]. Among these catalases, the typical monofunctional heme catalase DR1998 is the major catalase in D. radiodurans, and its expression is positively regulated by the transcriptional regulators DrRRA [14], OxyR [15], and PprM [16]. KatE2 also has extensive catalase activity, whereas DRA0146 does not exhibit catalase activity [17]. However, the biological functions and regulation of katE2 have not yet been investigated in detail.
Bacterial regulatory noncoding RNA (ncRNAs), also referred to as small regulatory RNAs (sRNAs), are the core component of the bacterial stress response network and are multifunctional regulators that allow bacteria to adapt to a complex environment [18]. Many ncRNAs are involved in the regulation of diverse physiological processes, such as the cell division process [19], stress responses [20], quorum sensing [21], and bacterial virulence [22,23]. At present, most ncRNAs that have been characterized are trans-encoded and play a positive or negative role in mRNA stability and translation efficiency [24,25,26]. As ncRNA-based regulation is hypothesized to be dynamic and efficient, it represents a useful mechanism for polyextremophilic organisms to adapt to various survival stresses.
At present, with the rapid development of advanced bioinformatics prediction and sequencing technology, more and more ncRNAs have been identified in almost all bacteria [27,28]. Up to now, most of the ncRNAs which have been characterized in physiological functions and regulatory mechanisms originated from intestinal bacteria, such as E. coli and Salmonella [24,29]. The oxidative stress-induced sRNA OxyS from E.coli was the first characterized ncRNA involved in oxidative stress response [30], and has a negative regulatory effect on the mRNAs of transcription factors such as FhlA, RpoS, and FlhDC, as well as some other proteins [31,32]. Recent studies revealed that an OxyS-induced molecular checkpoint relay leads to temporary cell cycle arrest to facilitate DNA damage repair, thereby increasing viability following stress [33]. The Pseudomonas stutzeri ncRNA NfiS positively regulates the oxidative stress response by targeting a catalase-encoding gene, katB [34]. However, the detailed function and regulation of ncRNAs in the Deinococcus genus are only beginning to be studied, even though the first ncRNA of D. radiodurans similar to Y RNA was discovered nearly 19 years ago [35,36]. The binding complexes of Y RNA and RSR, a Ro protein ortholog, contribute to UV irradiation resistance. At present, 41 potential ncRNAs have been identified in D. radiodurans by combining deep sequencing analysis and computational predictions [37], but their biological functions and precise molecular mechanisms have not been characterized in detail except for Dsr18 (DnrH), which positively influences heat tolerance by increasing the transcription of hsp20 mRNA [38]. One ncRNA named Dsr39 exhibited at least a 2-fold decrease in band intensity after 15-kGy irradiation analyzed by Northern blot [37]. In this study, we found that Dsr39 was highly expressed under oxidative stress induced by hydrogen peroxide, and the disruption of this ncRNA rendered D. radiodurans more sensitive to oxidative stress. Therefore, this ncRNA was renamed OsiR (oxidative stress-induced ncRNA). Our results indicate that OsiR act as a crucial regulator to positively regulate the oxidative stress resistance of D. radiodurans by directly base pairing with katE2 mRNA, a catalase gene. The significant roles of OsiR in the oxidative stress response highlight the importance of ncRNA-mediated regulation under stress conditions in the Deinococcus species.
2. Results
2.1. Expression of osiR Is Induced by Oxidative Stress
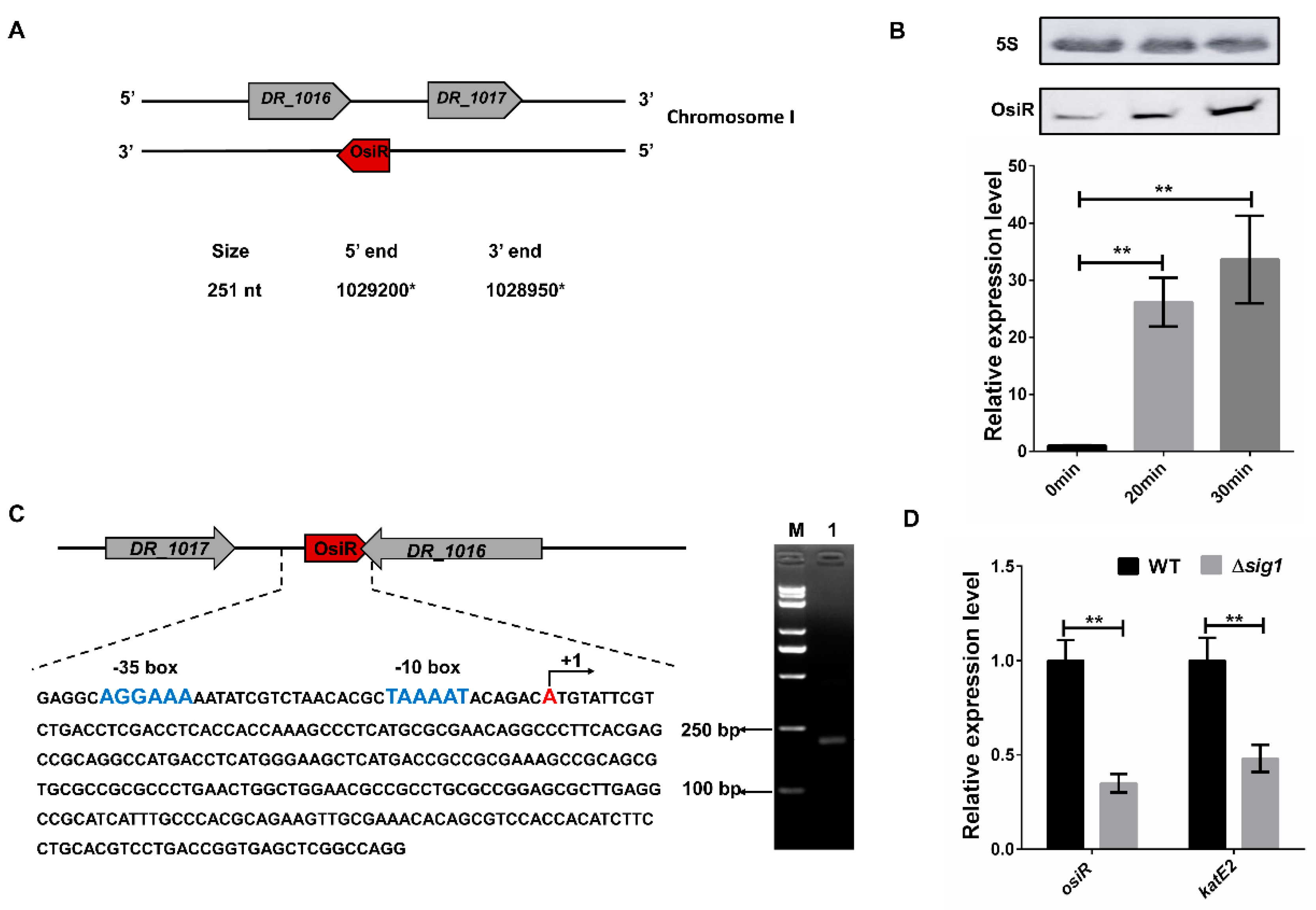
OsiR, which was initially identified in a previous report [37], is located in the intergenic region between DR_1016 and DR_1017 on chromosome I of D. radiodurans, with the 3’end overlapping with the 3’end of DR_1016 by 79 bp (Figure 1A). DR_1016 encodes a tRNA-modifying GTPase, and DR_1017 encodes an rRNA methylase. qRT-PCR and Northern blot analyses revealed that expression of osiR was significantly upregulated under oxidative stress conditions (Figure 1B), suggesting that it may play a physiological role in the oxidative stress response.
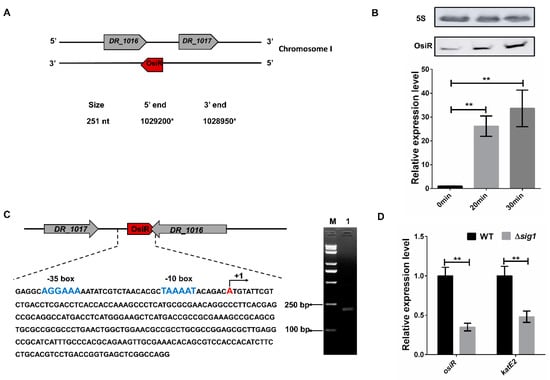
Figure 1.
Expression and genomic context of OsiR. (A) the genomic location of osiR on chromosome I [37]; (B) transcript levels of osiR detected by Northern blot and qRT-PCR in D. radiodurans. 0 min, 20 min, 30 min indicates the time of H2O2 treatment; (C) physical map and nucleotide sequence of osiR in D. radiodurans. (blue) −10 box and −35 box in the promoter of OsiR predicted by BPROM; (+1) transcription start site mapped by 5’RACE; M: Trans2K PlusII Marker; Lane 1: product of the second nested PCR for 5’RACE; (D) effect of a sig1 mutation on the expression of osiR and katE2. The qRT-PCR data shown are the averages of three biological replicates. Error bars represent standard deviations. The statistical significance of the difference was confirmed by Student’s t-tests (**, p < 0.01).
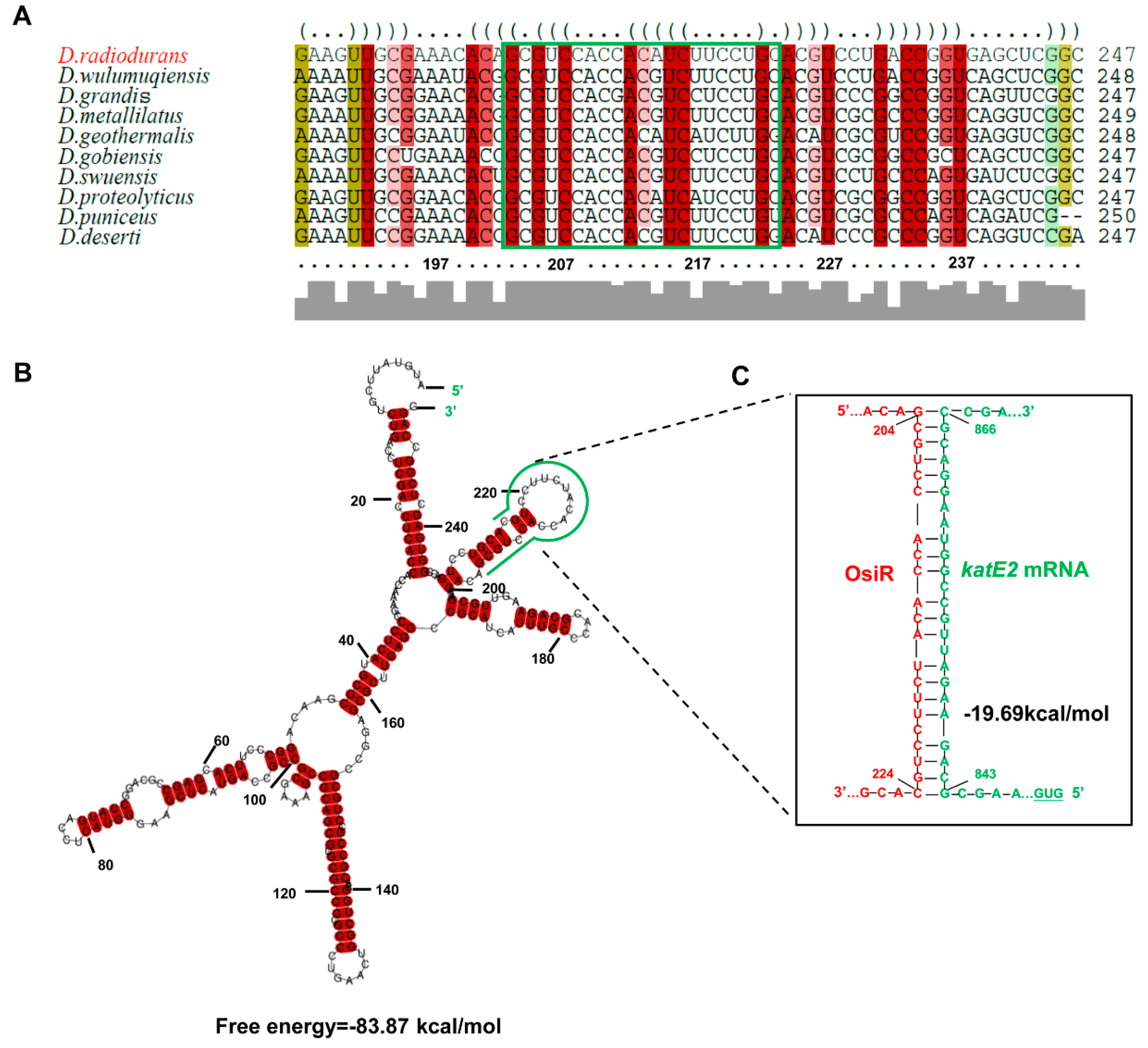
The full-length osiR is 251 nt [37]; the transcription start site of osiR determined by 5’RACE is an adenine A at position 1,029,200 of chromosome 1 (Figure 1A, C). In the promoter region of osiR, a −10 box and a −35 box were predicted by BPROM, as shown in Figure 1C; these motifs are usually recognized and bound by the σ70 factor. The results of qRT-PCR assay showed that the transcript level of osiR was significantly decreased in the absence of sig1, suggesting that the expression of osiR may be controlled by Sig1, a σ70 factor in D. radiodurans (Figure 1D). Multi-sequence alignment of osiR and its homologs underscores the high degree of conservation in a region of 169–251 nt (Figure 2A), implying that this region likely plays the same role among members of Deinococcus. The predicted secondary structure of OsiR contains two stem–loop structures in this region (Figure 2B). These results indicated that OsiR is a Deinococcus-specific regulatory ncRNA induced by oxidative stress and may play a positive role in oxidative stress response.
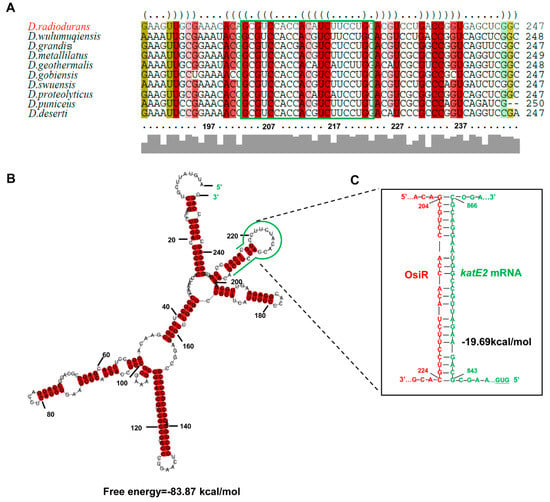
Figure 2.
Sequence conservation and structural features of OsiR. (A) multisequence alignment of osiR homologous genes analyzed by LocARNA [39]. The colors indicate that the structural conservation, compatible base pairs are colored; the saturation decreases with the number of incompatible base pairs. Thus, it indicates the structural conservation of the base pair, for which red indicates 100% of the structure sequences is identical. (B) The secondary structure of OsiR predicted by RNAalifold [40]. The potential interaction region of OsiR with katE2 mRNA is shown in green. The secondary structure stability of ncRNA is represented by the free energy values. (C) The interaction between OsiR and katE2 mRNA analyzed by IntaRNA. OsiR is shown in red, and the katE2 mRNA is shown in green.
2.2. OsiR Contributes to the Oxidative Stress Tolerance in D. radiodurans
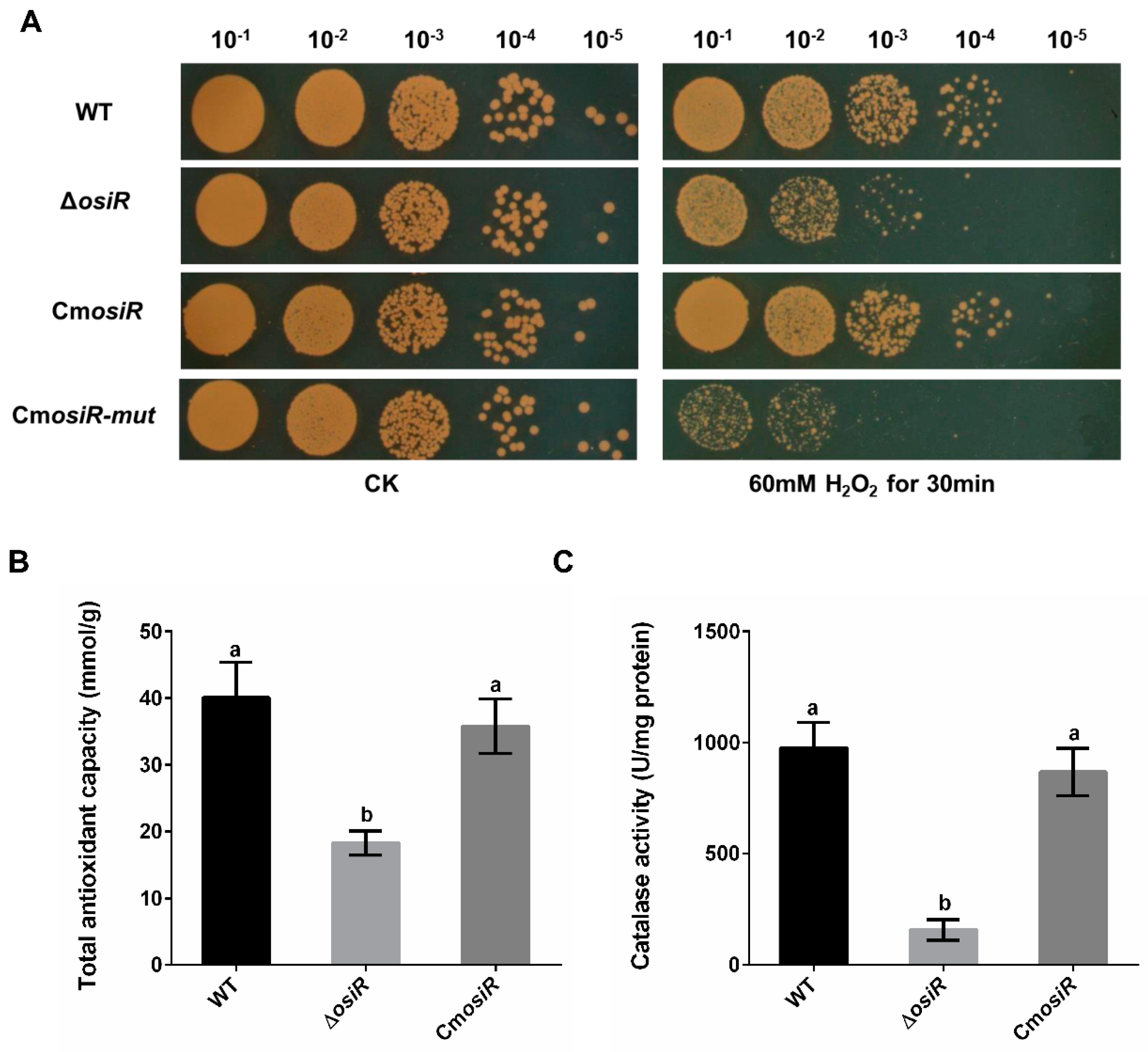
To further characterize the physiological role of OsiR in D. radiodurans under oxidative stress, the wild-type strain (WT), the osiR-deletion mutant strain (ΔosiR), and complementation strain (CmosiR) were exposed to 60 mM H2O2 for 30 min. Under normal growth conditions (CK), there was no significant difference in the survival status of these strains; however, under oxidative stress conditions (60 mM H2O2 for 30 min), the survival ability of ΔosiR decreased significantly compared to WT, while CmosiR had a similar survival ability to WT (Figure 3A). To further quantify the oxidative stress resistance of OsiR in D. radiodurans, the total antioxidant capacity (Figure 3B) and catalase activity of these strains (Figure 3C) were determined under oxidative stress; the results indicated that both total antioxidant capacity and catalase activity were significantly decreased in the absence of osiR. These results suggest that this ncRNA, as a positive regulator, plays an important role in the oxidative stress resistance in D. radiodurans, thus prompting us to search for target genes related to the oxidative stress response.
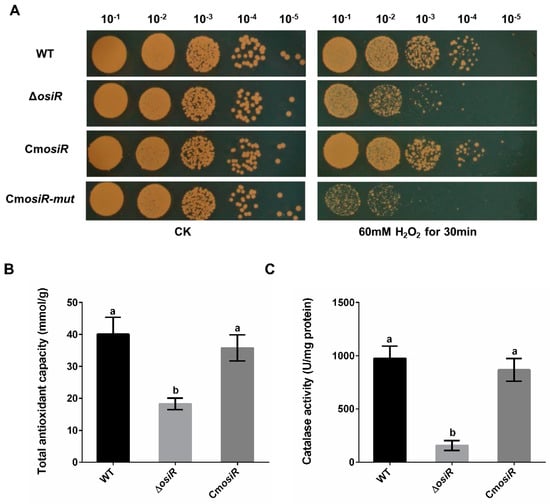
Figure 3.
Deletion of osiR reduces the oxidative stress tolerance in D. radiodurans. (A) survival phenotype plate assay under oxidative stress conditions (one of three independent replicates). Serial 10-fold dilutions of OD-standardized cultures were spotted onto TGY plates before and after exposure to 60 mM H2O2. WT, wild type; ΔosiR, osiR deletion mutant; CmosiR, complementary strain carrying wild-type osiR; CmosiR-mut, complementary strain carrying mutant osiR; CK, untreated culture control. Experiments were performed in triplicate. (B) and (C) effect of osiR mutation on the total antioxidant capacity and catalase activity of D. radiodurans. The values shown were derived from three independent experiments and error bars represent standard deviations. The statistical difference among the three samples (WT, ΔosiR, CmosiR) was determined by Tukey multiple comparisons (p < 0.05); the same letter indicates that the difference is not significant and different letters indicate significant differences.
2.3. katE2 Is a Target of OsiR in Response to Oxidative Stress
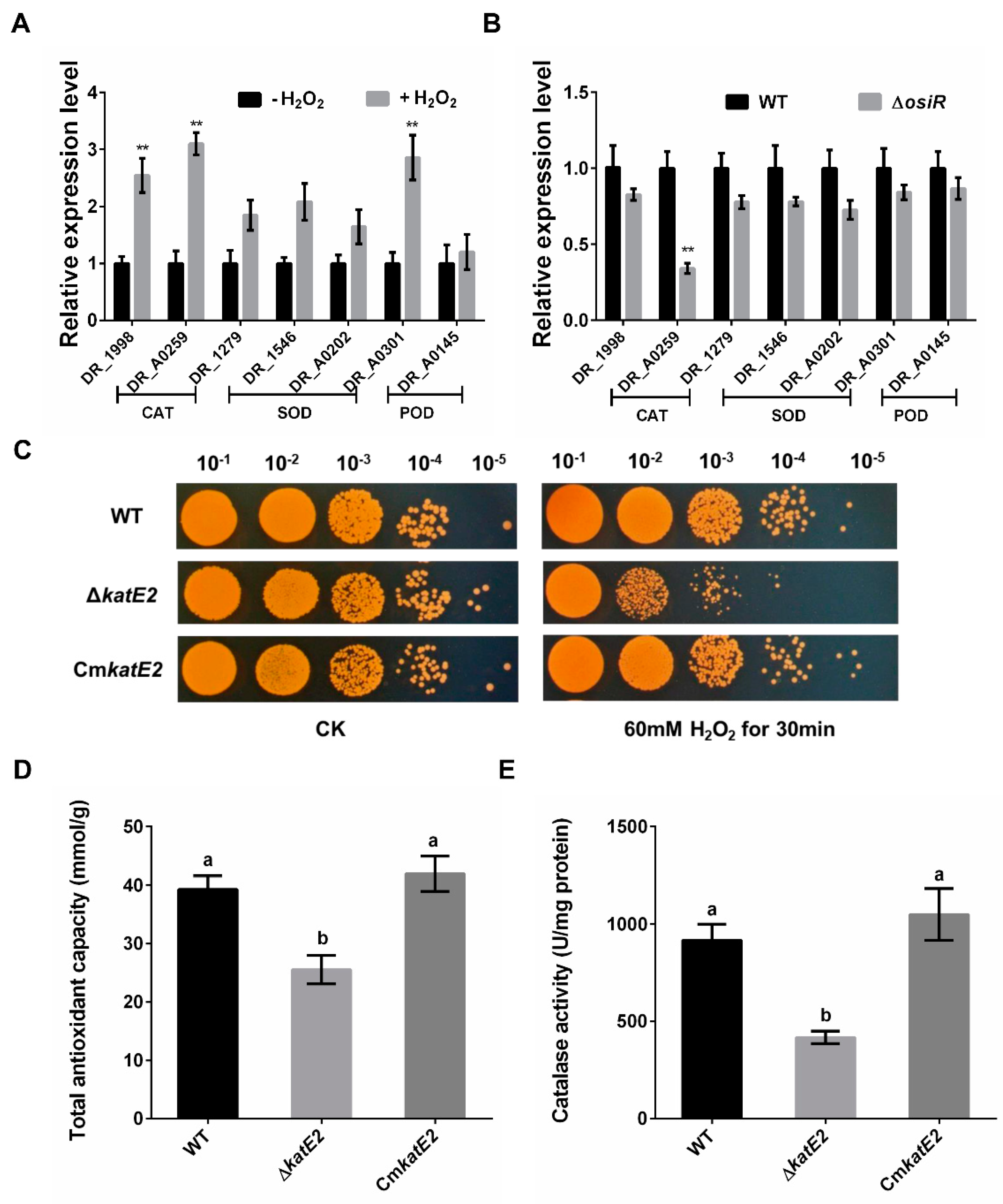
To identify potential targets of OsiR, qRT-PCR was used to determine the transcript levels of oxidative-related genes in D. radiodurans under oxidative stress conditions. The data showed that 3 genes, including katE2, exhibited a significant increase in expression after H2O2 shock (Figure 4A); however, only the katE2 transcript level was significantly downregulated in ΔosiR compared to that in WT (Figure 4B), suggesting that the expression of katE2 is OsiR dependent via an unknown mechanism. In addition, in silico (IntaRNA) analysis revealed a putative interaction between a typical stem–loop structure of OsiR (204–224 nt) and the CDS of the katE2 mRNA (843–866 nt), with an interaction energy of −19.69 kcal/mol (Figure 2C). Thus, it was speculated that katE2 is a potential target of OsiR under oxidative stress.
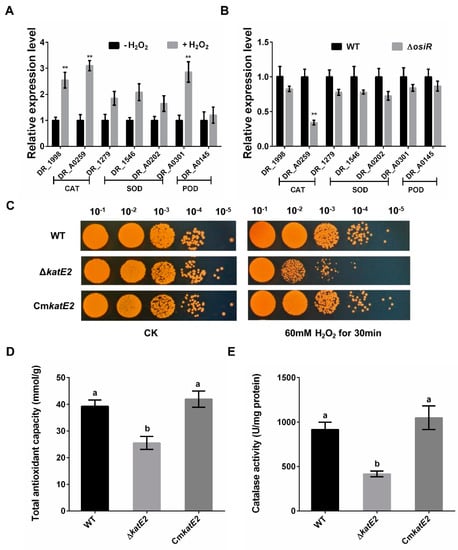
Figure 4.
Regulation of OsiR on genes related to oxidative stress resistance and functional analysis of katE2 under oxidative stress. (A) expression of seven selected oxidative stress–response genes of D. radiodurans grown in TGY with or without H2O2; (B) transcript levels of these genes of WT and ΔosiR strains grown in TGY with H2O2. All experiments were performed three times, and the results are represented as the mean ± standard deviation. Statistical analyses were performed using a Student’s t-test. The statistical significance is indicated as follows: **, p < 0.01; (C) survival phenotype plate assay of WT, ΔkatE2 and CmkatE2 under oxidative stress conditions; (D) and (E) effect of katE2 deletion on the total antioxidant capacity and catalase activity of D. radiodurans. The values shown were derived from three independent experiments, and error bars represent standard deviations. The statistical difference among the three samples (WT, ΔosiR, CmosiR) was determined by Tukey multiple comparisons (p < 0.05); the same letter indicates that the difference is not significant and different letters indicate significant differences.
katE2 encodes a KatE-type catalase that can protect cell components from oxidative damage under various stress conditions [11,16]. To investigate the biological function of katE2, a katE2-deletion mutant strain (ΔkatE2) and complementary strain (CmkatE2) were constructed, and oxidative stress tolerance was determined. The phenotypic results showed that ΔkatE2 exhibited higher sensitivity to oxidative stress than WT (Figure 4C). Consistent with the phenotypic results, the disruption of katE2 resulted in a significant decrease in the total antioxidant capacity and catalase activity in D. radiodurans (Figure 4D,E). In addition, expression of katE2 is also significantly decreased in Δsig1 deletion mutant (Figure 1D). Taken together, these data confirmed that katE2 is a potential target of OsiR and that the loss of katE2 significantly decreased the oxidative stress tolerance in D. radiodurans.
2.4. OsiR Positively Regulates the Expression of katE2
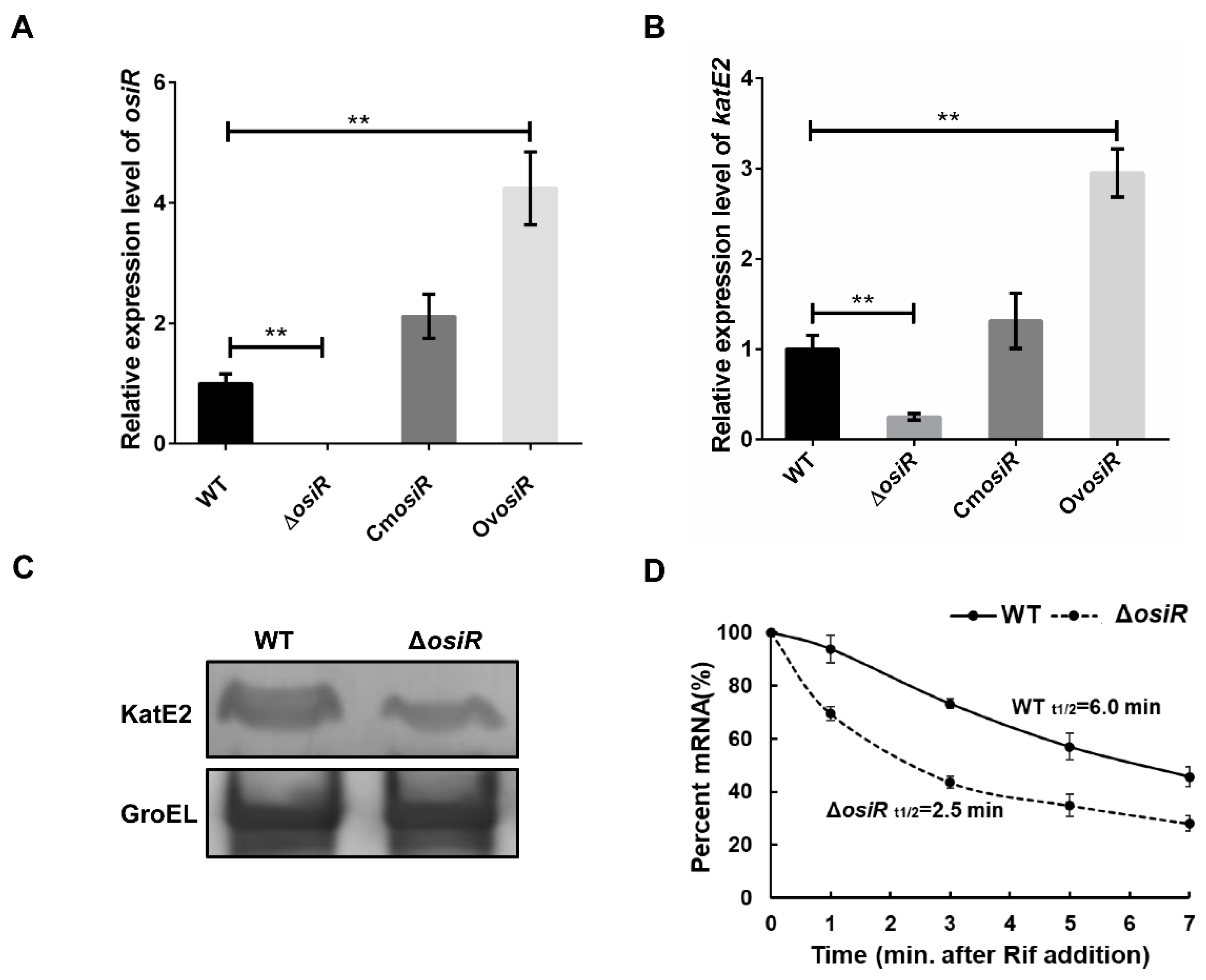
Positive regulation by ncRNA often occurs through the stabilization of transcripts, which leads to increased translation [41]. To further determine whether the presence of OsiR leads to an increased abundance of katE2 transcripts, the relative expression levels of osiR and katE2 in WT, ΔosiR, CmosiR and OvosiR under oxidative stress (60 mM H2O2 for 30 min) were examined by qRT-PCR. The data indicated that the deletion of osiR resulted in a significant reduction in katE2 transcript level, while the overexpression of osiR significantly increased the katE2 transcript level (Figure 5A, B). Western blot analysis showed a significant reduction in the KatE2 protein level in ΔosiR (Figure 5C). In addition, mRNA stability analysis indicated that the half-life of katE2 mRNA in WT and ΔosiR were 6.0 min and 2.5 min, respectively (Figure 5D). These results led to a hypothesis that, in the absence of OsiR, the katE2 transcript is rapidly degraded, leading to a decreased KatE2 protein production and oxidative stress tolerance in D. radiodurans.
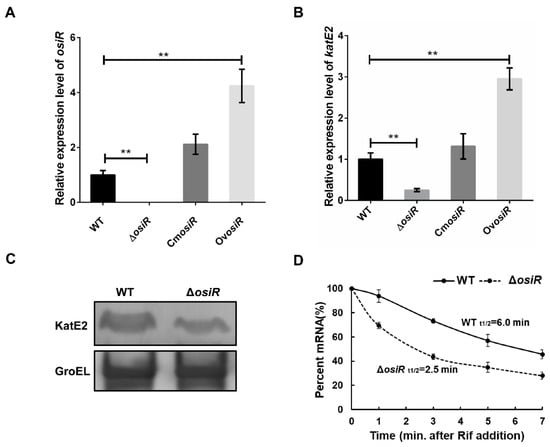
Figure 5.
Deletion of osiR leads to a decrease in katE2 expression. (A) and (B) qRT-PCR analysis of osiR and katE2 in WT, ΔosiR, CmosiRand OvosiR. All experiments were performed three times, and the results are represented as the mean ± standard deviation. Statistical analyses were performed using a Student’s t-test. Statistical significance is indicated as follows: **, p value < 0.01. (C) Western blot analysis of the KatE2 protein in WT and ΔosiR. GroEl protein served as a loading control; (D) determination of the katE2 mRNA half-life in WT and ΔosiR under oxidative stress conditions. The data shown are the averages of three biological replicates. Error bars represent standard deviations.
2.5. OsiR Directly Base Pairs With katE2 mRNA to Enhance Oxidative Stress Tolerance in D. radiodurans
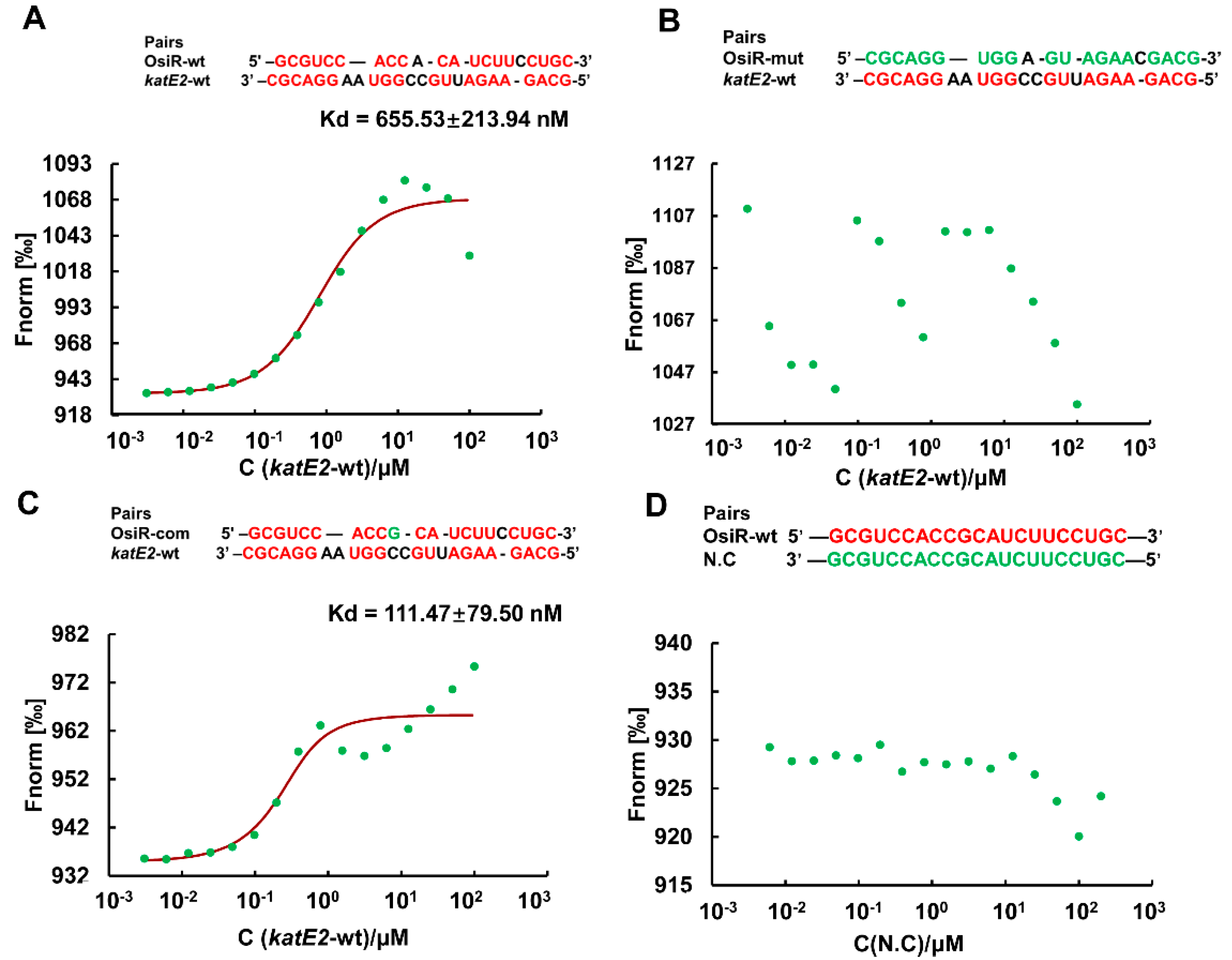
Taking the above results together, it was concluded that katE2 is a target of OsiR during the regulation of oxidative stress tolerance in D. radiodurans. ncRNA-mediated regulation of gene expression usually occurs by direct base pairing, and the IntaRNA program predicted a ncRNA-mRNA base pairing interaction between OsiR and the katE2 mRNA (Figure 2C). To verify this interaction, MST analysis, which allows for the sensitive measurement of molecular interactions in solution, was performed to detect the binding affinity between OsiR and the katE2 mRNA. The MST results showed that OsiR-wt bound katE2-wt in a normal manner with a dissociation constant (Kd) of 655.53 ± 213.94 nM, suggesting a strong interaction (Figure 6A). After replacing 19 bases in the base-pairing region of OsiR, there is no binding affinity between OsiR-mut and katE2-wt (Figure 6B). In addition, the OsiR-com containing compensatory mutations in the base-pairing region bound to katE2-wt in a high match level (Kd = 111.47 ± 79.50 nM) (Figure 6C), suggesting a stronger interaction. The negative control as shown in Figure 6D exhibited no binding curve between OisR-wt and N.C which contains the completely mismatched base complementary region with OsiR. Together, these data strongly suggest that OsiR is capable of interacting with the katE2 mRNA via direct base pairing and the 19-nt of OsiR is very important for the binding of OsiR and katE2 mRNA.
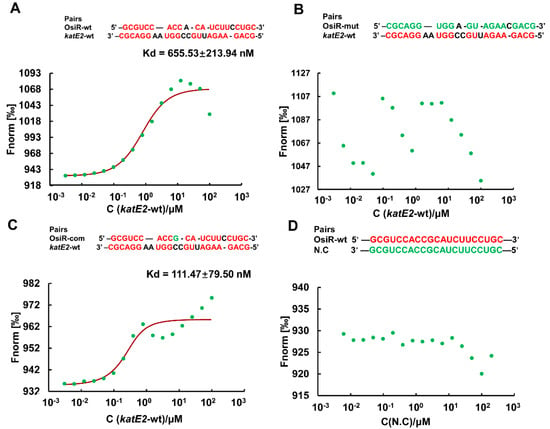
Figure 6.
The binding affinity of OsiR to the katE2 mRNA by MST analysis in vitro. Pairing nucleotides are shown in red. Point mutations introduced into synthesized oligonucleotide derivatives are shown in green. The Kd value reflects the binding affinity between two molecular fragments. wt: wild type; mut: mismatch mutation; com: compensatory mutation; N.C: negative control. (A) OsiR-wt/katE2-wt; (B) OsiR-mut/katE2-wt; (C) OsiR-com/ katE2-wt; (D) OsiR-wt/N.C
To further determine the effect of this base-pairing region of OsiR interacting with katE2 mRNA as confirmed by MST on the physiological function of OsiR, the oxidative stress tolerance of the ΔosiR complemented with mutated osiR (CmosiR-mut) was assessed. As shown in Figure 3A, the oxidative stress tolerance of ΔosiR was almost fully complemented by WT osiR but hardly complemented by the mutated osiR. These results strongly indicate that the 19-nt base-pair located on the stem–loop structure of OsiR, complementarity with katE2 mRNA, is functionally required for OsiR to positively regulate the oxidative stress tolerance in D. radiodurans.
3. Discussion
D. radiodurans is unparalleled among all known organisms with regard to extreme resistance to oxidative stress-generating agents which affects all cellular macromolecules [7,10]. More than a hundred genes have been identified to contribute to oxidative stress tolerance in D. radiodurans; however, to date, the regulatory mechanism underlying the extreme oxidative stress tolerance in D. radiodurans has not been fully elucidated [5,11]. ncRNA-mediated regulation plays an important role in stress adaptation and gene regulation in almost all bacteria [20,42,43]. Furthermore, post-transcriptional regulation by ncRNAs enables bacteria to respond to the signals in an extremely flexible and sensitive manner [44,45]. Over the last few years, an increasing number of bacterial ncRNAs have been identified by combining RNA-sequences and bioinformatics [27,28,36]; however, until now, there are very few studies unravelling the biological roles and/or identifying the target genes of ncRNAs in extremophilic bacterium D. radiodurans. To our knowledge, the research on the specific molecular regulatory mechanism of ncRNA in regulating oxidative stress resistance in D. radiodurans has not been reported. Our work shows that OsiR could be a novel Deinococcus-specific ncRNA that positively regulates oxidative stress tolerance via direct base-pairing with the katE2 mRNA in D. radiodurans. As indicated by Northern blot, qRT-PCR, and physiological phenotype analyses, the osiR gene was highly expressed under oxidative stress condition from a Sig1-dependent promoter. Moreover, deletion of osiR led to a significant decrease in oxidative stress tolerance and catalase activity in D. radiodurans.
KatEs are a kind of common catalase that detoxifies H2O2 widely distributed in Deinococcus genus (Table S1). D. radiodurans genomes encoded two KatE-type catalases (KatE1 and KatE2) to cope with oxidative stress [17]. Here, our results clearly demonstrate that katE2 mRNA is a target of OsiR under oxidative stress condition and that the expression of katE2 is positively regulated by OsiR at the posttranscriptional level. MST in vitro further demonstrated that the effect of OsiR on katE2 expression was achieved through a direct interaction mediated by base pairing. Furthermore, secondary structure analysis of the katE2 mRNA predicted a hairpin structure in the coding region downstream of the start codon, and such a hairpin structure is generally believed to affect translation efficiency. Interaction between OsiR with katE2 mRNA promotes the ribosome binding and progression (Figure S1). This is a novel regulatory mechanism different from that of katE1, the expression of which is controlled at the transcriptional level by protein regulators such as DrRRA [14], OxyR, and PprM [16], suggesting that the coordination of katE1 and katE2 expression may be an efficient strategy for ROS scavenging in D. radiodurans. In addition, deletion of katE2 also significantly decreased oxidative stress resistance as well as catalase activities in D. radiodurans. qRT-PCR analysis indicated that the transcription of katE2 was significantly induced by oxidative stress, again confirming the role of katE2 in oxidative stress tolerance.
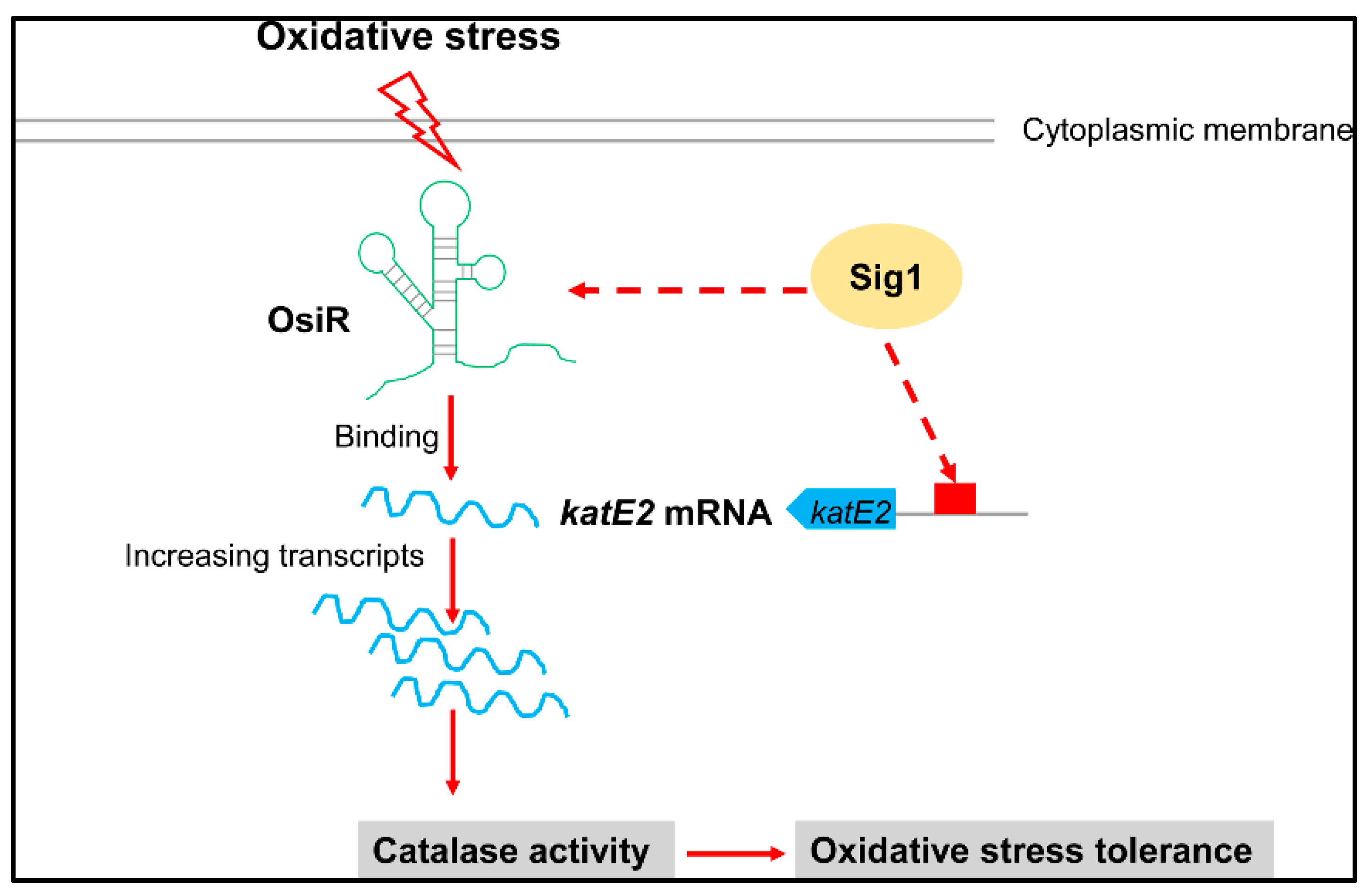
Taken together, our work indicates that OsiR is a novel ncRNA involved in the oxidative stress response in D. radiodurans. The working model as shown in Figure 7 illustrates the regulatory mechanism of OsiR with regard to how it regulates oxidative stress tolerance in D. radiodurans. At the transcriptional level of global regulation, the expression of osiR and katE2 depends on Sig1; at the posttranscriptional level, OsiR promotes expression of katE2 by increasing the stability of katE2 mRNA, and ultimately enhances the catalase activity and oxidative stress tolerance of D. radiodurans. Whether OsiR directly regulates the expression of other oxidative-related genes under various physiological processes remains to be further explored. Future research should help us understand why this bacterium is extremely resistant to ionizing radiation and oxidative stress. Our work presented herein exemplifies the growing appreciation of the importance of ncRNAs in Deinococcus and further expands the list of ncRNAs known to play a role in oxidative stress response.
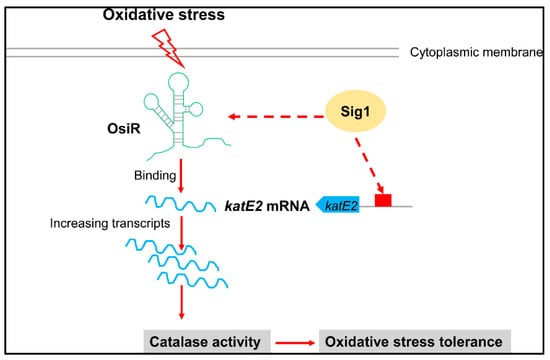
Figure 7.
Model for the OsiR-mediated regulatory network in the oxidative stress response in D. radiodurans. OsiR acts as a riboregulatory to contribute to oxidative stress tolerance and catalase activity via direct base pairing with katE2 mRNA or via indirect transcriptional activation of Sig1. For more details, see the results and discussion.
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
All bacterial strains and plasmids used in this study are described in Table 1. D. radiodurans was obtained from China General Microbiological Culture Collection Center (CGMCC 1.633, Beijing, China). D. radiodurans and derivatives were cultured in liquid TGY medium (1% tryptone, 0.1% glucose, and 0.5% yeast extract) at 30 °C or cultured on solid TGY medium with 1.5% agar. When required, kanamycin and chloromycetin were added to final concentrations of 20, 3.4 µg/mL, respectively. E. coli strains were cultured in LB medium.

Table 1.
Strains and plasmids used in this study.
4.2. Nucleotide Sequence
4.3. Construction of the Deletion Mutants and Complementation Strains
The osiR and katE2 deletion mutants were constructed by a double crossover recombination of a kanamycin (kan) resistance cassette into the genome as described previously [46,47]. PCR primers were designed according to the sequence of osiR in the genome. The primers OsiR-P1 and OsiR-P2 were used to amplify the upstream DNA fragment of osiR (432 bp). The kan resistance gene (NptII) was amplified from pKatAPH3 by the primers OsiR-P3 and OsiR-P4 (946 bp). The primers OsiR-P5 and OsiR-P6 were used to amplify the downstream DNA fragment of osiR (503 bp). The three amplified DNA fragments were used as templates for the overlap reaction and the resulting PCR fragment (1829 bp) was directly transformed into D. radiodurans. The single colony with kanamycin (20 µg/mL) resistance was selected. Finally, PCR and DNA sequencing were used to verify that the osiR gene was knocked out. The successfully constructed mutant was named ΔosiR.
DNA fragments containing the osiR gene and its promoter region were amplified by PCR. The PCR products were digested with HindIII/BamHI and ligated to the plasmid pRADZ3 to generate the complementation plasmid pRADZ3-osiR. pRADZ3-osiR was directly transformed into ΔosiR and D. radiodurans to generate the complementary strain CmosiR and the overexpressing strain OvosiR, which were selected with 20 µg/mL kanamycin and 3.4 µg/mL chloramphenicol. The strains were confirmed by PCR and DNA sequencing. The katE2-deletion mutant and complementary strain were constructed in the same way.
4.4. Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated with a PureLink RNA Mini Kit (Invitrogen, Thermo Fisher, Waltham, MA, United States) and was reverse-transcribed into cDNA with the PrimeScriptTM RT reagent kit. qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Vazyme Biotech, NanJing, China) with an AB7500 Fast Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The 16S rRNA gene (DR_r06) was used as the endogenous reference control to normalize the expression of target genes in each cDNA sample. The relative expression levels of genes were calculated by the comparative threshold cycle (2−ΔΔCT) method. All qRT-PCR experiments were performed with three biological replicates.
4.5. 5‘RACE
The transcriptional start site of the osiR gene was determined using a 5’ rapid amplification of cDNA ends (5’RACE) kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. In short, total RNA was isolated, and the first-strand of cDNA was generated using a specific primer (GSP1) for the osiR gene. The original cDNA was purified and the 3’ end of the cDNA was tailed with dATP by the recombinant terminal transferase, and dA-tailed cDNA amplification was performed using the specific primer GSP2 and the anchor primer. Then, the second amplification round was performed using the specific primer GSP3. The PCR products were cloned into the pMD18-T vector (Takara Bio, Dalian, China) and sequenced to map the 5’ ends of the transcript. The primer sequences of GSP1, GSP2, and GSP3 are listed in Table S2.
4.6. Northern Blot Analysis
Northern blot was performed following the previous reports [38,48]. Total RNA was isolated from WT D. radiodurans grown under oxidative stress and normal conditions. The RNA samples (15 µg) were first denatured at 90 °C for 10 min and then separated on an 8% urea polyacrylamide gel and electroblotted onto a positively charged nylon membrane. Digoxigenin-labeled DNA probes (30 nt) were synthesized (Sangon Biotech, Shanghai, China) and used for hybridization overnight at 42 °C. The membrane was incubated in blocking buffer for 2–3 h at room temperature, placed in blocking buffer with an anti-DIG antibody for 30 min and washed in DIG washing buffer 3–4 times. Finally, the membrane was incubated in detection buffer for 5–15 min and in CSPD (chemiluminescence substrate) solution for 15–30 min in the dark. Band intensity analyses were performed using an Amersham imager 600 RGE (GE Healthcare, Pittsburgh, USA).
4.7. Western Blot Analysis
Western blot was performed as previously described [16]. To investigate the KatE2 protein level in WT and ΔosiR under oxidative stress conditions, bacterial cells (OD600 = 0.8, treated with 60 mM H2O2 for 30 min) were harvested by centrifugation at 4 °C for 7 min. The cells were washed twice and resuspended in 1 × PBS and disrupted ultrasonically on ice with an ultrasonic processor. The cell-free extracts were collected by centrifugation, and the protein concentration was determined using the Bradford colorimetric assay with bovine serum albumin (BSA) as the standard. A 20 µL aliquot of each sample was heated at 10 °C for 8 min, subjected to 12% SDS-PAGE and transferred onto a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was incubated with a rabbit anti-KatE2 polyclonal antibody (1:2500). D. radiodurans GroEL as a control was detected using E. coli GroEL antiserum (1:2500). Peroxidase-conjugated goat anti-rabbit IgG (1:5000) was used as the secondary antibody. Chemiluminescent signals on the PVDF membrane were visualized using an Amersham imager 600 RGE (GE Healthcare, Pittsburgh, USA).
4.8. Oxidative Stress Survival Assays
Oxidative stress survival assays were carried out according to previous methods [46,48]. Strains were grown overnight in TGY broth at 30 °C and were transferred into fresh TGY broth up to OD600 = 0.8; 60 mM H2O2 was added to the medium for 30 min. Next, 8 µL of serial 1 × PBS 10-fold dilutions of OD-standardized cultures were spotted onto TGY plates, which were incubated at 30 °C for 3 days before colony growth was observed. All the assays were performed in triplicate.
4.9. Half-Life Experiment
Measurement of the katE2 mRNA half-life was performed as described previously [34]. To determine the half-life of katE2 mRNA under oxidative stress, 60 mM H2O2 was added to TGY broth when cells were cultured to OD600 = 0.8. Rifampin (40 mg/mL) was added immediately after H2O2 treatment, and 2 mL samples were collected at different time points (0, 1, 2, 3, 4, and 5 min). Subsequently, 4 mL RNAlater (Sigma, Saint Louis, MO, USA) was added to protect RNA, and the samples were incubated at room temperature for 5 min and centrifuged for 2 min at 12,000 rpm. The pellets were quickly frozen in liquid nitrogen. Total RNA was isolated and cDNA was synthesized and used to estimate katE2 mRNA levels by qRT-PCR. Data are presented as the percentage of katE2 mRNA levels relative to the amount of these mRNAs at time point zero. The data are the mean of three biological repeats.
4.10. Determination of Total Antioxidant Capacity and Catalase Activity
The total antioxidant capacity and catalase activities of samples were determined using the total antioxidant capacity kit and catalase activity kit (Beyotime Institute of Biotechnology, Jiangsu, China) according to the instructions. The principle of the FRAP method for determination of total antioxidant capacity is that Ferric-tripyridyltriazine (Fe3+-TPTZ) can be reduced by antioxidants under acidic conditions to produce blue Fe2+-TPTZ, and then the total antioxidant capacity in the sample can be determined by measuring the blue Fe2+-TPTZ at 593 nm. All bacterial cells (OD600 = 0.8, treated with 60 mM H2O2 for 30 min) were harvested by centrifugation for 7 min at 12,000 rpm, washed and resuspended in 1 × PBS, and disrupted on ice with an ultrasonic processor. The clear supernatants were collected by centrifugation at 13,000× g at 4 °C for 15 min. Total protein concentrations were determined by the Bradford method using BSA as the standard. The next operation steps were completed according to the instructions of total antioxidant capacity test kit. The catalase activity assays were performed according to the kit protocol. Samples were prepared and added to a 96-well plate and incubated at 25 °C for 15–45 min, and the absorbance was determined at 520 nm. For total antioxidant capacity, a standard curve was generated using the FeSO4•7H2O provided in the kit. Next, the 180 µL FRAP solution was added to a 96-well plate, and different samples, control, and FeSO4 standard solution were added; the plates were incubated at 37 °C for 5 min, and absorbance was determined at 593 nm. The total antioxidant capacity of the sample was calculated according to the standard curve. The experiments were performed three times independently.
4.11. Microscale Thermophoresis Measurements (MST)
MST experiments were performed according to previous reports [34,48]. A set of full-length ncRNAs containing WT or derivative OsiR for MST was transcribed in vitro by GenePharma using a MAXIscript kit (Thermo Fisher, MA, USA). Another set of 6-FAM-labeled 70-nt single-stranded RNAs (ssRNAs) containing WT katE2 mRNA (N- katE2 competitors) was synthesized by GenePharma (GenePharma, Jiangsu, China), as listed in Table S3. Four microliters of sample containing 250 nM labeled probe and increasing concentrations of a non-labeled competitor (from 10 nM to 340 µM) were loaded into standard treated silicon capillaries (Monolith NT.115 series capillaries; catalog no. MO-K002). Measurements were carried out using a Monolith NT.115 instrument (NanoTemper Technologies, Munich, Germany) at 25 °C in diethyl pyrocarbonate (DEPC)-treated water with 40% excitation power and medium MST-Power. The dissociation constant (Kd) was calculated as described previously [49]. Data analyses were performed using Nanotemper Analysis software (NanoTemper Technologies, Munich, Germany).
5. Patents
201810666398.0; 201810666544.X.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/9/3200/s1. Supplementary Figure S1: Schematic of the interaction between OsiR and katE2 mRNA. Supplementary Table S1: Distribution of homologous genes of katE1 and katE2 in Deinococcus species. Supplementary Table S2: Primers used in this study. Supplementary Table S3: Synthesized ssRNA oligonucleotide derivatives for MST.
Author Contributions
Conceptualization, L.G., M.L., and M.C.; methodology, L.G., X.C., Y.T., Y.Y., Y.Z., Z.Z., W.Z., M.L., and M.C.; formal analysis, L.G., X.C., Y.T., Y.Y., Y.Z., Z.Z., W.Z., M.L., and M.C.; resources, L.G.; data curation, L.G. and M.C.; writing—original draft preparation, L.G.; writing—review and editing, M.L. and M.C.; supervision, M.C.; project administration, M.L. and M.C.; funding acquisition, Z.Z., M.L., and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Agriculture Transgenic Program (No. 2019ZX08010-004; 2016ZX08009003-002) and the National Natural Science Foundation of China (No. 31570080, No. 31930004, No. 31770067).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Battista, J.R.; Earl, A.M.; Park, M.J. Why is Deinococcus Radiodurans so Resistant to Ionizing Radiation? Trends Microbiol. 1999, 7, 362–365. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Tatusov, R.L.; Minton, K.W.; Koonin, E.V.; Daly, M.J. Genome of the Extremely Radiation-Resistant Bacterium Deinococcus Radiodurans Viewed from the Perspective of Comparative Genomics. Microbiol. Mol. Biol. Rev. 2001, 65, 44–79. [Google Scholar] [CrossRef] [PubMed]
- Englander, J.; Klein, E.; Brumfeld, V.; Sharma, A.K.; Doherty, A.J.; Minsky, A. DNA Toroids: Framework for DNA Repair in Deinococcus Radiodurans and in Germinating Bacterial Spores. J. Bacteriol. 2004, 186, 5973–5977. [Google Scholar] [CrossRef] [PubMed]
- Slade, D.; Radman, M. Oxidative Stress Resistance in Deinococcus Radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [PubMed]
- Agapov, A.A.; Kulbachinskiy, A.V. Mechanisms of Stress Resistance and Gene Regulation in the Radioresistant Bacterium Deinococcus radiodurans. Biochem. (Moscow) 2015, 80, 1201–1216. [Google Scholar] [CrossRef] [PubMed]
- D’Autreaux, B.; Toledano, M.B. ROS as Signalling Molecules: Mechanisms that Generate Specificity in ROS Homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.J. A New Perspective On Radiation Resistance Based on Deinococcus Radiodurans. Nat. Rev. Microbiol. 2009, 7, 237–245. [Google Scholar] [CrossRef]
- Imlay, J.A. Cellular Defenses Against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef]
- Mishra, S.; Imlay, J. Why Do Bacteria Use so Many Enzymes to Scavenge Hydrogen Peroxide? Arch. Biochem. Biophys. 2012, 525, 145–160. [Google Scholar] [CrossRef]
- Battista, J.R. Against All Odds: The Survival Strategies of Deinococcus Radiodurans. Annu. Rev. Microbiol. 1997, 51, 203–224. [Google Scholar] [CrossRef]
- Lim, S.; Jung, J.H.; Blanchard, L.; de Groot, A. Conservation and Diversity of Radiation and Oxidative Stress Resistance Mechanisms in Deinococcus Species. Fems. Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Wu, Y.; Sheng, D.; Zheng, Z.; Gao, G.; Hua, Y. Chemiluminescence Assay for Reactive Oxygen Species Scavenging Activities and Inhibition on Oxidative Damage of DNA in Deinococcus Radiodurans. Lumin. 2004, 19, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Schellhorn, H.E. Induction of Resistance to Hydrogen Peroxide and Radiation in Deinococcus Radiodurans. Can. J. Microbiol. 1995, 41, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, G.; Chen, H.; Zhao, Y.; Xu, N.; Tian, B.; Hua, Y. DrRRA: A Novel Response Regulator Essential for the Extreme Radioresistance of Deinococcus Radiodurans. Mol. Microbiol. 2008, 67, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xu, G.; Zhao, Y.; Tian, B.; Lu, H.; Yu, X.; Xu, Z.; Ying, N.; Hu, S.; Hua, Y. A Novel OxyR Sensor and Regulator of Hydrogen Peroxide Stress with One Cysteine Residue in Deinococcus Radiodurans. PLoS ONE 2008, 3, e1602. [Google Scholar] [CrossRef]
- Jeong, S.W.; Seo, H.S.; Kim, M.K.; Choi, J.I.; Lim, H.M.; Lim, S. PprM is Necessary for Up-Regulation of katE1, Encoding the Major Catalase of Deinococcus Radiodurans, under Unstressed Culture Conditions. J. Microbiol. 2016, 54, 426–431. [Google Scholar] [CrossRef]
- Jeong, S.W.; Jung, J.H.; Kim, M.K.; Seo, H.S.; Lim, H.M.; Lim, S. The Three Catalases in Deinococcus Radiodurans: Only Two Show Catalase Activity. Biochem. Biophys. Res. Commun. 2016, 469, 443–448. [Google Scholar] [CrossRef]
- Nitzan, M.; Rehani, R.; Margalit, H. Integration of Bacterial Small RNAs in Regulatory Networks. Annu. Rev. Biophys. 2017, 46, 131–148. [Google Scholar] [CrossRef]
- Bouche, F.; Bouche, J.P. Genetic Evidence that DicF, a Second Division Inhibitor Encoded by the Escherichia coli dicB Operon, is Probably RNA. Mol. Microbiol. 1989, 3, 991–994. [Google Scholar] [CrossRef]
- Holmqvist, E.; Wagner, E.G.H. Impact of Bacterial sRNAs in Stress Responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef]
- Papenfort, K.; Silpe, J.E.; Schramma, K.R.; Cong, J.P.; Seyedsayamdost, M.R.; Bassler, B.L. A Vibrio Cholerae Autoinducer-Receptor Pair that Controls Biofilm Formation. Nat. Chem. Biol. 2017, 13, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.F.; Micheva-Viteva, S.; Li, N.; Hong-Geller, E. Small RNA-mediated Regulation of Host-Pathogen Interactions. Virulence 2013, 4, 785–795. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Papenfort, K.; Vogel, J. Small RNA Functions in Carbon Metabolism and Virulence of Enteric Pathogens. Front Cell Infect. Microbiol. 2014, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea: Who They are, What They Do, and How They Do It. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Vogel, J. Multiple Target Regulation by Small Noncoding RNAs Rewires Gene Expression at the Post-Transcriptional Level. Res. Microbiol. 2009, 160, 278–287. [Google Scholar] [CrossRef]
- Lu, X.; Goodrich-Blair, H.; Tjaden, B. Assessing Computational Tools for the Discovery of Small RNA Genes in Bacteria. RNA 2011, 17, 1635–1647. [Google Scholar] [CrossRef]
- Waters, L.S.; Storz, G. Regulatory RNAs in Bacteria. Cell 2009, 136, 615–628. [Google Scholar] [CrossRef]
- Lalaouna, D.; Simoneau-Roy, M.; Lafontaine, D.; Masse, E. Regulatory RNAs and Target mRNA Decay in Prokaryotes. Biochim. Et Biophys. Acta (BBA)—Gene Regul. Mech. 2013, 1829, 742–747. [Google Scholar] [CrossRef]
- Altuvia, S.; Weinstein-Fischer, D.; Zhang, A.; Postow, L.; Storz, G. A Small, Stable RNA Induced by Oxidative Stress: Role as a Pleiotropic Regulator and Antimutator. Cell 1997, 90, 43–53. [Google Scholar] [CrossRef]
- De Lay, N.; Gottesman, S. A Complex Network of Small Non-Coding RNAs Regulate Motility in Escherichia Coli. Mol. Microbiol. 2012, 86, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Tjaden, B.; Goodwin, S.S.; Opdyke, J.A.; Guillier, M.; Fu, D.X.; Gottesman, S.; Storz, G. Target Prediction for Small, Noncoding RNAs in Bacteria. Nucleic Acids Res. 2006, 34, 2791–2802. [Google Scholar] [CrossRef] [PubMed]
- Barshishat, S.; Elgrably-Weiss, M.; Edelstein, J.; Georg, J.; Govindarajan, S.; Haviv, M.; Wright, P.R.; Hess, W.R.; Altuvia, S. OxyS Small RNA Induces Cell Cycle Arrest to Allow DNA Damage Repair. EMBO J. 2018, 37, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhan, Y.; Yan, Y.; Liu, Y.; Hu, G.; Wang, S.; Yang, H.; Qiu, X.; Liu, Y.; Li, J.; et al. The Pseudomonas stutzeri-Specific Regulatory Noncoding RNA NfiS Targets katB mRNA Encoding a Catalase Essential for Optimal Oxidative Resistance and Nitrogenase Activity. J. Bacteriol. 2019, 201, e00334-19. [Google Scholar] [CrossRef]
- Chen, X.; Quinn, A.M.; Wolin, S.L. Ro Ribonucleoproteins Contribute to the Resistance of Deinococcus Radiodurans to Ultraviolet Irradiation. Genes Dev. 2000, 14, 777–782. [Google Scholar] [CrossRef]
- Chen, X.; Taylor, D.W.; Fowler, C.C.; Galan, J.E.; Wang, H.W.; Wolin, S.L. An RNA Degradation Machine Sculpted by Ro Autoantigen and Noncoding RNA. Cell 2013, 153, 166–177. [Google Scholar] [CrossRef]
- Tsai, C.H.; Liao, R.; Chou, B.; Contreras, L.M. Transcriptional Analysis of Deinococcus Radiodurans Reveals Novel Small RNAs that are Differentially Expressed under Ionizing Radiation. Appl. Environ. Microbiol. 2015, 81, 1754–1764. [Google Scholar] [CrossRef]
- Xue, D.; Chen, Y.; Li, J.; Han, J.; Liu, Y.; Jiang, S.; Zhou, Z.; Zhang, W.; Chen, M.; Lin, M.; et al. Targeting Hsp20 Using the Novel Small Non-Coding RNA DnrH Regulates Heat Tolerance in Deinococcus radiodurans. Front. Microbiol. 2019, 10, 2354. [Google Scholar] [CrossRef]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate Boundary Prediction and Improved Detection of Structural RNAs. RNA 2012, 18, 900–914. [Google Scholar] [CrossRef]
- Bernhart, S.H.; Hofacker, I.L.; Will, S.; Gruber, A.R.; Stadler, P.F. RNAalifold: Improved Consensus Structure Prediction for RNA Alignments. BMC Bioinform. 2008, 9, 474. [Google Scholar] [CrossRef]
- Zapf, R.L.; Wiemels, R.E.; Keogh, R.A.; Holzschu, D.L.; Howell, K.M.; Trzeciak, E.; Caillet, A.R.; King, K.A.; Selhorst, S.A.; Naldrett, M.J.; et al. The Small RNA Teg41 Regulates Expression of the Alpha Phenol-Soluble Modulins and is Required for Virulence in Staphylococcus aureus. mBio 2019, 10, e02484-18. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.V.; Roberts, J.T.; Patterson, D.G.; Coley, A.B.; Allred, J.A.; Denner, J.M.; Johnson, J.P.; Mullen, G.E.; O’Neal, T.K.; Smith, J.T.; et al. Novel Small RNA (sRNA) Landscape of the Starvation-Stress Response Transcriptome of Salmonella Enterica Serovar Typhimurium. RNA Biol. 2016, 13, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Verneuil, N.; Hartke, A.; Giard, J.C. Physiological Roles of Small RNA Molecules. Microbiology 2014, 160, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Little, R.H.; Malone, J.G. Quick Change: Post-Transcriptional Regulation in Pseudomonas. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Lenz, D.H.; Mok, K.C.; Lilley, B.N.; Kulkarni, R.V.; Wingreen, N.S.; Bassler, B.L. The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio Harveyi and Vibrio Cholerae. Cell 2004, 118, 69–82. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, J.; Liu, X.; Liu, Y.; Guo, C.; Zhang, L.; Han, J.; Wu, X.; Xue, D.; Gomaa, A.E.; et al. DrwH, a Novel WHy Domain-Containing Hydrophobic LEA5C Protein from Deinococcus radiodurans, Protects Enzymatic Activity under Oxidative Stress. Sci. Rep. 2017, 7, 9281. [Google Scholar] [CrossRef]
- Appukuttan, D.; Seo, H.S.; Jeong, S.; Im, S.; Joe, M.; Song, D.; Choi, J.; Lim, S. Expression and Mutational Analysis of DinB-Like Protein DR0053 in Deinococcus radiodurans. PLoS ONE 2015, 10, e118275. [Google Scholar] [CrossRef]
- Zhan, Y.; Yan, Y.; Deng, Z.; Chen, M.; Lu, W.; Lu, C.; Shang, L.; Yang, Z.; Zhang, W.; Wang, W.; et al. The Novel Regulatory ncRNA, NfiS, Optimizes Nitrogen Fixation Via Base Pairing with the Nitrogenase Gene nifK mRNA in Pseudomonas Stutzeri A1501. Proc. Natl. Acad. Sci. USA 2016, 113, E4348–E4356. [Google Scholar] [CrossRef]
- Lippok, S.; Seidel, S.A.; Duhr, S.; Uhland, K.; Holthoff, H.P.; Jenne, D.; Braun, D. Direct Detection of Antibody Concentration and Affinity in Human Serum Using Microscale Thermophoresis. Anal. Chem. 2012, 84, 3523–3530. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).