Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field
Abstract
1. Introduction
2. Results
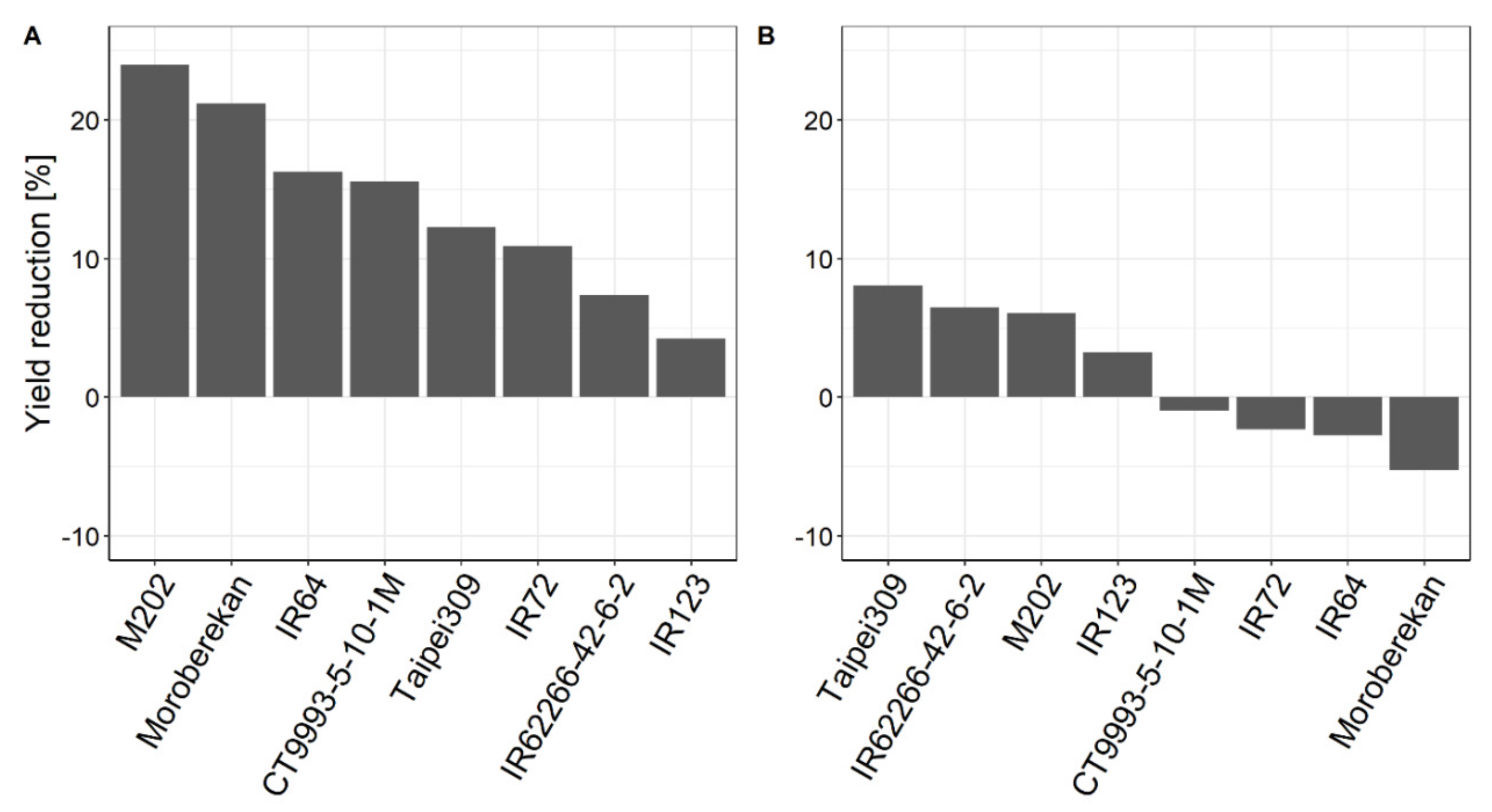
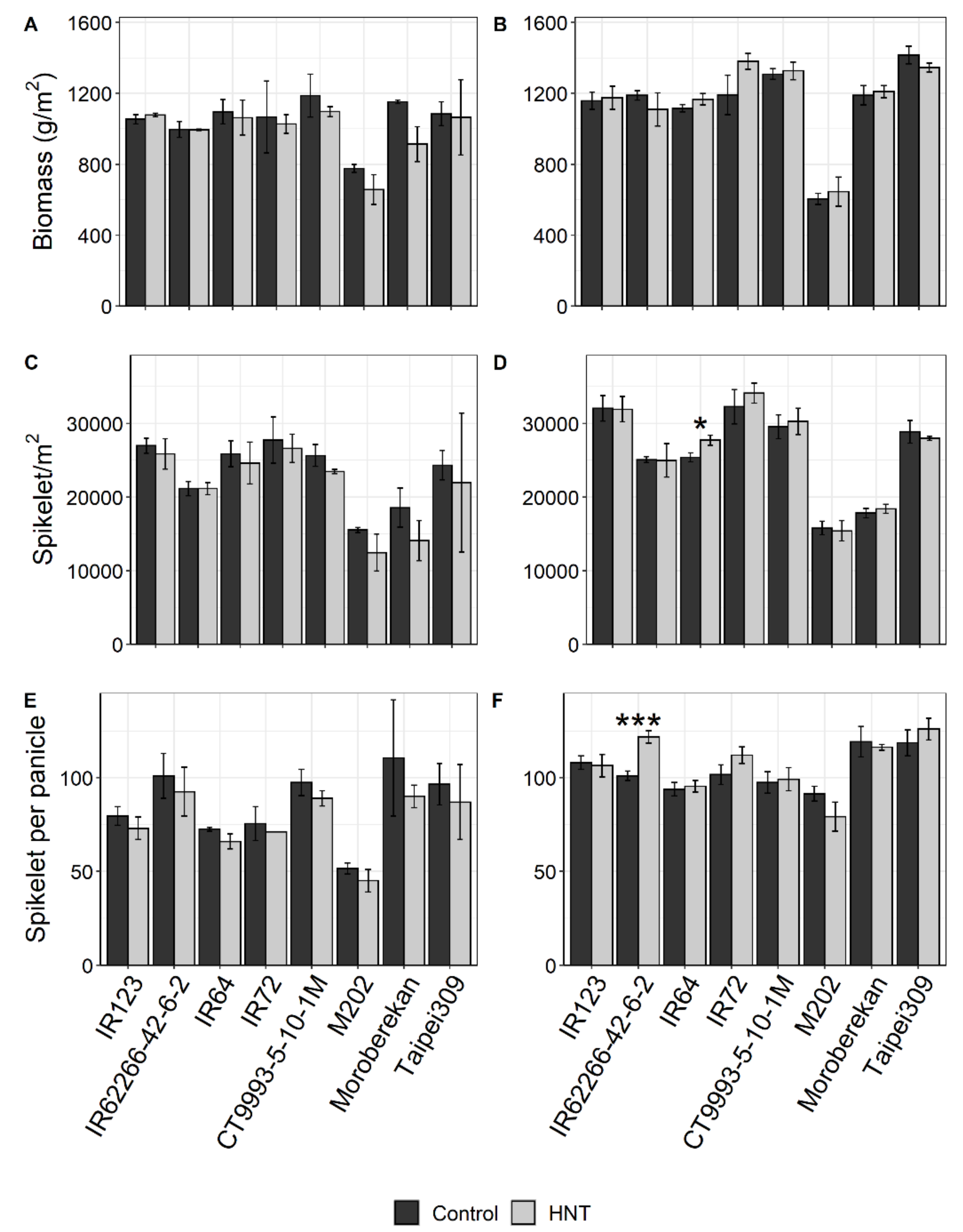
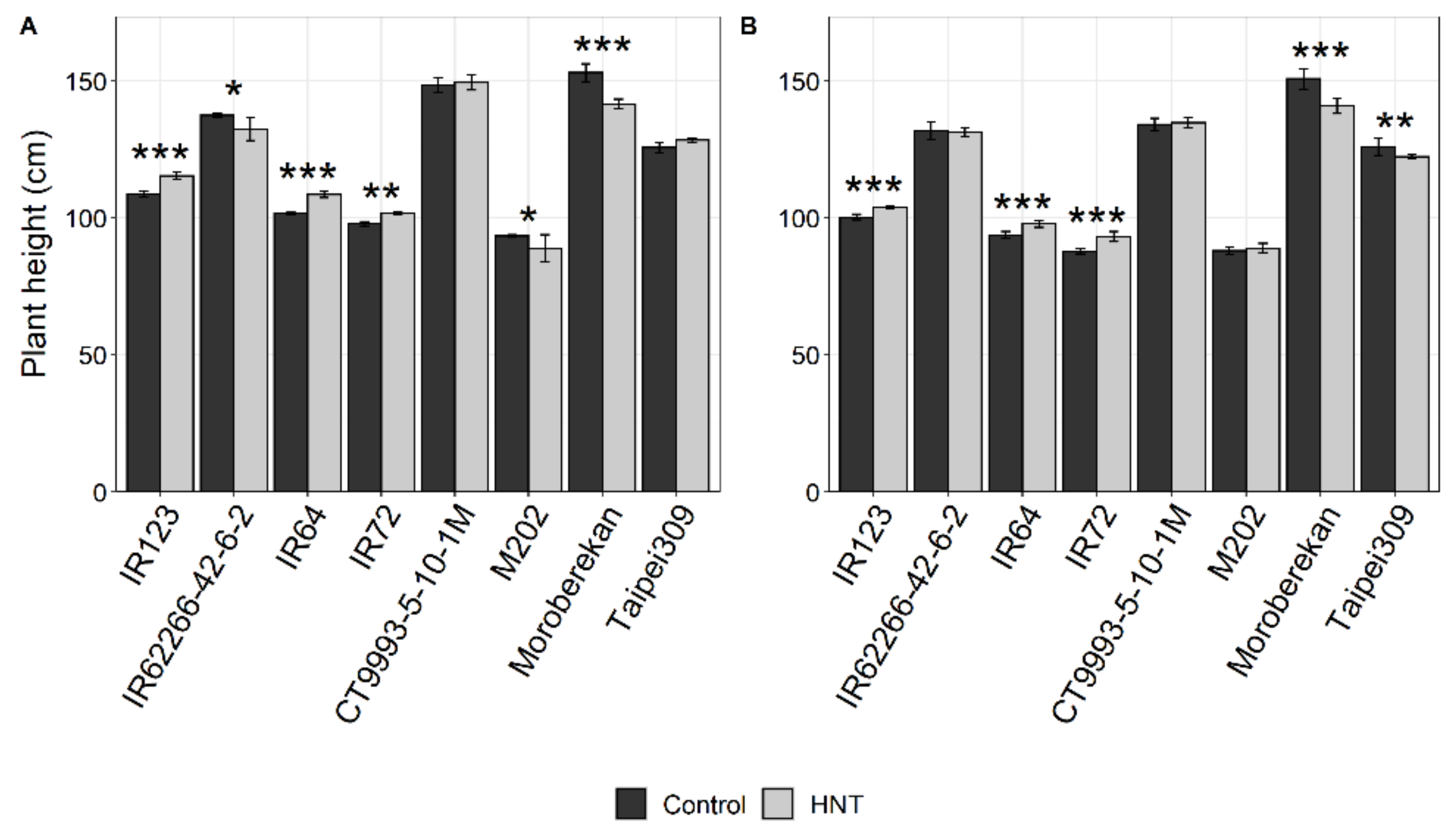
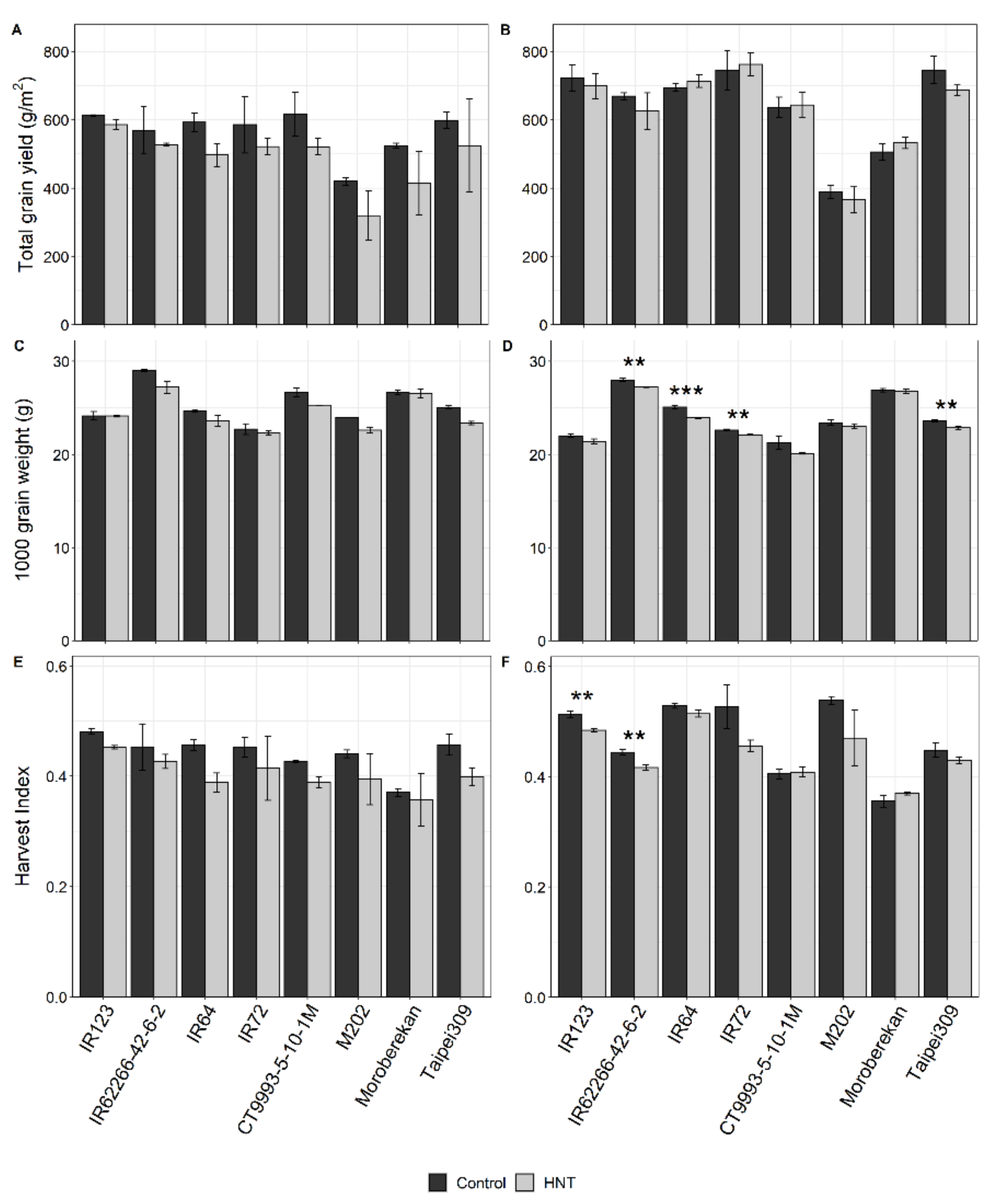
2.1. Influence of HNT on Agronomic Parameters
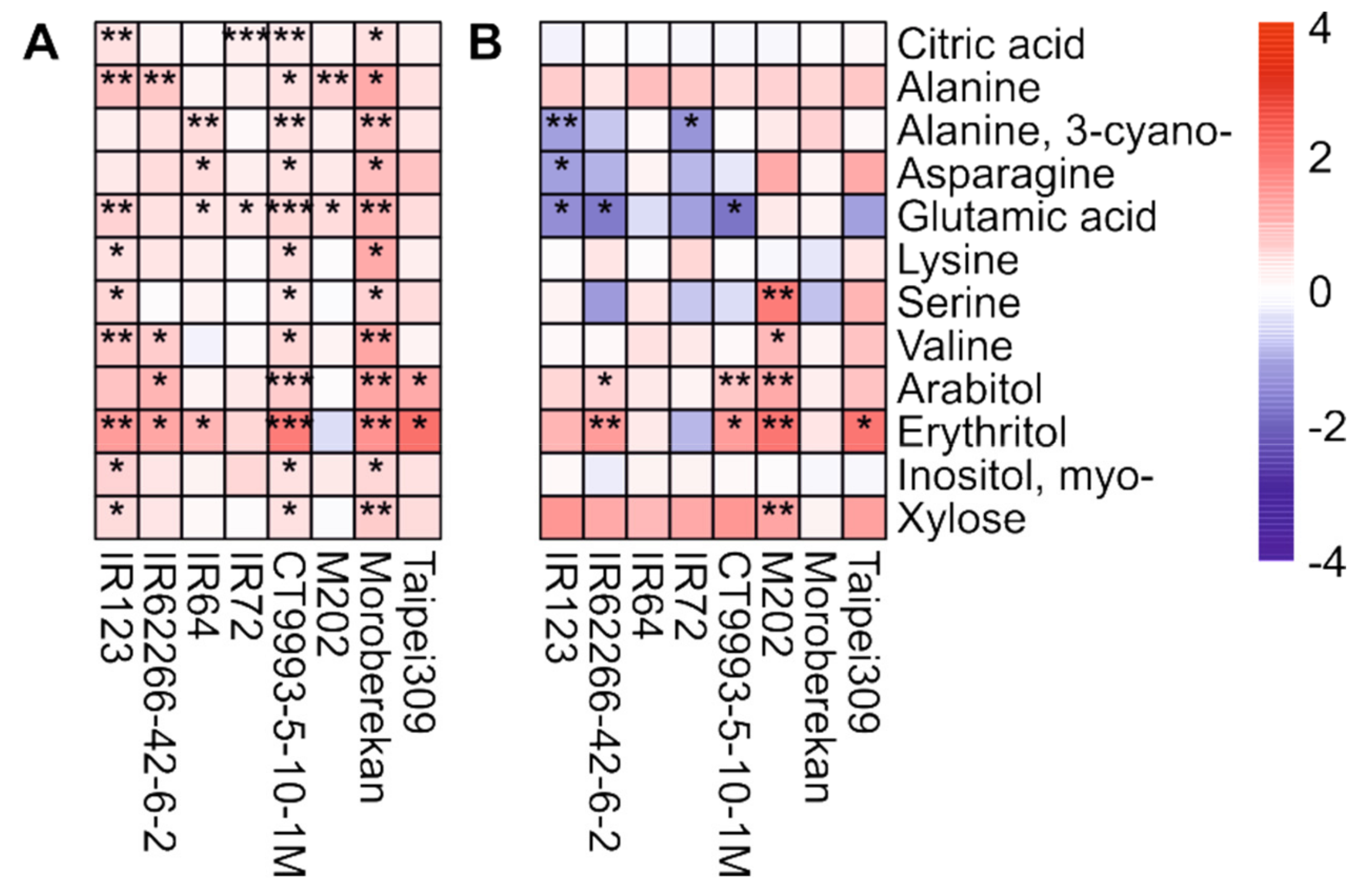
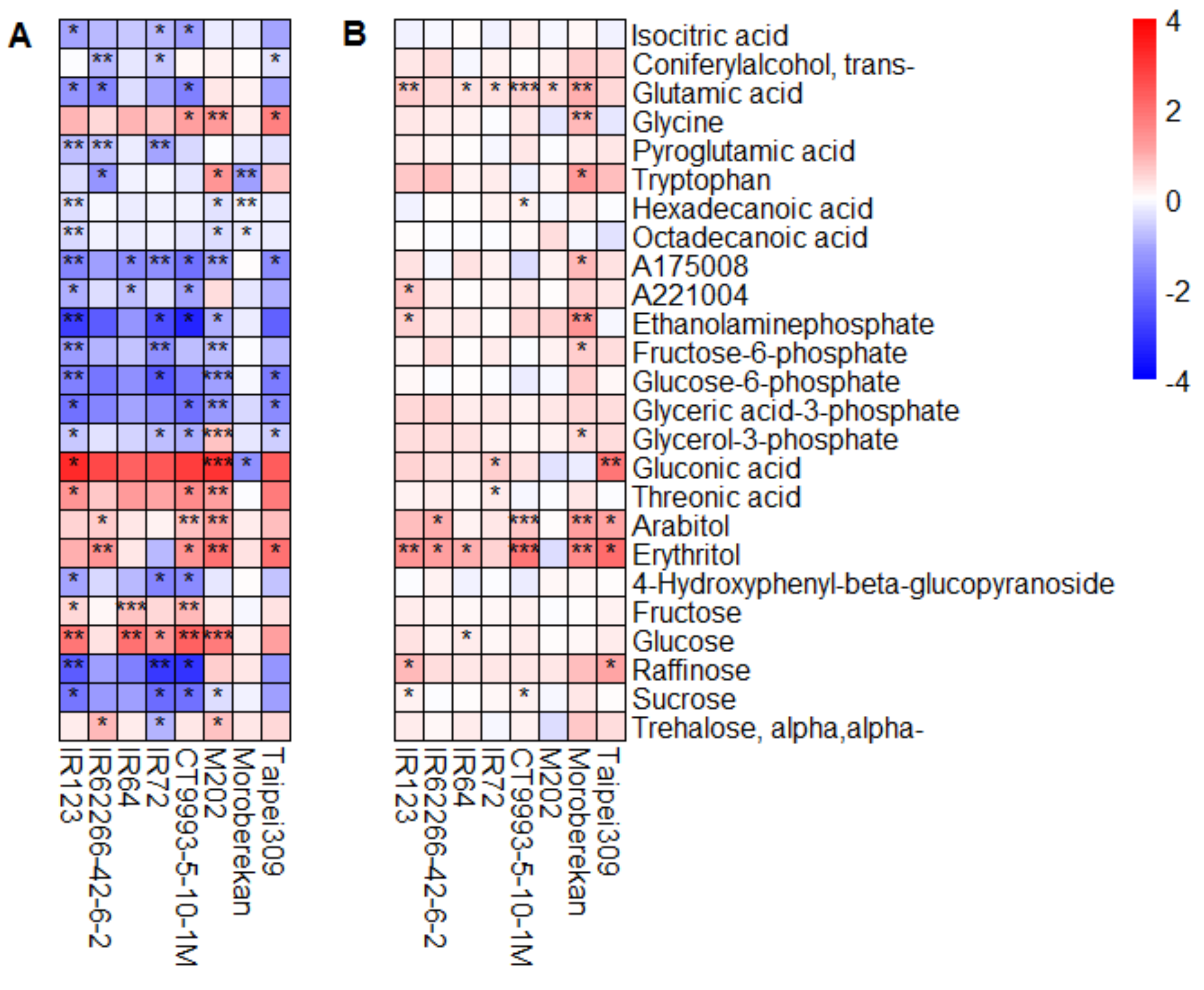
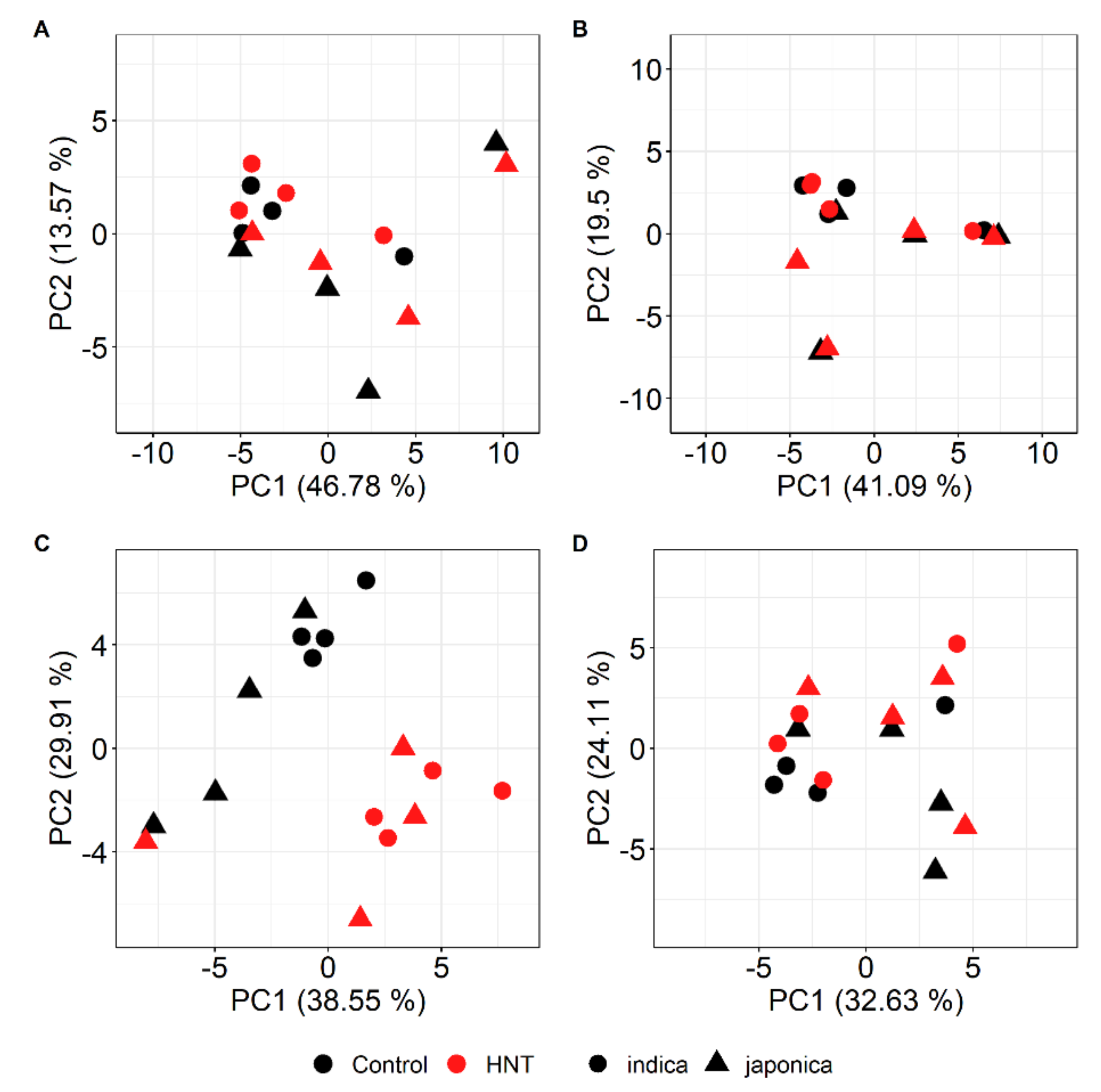
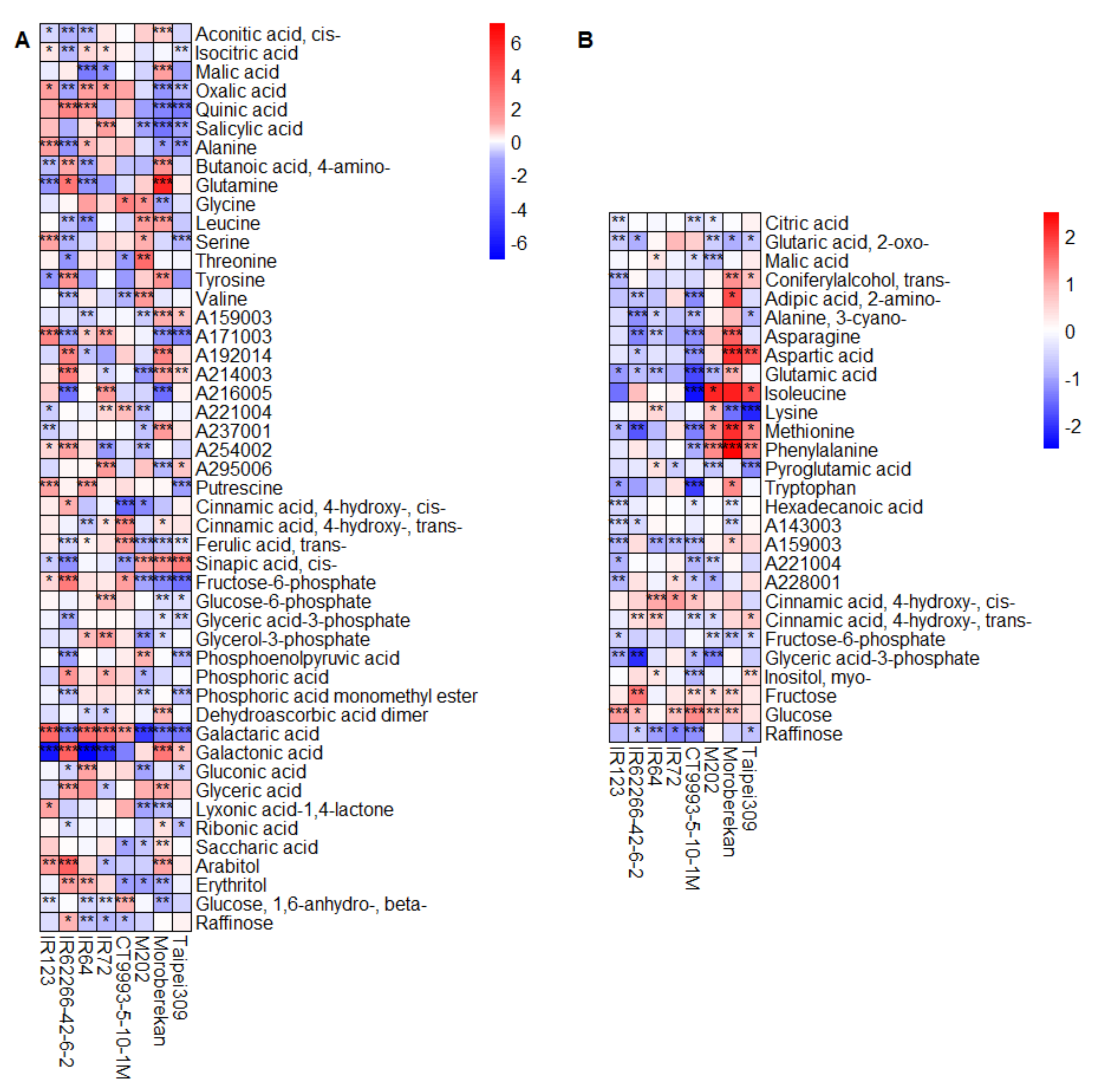
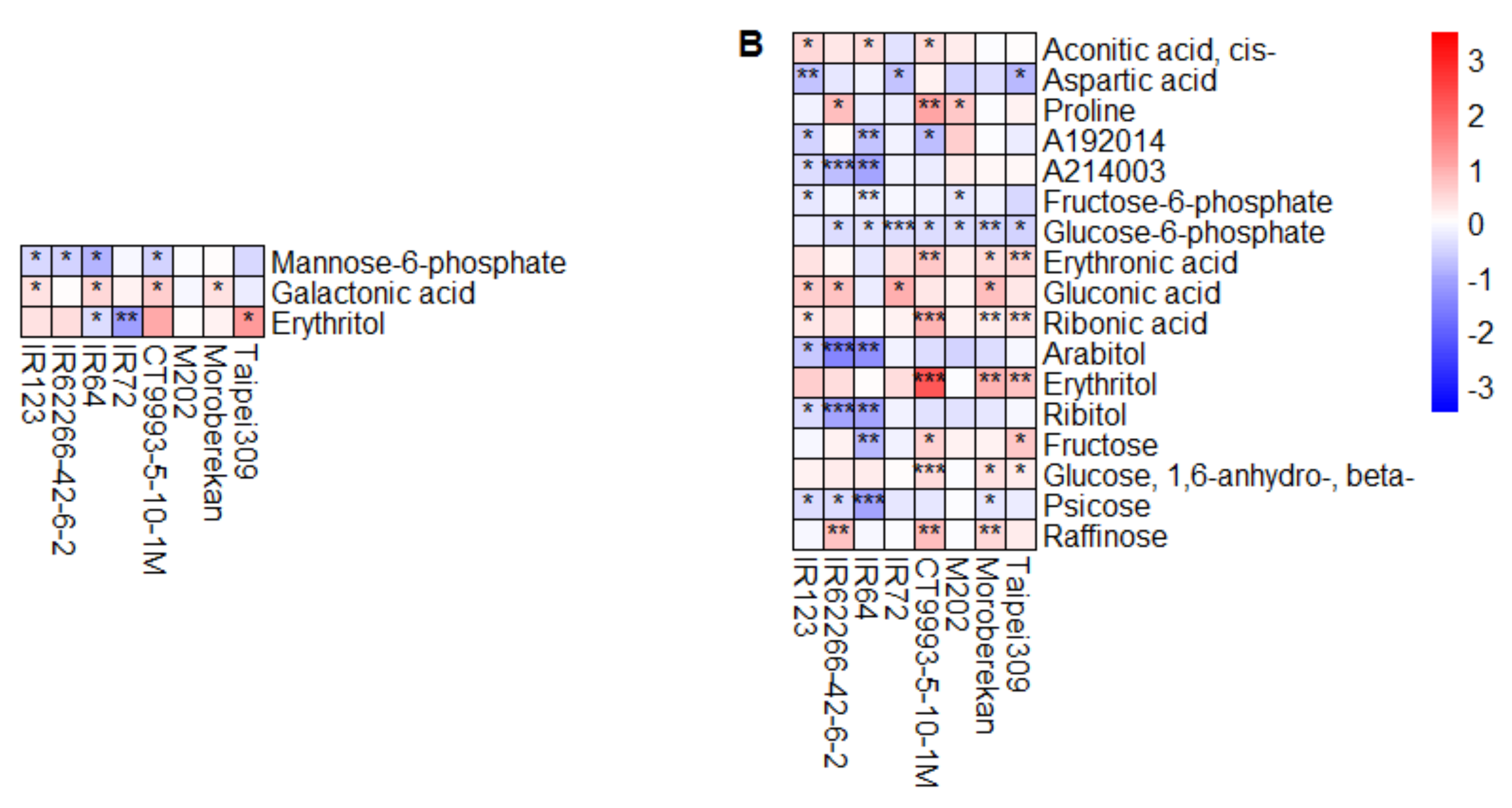
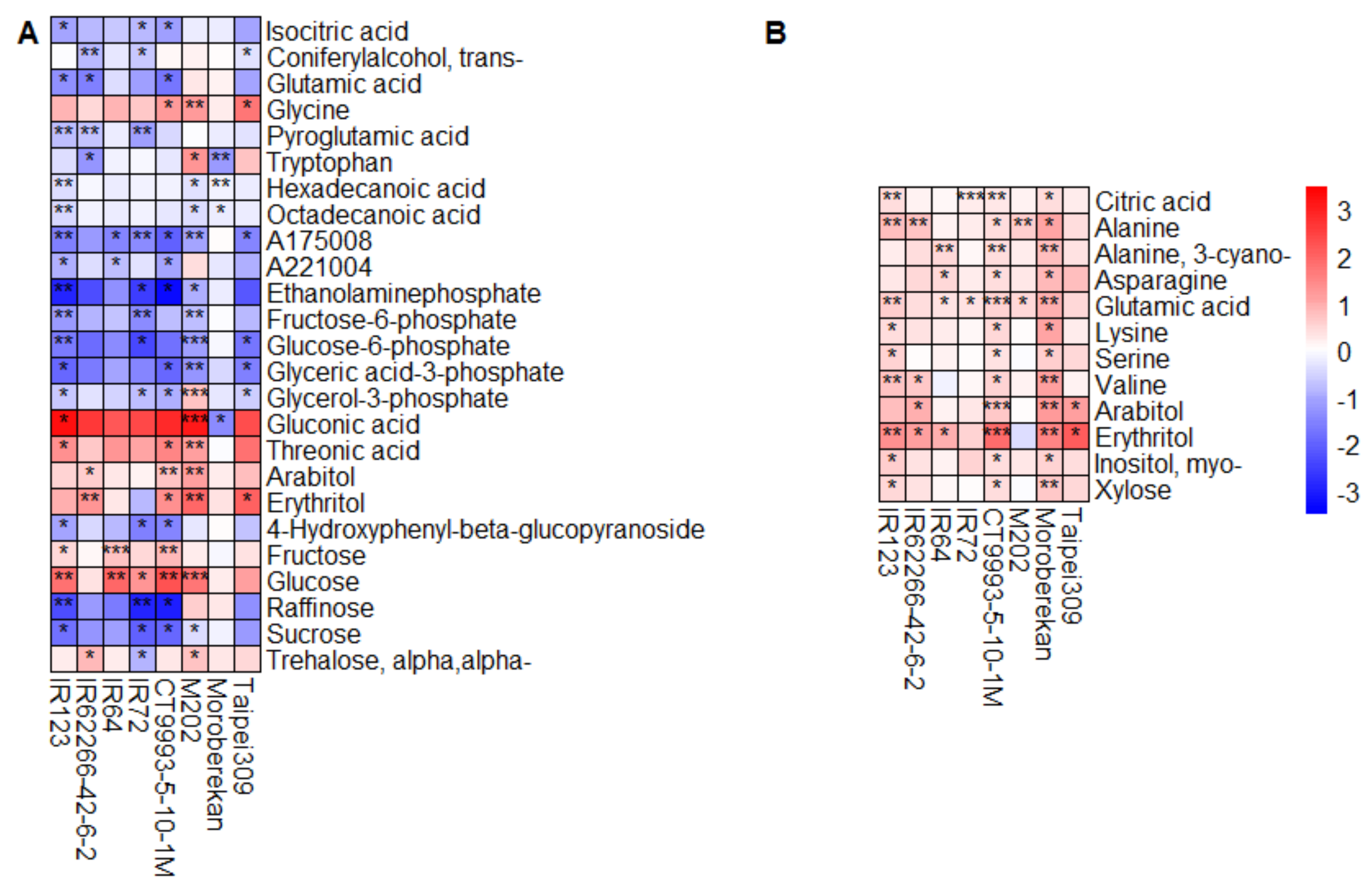
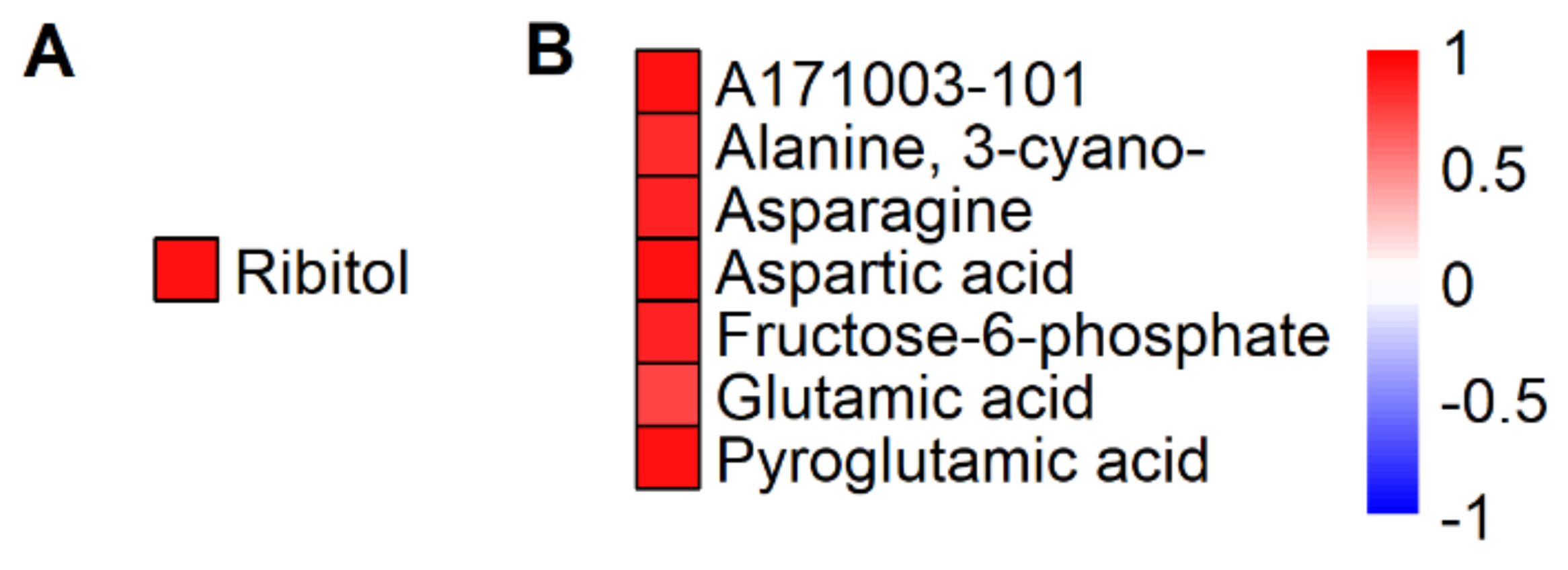
2.2. HNT’s Effects on the Metabolome Are More Pronounced in Panicles Than in Flag Leaves
3. Discussion
4. Materials and Methods
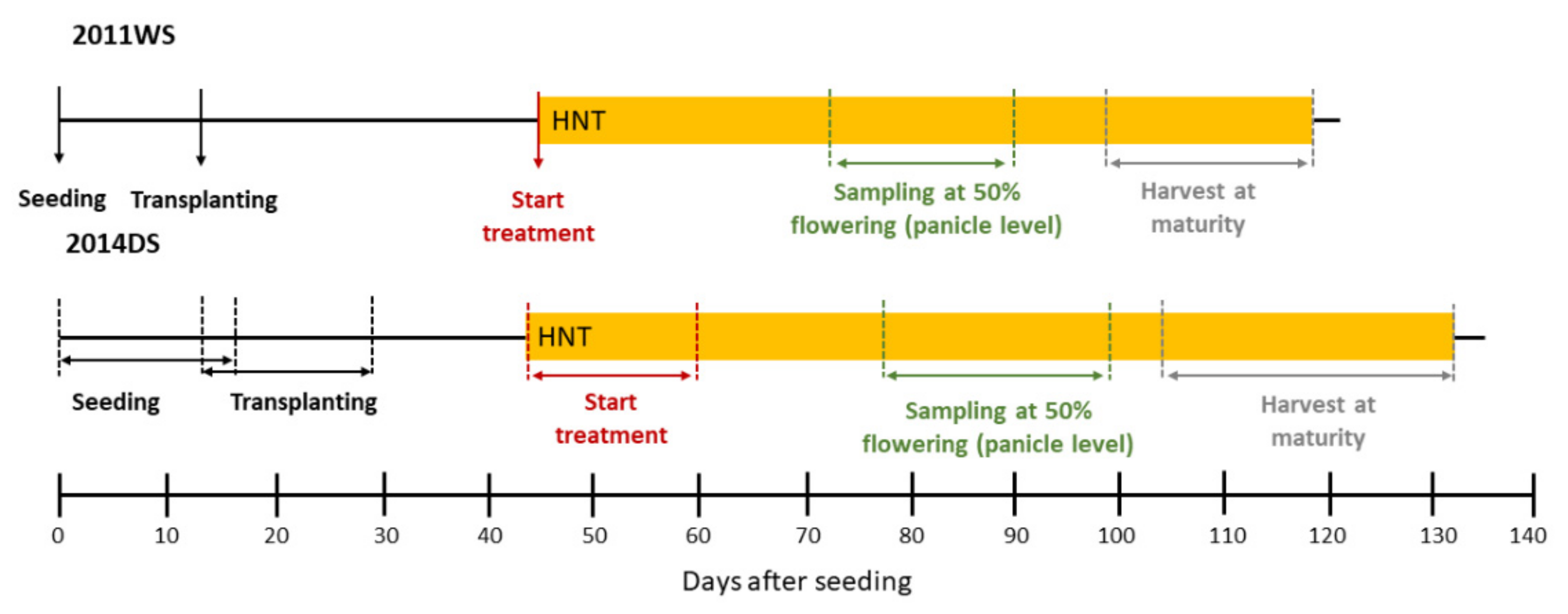
4.1. Plant Material, Cultivation and HNT Stress Treatment
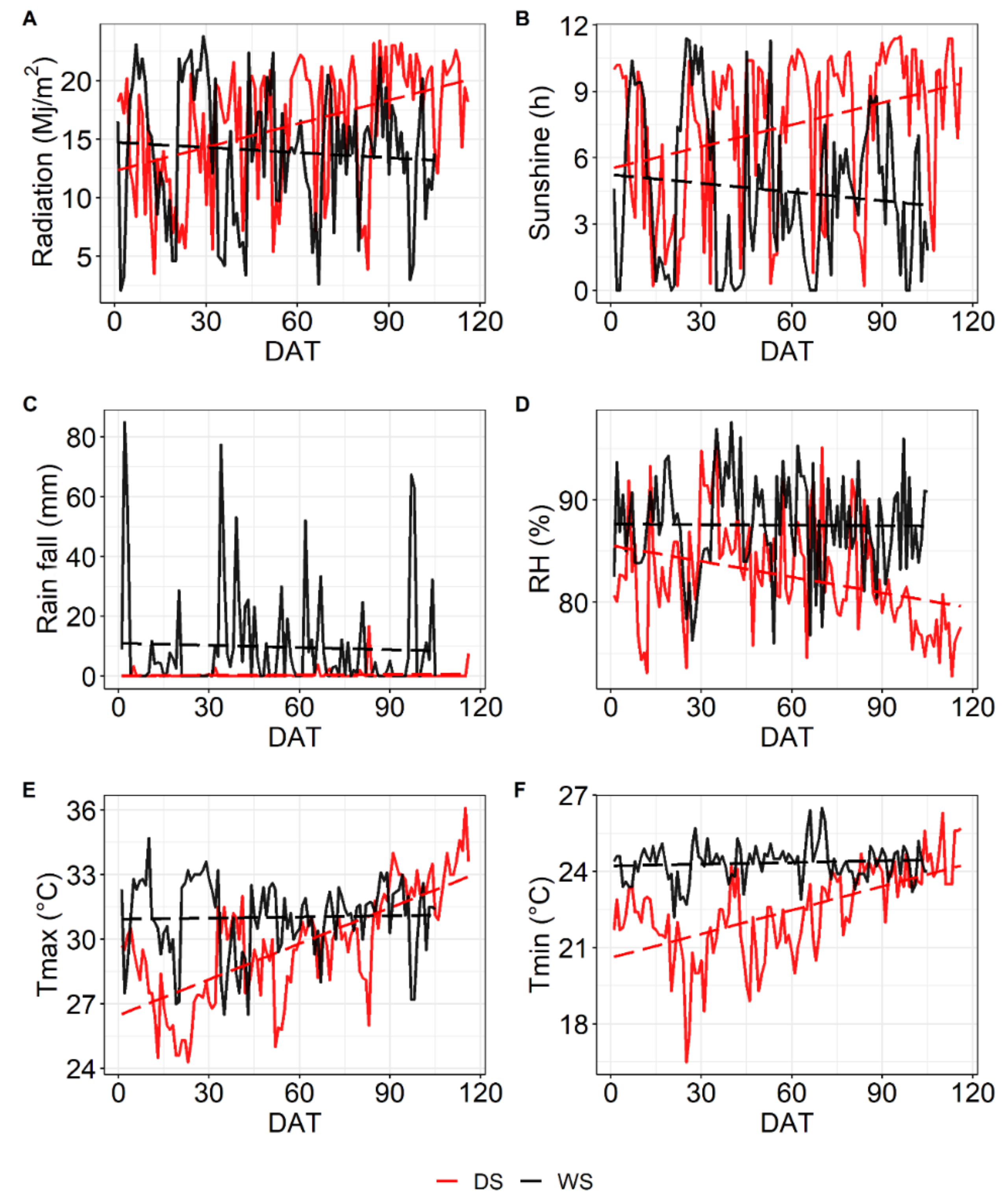
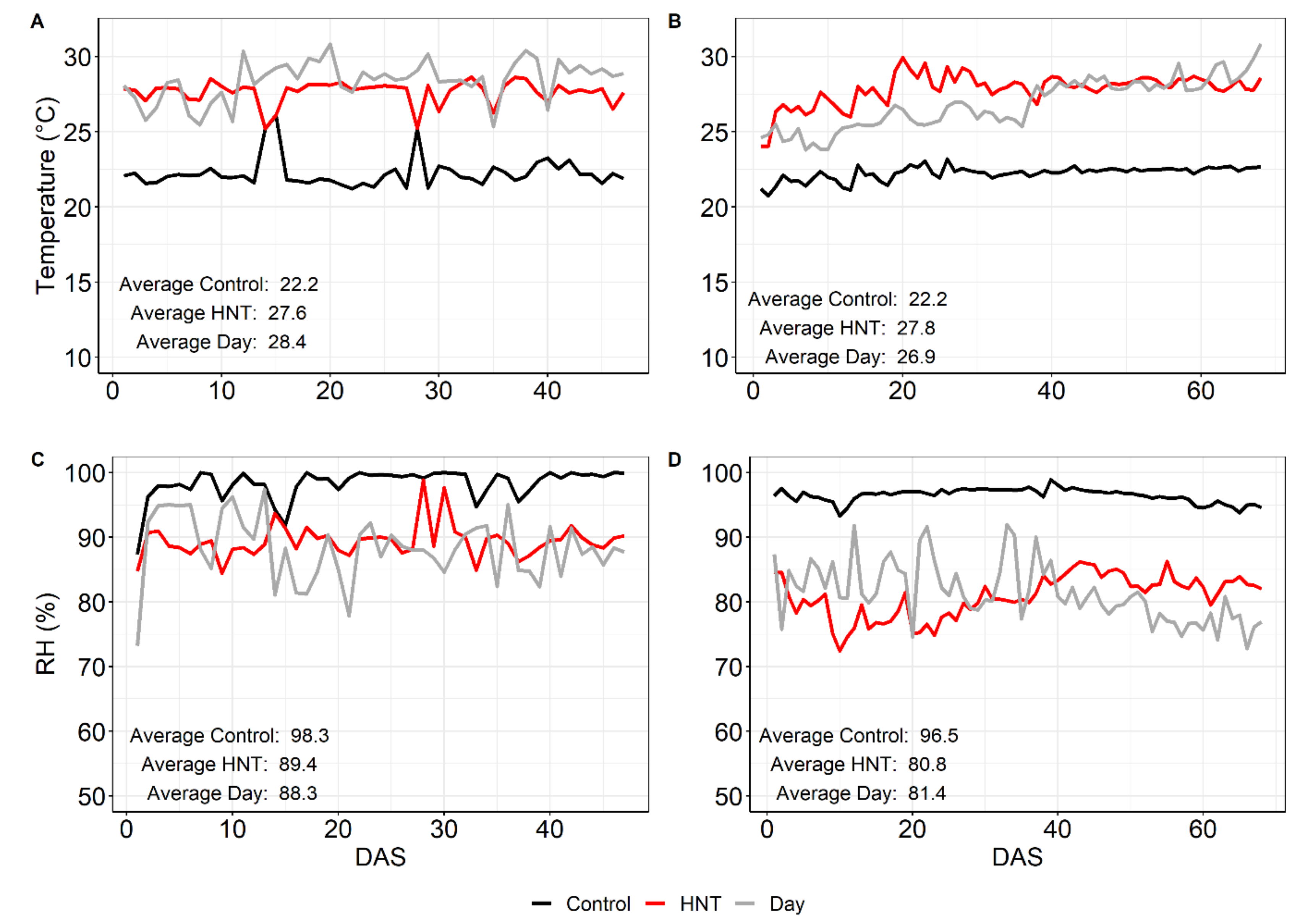
4.2. Weather Data
4.3. Growth Analysis, Grain Yield and Yield Components
4.4. Metabolite Profiling and Data Processing
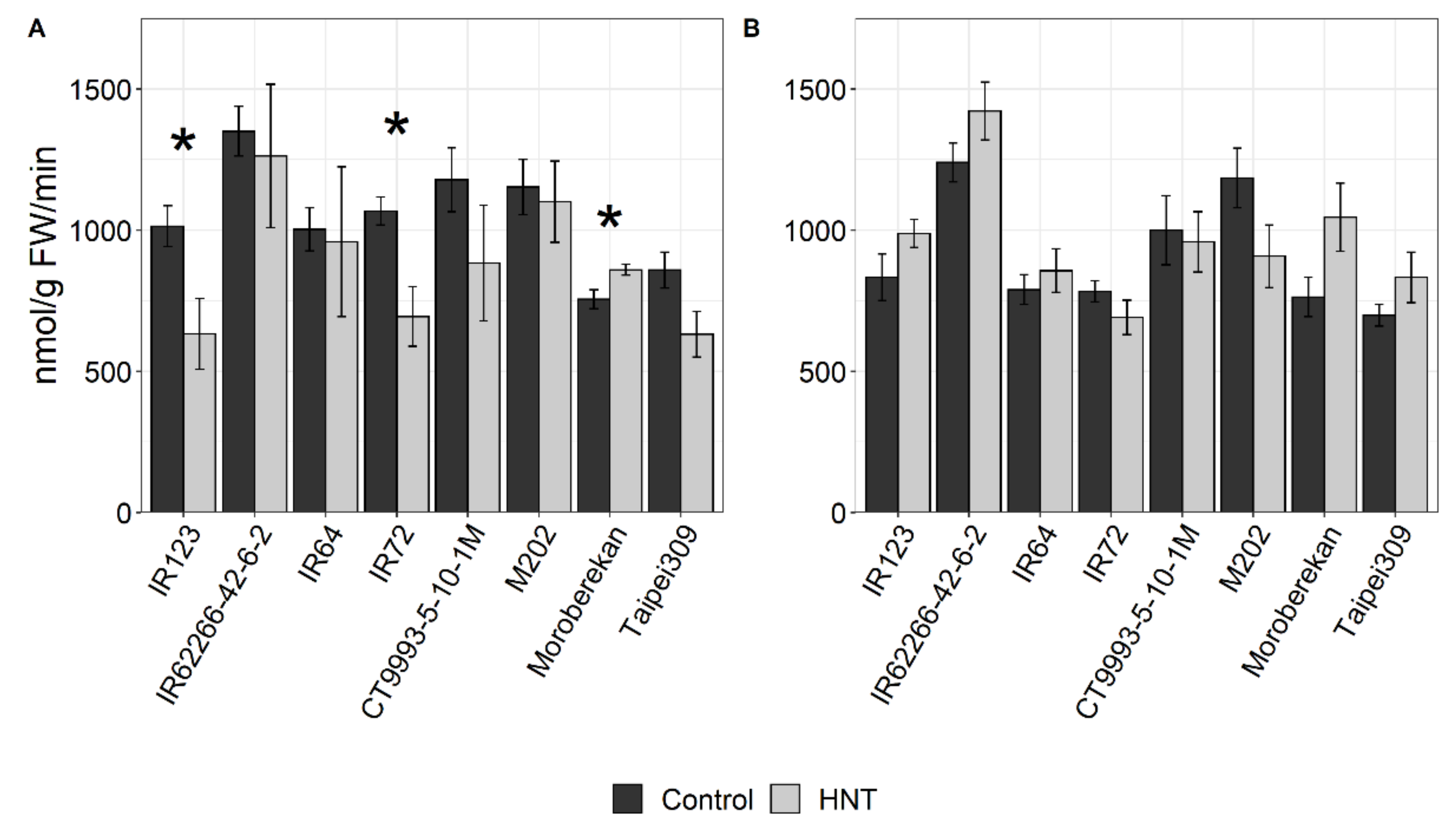
4.5. Enzyme Activity
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | 1-aminocyclopropane-1-carboxylic acid |
| AlaAT | Alanine aminotransferase |
| DAS | Days after stress |
| DAT | Days after transplanting |
| DS | Dry season |
| DTR | Diurnal temperature range |
| FC | Fold change |
| G | Genotype |
| GABA | 4-amino butanoic acid |
| GC-MS | Gas chromatography – Mass spectrometry |
| HCN | Hydrogen cyanide |
| HNT | High night temperature |
| IRRI | International Rice Research Institute |
| PC | Principal Component |
| PCA | Principal Component Analysis |
| RH | Relative humidity |
| S | Season |
| T | Treatment |
| TCA | Tricarboxylic acid |
| WS | Wet season |
Appendix A
References
- GRISP. Global Rice Science Partnership; International Rice Research Institute: Los Baños, Philippines, 2013. [Google Scholar]
- FAO. FAOSTAT Database 2009. Available online: http://faostat.fao.org/ (accessed on 23 October 2019).
- IPCC. AR5 Climate Change 2014: Impacts, Adaptation, and Vulnerability; Cambridge Univ. Press: Cambridge, UK, 2014. [Google Scholar]
- FAO. FAOSTAT Database 2014. Available online: http://faostat.fao.org/ (accessed on 2 November 2019).
- Easterling, D.R.; Horton, B.; Jones, P.D.; Peterson, T.C.; Karl, T.R.; Parker, D.E.; Salinger, M.J.; Razuvayev, V.; Plummer, N.; Jamason, P.; et al. Maximum and minimum temperature trends for the globe. Science 1997, 277, 364–367. [Google Scholar] [CrossRef]
- Donat, M.G.; Alexander, L.V. The shifting probability distribution of global daytime and night-time temperatures. Geophys. Res. Lett. 2012, 39, L14707. [Google Scholar] [CrossRef]
- Vose, R.S.; Easterling, D.R.; Gleason, B. Maximum and minimum temperature trends for the globe: An update through 2004. Geophys. Res. Lett. 2005, 32, L23822. [Google Scholar] [CrossRef]
- Sillmann, J.; Kharin, V.V.; Zhang, X.; Zwiers, F.W.; Bronaugh, D. Climate extremes indices in the CMIP5 multimodel ensemble: Part 1. Model evaluation in the present climate. J. Geophys. Res. Atmos. 2013, 118, 1716–1733. [Google Scholar] [CrossRef]
- Davy, R.; Esau, I.; Chernokulsky, A.; Outten, S.; Zilitinkevich, S. Diurnal asymmetry to the observed global warming. Int. J. Climatol. 2017, 37, 79–93. [Google Scholar] [CrossRef]
- Peng, S.; Piao, S.; Ciais, P.; Myneni, R.B.; Chen, A.; Chevallier, F.; Dolman, A.J.; Janssens, I.A.; Peñuelas, J.; Zhang, G.; et al. Asymmetric effects of daytime and night-time warming on Northern Hemisphere vegetation. Nature 2013, 501, 88–92. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, G. Change in extreme temperature event frequency over mainland China, 1961−2008. Clim. Res. 2011, 50, 125–139. [Google Scholar] [CrossRef]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef]
- Zhao, X.; Fitzgerald, M. Climate change: Implications for the yield of edible rice. PLoS ONE 2013, 8, e66218. [Google Scholar] [CrossRef]
- Rao, B.B.; Chowdary, S.P.; Sandeep, V.M.; Rao, V.U.M.; Venkateswarlu, B. Rising minimum temperature trends over India in recent decades: Implications for agricultural production. Glob. Planet. Chang. 2014, 117, 1–8. [Google Scholar] [CrossRef]
- Padma Kumari, B.; Londhe, A.L.; Daniel, S.; Jadhav, D.B. Observational evidence of solar dimming: Offsetting surface warming over India. Geophys. Res. Lett. 2007, 34, L21810. [Google Scholar] [CrossRef]
- Ambardekar, A.A.; Siebenmorgen, T.J.; Counce, P.A.; Lanning, S.B.; Mauromoustakos, A. Impact of field-scale nighttime air temperatures during kernel development on rice milling quality. Field Crops Res. 2011, 122, 179–185. [Google Scholar] [CrossRef]
- Mohammed, A.R.; Tarpley, L. Effects of night temperature, spikelet position and salicylic acid on yield and yield-related parameters of rice (Oryza sativa L.) plants. J. Agron. Crop Sci. 2011, 197, 40–49. [Google Scholar] [CrossRef]
- Nagarajan, S.; Jagadish, S.V.K.; Prasad, A.S.H.; Thomar, A.K.; Anand, A.; Pal, M.; Agarwal, P.K. Local climate affects growth, yield and grain quality of aromatic and non-aromatic rice in northwestern India. Agric. Ecosyst. Environ. 2010, 138, 274–281. [Google Scholar] [CrossRef]
- Shi, W.; Yin, X.; Struik, P.C.; Solis, C.; Xie, F.; Schmidt, R.C.; Huang, M.; Zou, Y.; Ye, C.; Jagadish, S.V.K. High day- and night-time temperatures affect grain growth dynamics in contrasting rice genotypes. J. Exp. Bot. 2017, 68, 5233–5245. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Yin, X.; Struik, P.C.; Xie, F.; Schmidt, R.C.; Jagadish, K.S.V. Grain yield and quality responses of tropical hybrid rice to high night-time temperature. Field Crops Res. 2016, 190, 18–25. [Google Scholar] [CrossRef]
- Lanning, S.B.; Siebenmorgen, T.J.; Counce, P.A.; Ambardekar, A.A.; Mauromoustakos, A. Extreme nighttime air temperatures in 2010 impact rice chalkiness and milling quality. Field Crops Res. 2011, 124, 132–136. [Google Scholar] [CrossRef]
- Morita, S.; Yonemaru, J.; Takanashi, J. Grain growth and endosperm cell size under high night temperatures in rice (Oryza sativa L.). Ann. Bot. 2005, 95, 695–701. [Google Scholar] [CrossRef]
- Rehmani, M.I.A.; Wei, G.; Hussain, N.; Ding, C.; Li, G.; Liu, Z.; Wang, S.; Ding, Y. Yield and quality responses of two indica rice hybrids to post-anthesis asymmetric day and night open-field warming in lower reaches of Yangtze River delta. Field Crops Res. 2014, 156, 231–241. [Google Scholar] [CrossRef]
- Lobell, D.B.; Ortiz-Monasterio, J.I.; Asner, G.P.; Matson, P.A.; Naylor, R.L.; Falcon, W.P. Analysis of wheat yield and climatic trends in Mexico. Field Crops Res. 2005, 94, 250–256. [Google Scholar] [CrossRef]
- Shi, W.; Muthurajan, R.; Rahman, H.; Selvam, J.; Peng, S.; Zou, Y.; Jagadish, K.S. Source-sink dynamics and proteomic reprogramming under elevated night temperature and their impact on rice yield and grain quality. New Phytol. 2013, 197, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Q.; Peng, S.; Zou, Y.; Chen, S.; Shi, W.; Qin, J.; Laza, M.R.C. Effects of high night temperature on yield and agronomic traits of irrigated rice under field chamber system condition. Aust. J. Crop Sci. 2013, 7, 7–13. [Google Scholar]
- Yang, Z.; Zhang, Z.; Zhang, T.; Fahad, S.; Cui, K.; Nie, L.; Peng, S.; Huang, J. The effect of season-long temperature increases on rice cultivars grown in the central and Southern regions of China. Front. Plant Sci. 2017, 8, 1908. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, U.; Li, X.; Köhl, K.I.; van Dongen, J.T.; Hincha, D.K.; Zuther, E. Differential physiological responses of different rice (Oryza sativa) cultivars to elevated night temperature during vegetative growth. Funct. Plant Biol. 2014, 41, 437–448. [Google Scholar] [CrossRef]
- Bahuguna, R.N.; Solis, C.A.; Shi, W.; Jagadish, K.S.V. Post-flowering night respiration and altered sink activity account for high night temperature-induced grain yield and quality loss in rice (Oryza sativa L.). Physiol. Plant. 2017, 159, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xia, J.; Liu, L.; Wan, S. Global patterns of the responses of leaf-level photosynthesis and respiration in terrestrial plants to experimental warming. J. Plant Ecol. 2013, 6, 437–447. [Google Scholar] [CrossRef]
- Mohammed, R.; Cothren, J.T.; Tarpley, L. High night temperature and abscisic acid affect rice productivity through altered photosynthesis, respiration and spikelet fertility. Crop Sci. 2013, 53, 2603–2612. [Google Scholar] [CrossRef]
- Dong, W.; Chen, J.; Wang, L.; Tian, Y.; Zhang, B.; Lai, Y.; Meng, Y.; Qian, C.; Guo, J. Impacts of nighttime post-anthesis warming on rice productivity and grain quality in East China. Crop J. 2014, 2, 63–69. [Google Scholar] [CrossRef]
- Fernie, A.R.; Schauer, N. Metabolomics-assisted breeding: A viable option for crop improvement? Trends Genet. 2009, 25, 39–48. [Google Scholar] [CrossRef]
- Oikawa, A.; Matsuda, F.; Kusano, M.; Okazaki, Y.; Saito, K. Rice Metabolomics. Rice 2008, 1, 63–71. [Google Scholar] [CrossRef]
- Nadella, K.D.; Marla, S.S.; Kumar, P.A. Metabolomics in agriculture. Omics A J. Integr. Biol. 2012, 16, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Krumsiek, J.; Bartel, J.; Theis, F.J. Computational approaches for systems metabolomics. Curr. Opin. Biotechnol. 2016, 39, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Zuther, E.; Koehl, K.; Kopka, J. Comparative metabolome analysis of the salt response in breeding cultivars of rice. In Advances in Molecular Breeding toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 285–315. [Google Scholar] [CrossRef]
- Liu, D.; Ford, K.L.; Roessner, U.; Natera, S.; Cassin, A.M.; Patterson, J.H.; Bacic, A. Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics 2013, 13, 2046–2062. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, W.; Zhang, F.; Deng, J.; Li, Z.; Fu, B. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE 2014, 9, e108020. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Park, W.J.; Kim, B.G.; Yoon, I.S. Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef]
- Ma, N.L.; Che Lah, W.A.; Abd Kadir, N.; Mustaqim, M.; Rahmat, Z.; Ahmad, A.; Lam, S.D.; Ismail, M.R. Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections. PLoS ONE 2018, 13, e0192732. [Google Scholar] [CrossRef]
- Baldoni, E.; Mattana, M.; Locatelli, F.; Consonni, R.; Cagliani, L.R.; Picchi, V.; Abbruscato, P.; Genga, A. Analysis of transcript and metabolite levels in Italian rice (Oryza sativa L.) cultivars subjected to osmotic stress or benzothiadiazole treatment. Plant Physiol. Biochem. 2013, 70, 492–503. [Google Scholar] [CrossRef]
- Degenkolbe, T.; Do, P.T.; Kopka, J.; Zuther, E.; Hincha, D.K.; Köhl, K.I. Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS ONE 2013, 8, e63637. [Google Scholar] [CrossRef]
- Li, X.; Lawas, L.M.F.; Malo, R.; Glaubitz, U.; Erban, A.; Mauleon, R.; Heuer, S.; Zuther, E.; Kopka, J.; Hincha, D.K.; et al. Metabolic and transcriptomic signatures of rice floral organs reveal sugar starvation as a factor in reproductive failure under heat and drought stress. Plant Cell Environ. 2015, 38, 2171–2192. [Google Scholar] [CrossRef]
- Shu, L.; Lou, Q.; Ma, C.; Ding, W.; Zhou, J.; Wu, J.; Feng, F.; Lu, X.; Luo, L.; Xu, G. Genetic, proteomic and metabolic analysis of the regulation of energy storage in rice seedlings in response to drought. Proteomics 2011, 11, 4122–4138. [Google Scholar] [CrossRef]
- Nam, K.H.; Shin, H.J.; Pack, I.S.; Park, J.H.; Kim, H.B.; Kim, C.G. Metabolomic changes in grains of well-watered and drought-stressed transgenic rice. J. Sci. Food Agric. 2016, 96, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Barnaby, J.Y.; Rohila, J.S.; Henry, C.G.; Sicher, R.C.; Reddy, V.R.; McClung, A.M. Physiological and metabolic responses of rice to reduced soil moisture: Relationship of water stress tolerance and grain production. Int. J. Mol. Sci. 2019, 20, 1846. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, H.; Hakata, M. Atlas of rice grain filling-related metabolism under high temperature: Joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010, 51, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Lawas, L.M.F.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice cultivars with different tolerance to combined drought and heat stress under field conditions. Gigascience 2019, 8, giz050. [Google Scholar] [CrossRef]
- Lawas, L.M.F.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice source and sink organs during recovery from combined drought and heat stress in the field. Gigascience 2019, 8, giz102. [Google Scholar] [CrossRef]
- Glaubitz, U.; Erban, A.; Kopka, J.; Hincha, D.K.; Zuther, E. High night temperature strongly impacts TCA cycle, amino acid and polyamine biosynthetic pathways in rice in a sensitivity-dependent manner. J. Exp. Bot. 2015, 66, 6385–6397. [Google Scholar] [CrossRef]
- Glaubitz, U.; Li, X.; Schaedel, S.; Erban, A.; Sulpice, R.; Kopka, J.; Hincha, D.K.; Zuther, E. Integrated analysis of rice transcriptomic and metabolomic responses to elevated night temperatures identifies sensitivity- and tolerance-related profiles. Plant Cell Environ. 2017, 40, 121–137. [Google Scholar] [CrossRef]
- Dhatt, B.K.; Abshire, N.; Paul, P.; Hasanthika, K.; Sandhu, J.; Zhang, Q.; Obata, T.; Walia, H. Metabolic dynamics of developing rice seeds under high night-time temperature stress. Front. Plant Sci. 2019, 10, 1443. [Google Scholar] [CrossRef]
- Van Oort, P.A.J.; Zwart, S.J. Impacts of climate change on rice production in Africa and causes of simulated yield changes. Glob. Chang. Biol. 2018, 24, 1029–1045. [Google Scholar] [CrossRef]
- Good, A.G.; Muench, D.G. Long-term anaerobic metabolism in root tissue (Metabolic products of pyruvate metabolism). Plant Physiol. 1993, 101, 1163–1168. [Google Scholar] [CrossRef]
- Shrawat, A.K.; Carroll, R.T.; DePauw, M.; Taylor, G.J.; Good, A.G. Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 2008, 6, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Ling, X.; Huang, J.; Peng, S. Meta-analysis and dose-response analysis of high temperature effects on rice yield and quality. Environ. Exp. Bot. 2017, 141, 1–9. [Google Scholar] [CrossRef]
- Jagadish, S.V.; Murty, M.V.; Quick, W.P. Rice responses to rising temperatures—challenges, perspectives and future directions. Plant Cell Environ. 2015, 38, 1686–1698. [Google Scholar] [CrossRef]
- Mohammed, A.R.; Tarpley, L. Effects of high night temperature and spikelet position on yield-related parameters of rice (Oryza sativa L.) plants. Eur. J. Agron. 2010, 33, 117–123. [Google Scholar] [CrossRef]
- Kanno, K.; Makino, A. Increased grain yield and biomass allocation in rice under cool night temperature. Soil Sci. Plant Nutr. 2010, 56, 412–417. [Google Scholar] [CrossRef]
- Coast, O.; Ellis, R.H.; Murdoch, A.J.; Quiñones, C.; Jagadish, K.S.V. High night temperature induces contrasting responses for spikelet fertility, spikelet tissue temperature, flowering characteristics and grain quality in rice. Funct. Plant Biol. 2015, 42, 149–161. [Google Scholar] [CrossRef]
- Liao, J.-L.; Zhou, H.-W.; Peng, Q.; Zhong, P.-A.; Zhang, H.-Y.; He, C.; Huang, Y.-J. Transcriptome changes in rice (Oryza sativa L.) in response to high night temperature stress at the early milky stage. Bmc Genom. 2015, 16, 18. [Google Scholar] [CrossRef]
- Dong, W.; Tian, Y.; Zhang, B.; Chen, J.; Zhang, W. Effects of asymmetric warming on grain quality and related key enzymes activities for japonica rice (Nanjing 44) under FATI facility. Acta Agron. Sin. 2011, 37, 832–841. [Google Scholar] [CrossRef]
- Chen, Y.; Murchie, E.H.; Hubbart, S.; Horton, P.; Peng, S. Effects of season-dependent irradiance levels and nitrogen-deficiency on photosynthesis and photoinhibition in field-grown rice (Oryza sativa). Physiol. Plant. 2003, 117, 343–351. [Google Scholar] [CrossRef]
- Impa, S.M.; Vennapusa, A.R.; Bheemanahalli, R.; Sabela, D.; Boyle, D.; Walia, H.; Jagadish, S.V.K. High night temperature induced changes in grain starch metabolism alters starch, protein, and lipid accumulation in winter wheat. Plant Cell Environ. 2020, 43, 431–447. [Google Scholar] [CrossRef]
- Good, A.G.; Johnson, S.J.; De Pauw, M.; Carroll, R.T.; Savidov, N.; Vidmar, J.; Lu, Z.; Taylor, G.; Stroeher, V. Engineering nitrogen use efficiency with alanine aminotransferase. Can. J. Bot. 2007, 85, 252–262. [Google Scholar] [CrossRef]
- Beatty, P.H.; Shrawat, A.K.; Carroll, R.T.; Zhu, T.; Good, A.G. Transcriptome analysis of nitrogen-efficient rice over-expressing alanine aminotransferase. Plant Biotechnol. J. 2009, 7, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, H.; Hirose, S.; Toki, S.; Akama, K.; Takaiwa, F. Molecular characterization of a gene for alanine aminotransferase from rice (Oryza sativa). Plant Mol. Biol. 1999, 39, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Liu, X.; Liu, F.; Ren, Y.; Wang, Y.; Zhu, J.; Teng, X.; Duan, E.; Wang, F.; Zhang, H.; et al. FLOURY ENDOSPERM12 encoding alanine aminotransferase 1 regulates carbon and nitrogen metabolism in rice. J. Plant Biol. 2019, 62, 61–73. [Google Scholar] [CrossRef]
- Beatty, P.H.; Carroll, R.T.; Shrawat, A.K.; Guevara, D.; Good, A.G. Physiological analysis of nitrogen-efficient rice overexpressing alanine aminotransferase under different N regimes. Botany 2013, 91, 866–883. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Valencia, M.O.; Ogawa, S.; Lu, Y.; Wu, L.; Downs, C.; Skinner, W.; Lu, Z.; Kridl, J.C.; Ishitani, M.; et al. Development and field performance of nitrogen use efficient rice lines for Africa. Plant Biotechnol. J. 2017, 15, 775–787. [Google Scholar] [CrossRef]
- Lai, K.W.; Yau, C.P.; Tse, Y.C.; Jiang, L.; Yip, W.K. Heterologous expression analyses of rice OsCAS in Arabidopsis and in yeast provide evidence for its roles in cyanide detoxification rather than in cysteine synthesis in vivo. J. Exp. Bot. 2009, 60, 993–1008. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Siegień, I.; Bogatek, R. Cyanide action in plants—from toxic to regulatory. Acta Physiol. Plant. 2006, 28, 483–497. [Google Scholar] [CrossRef]
- Helliwell, E.E.; Wang, Q.; Yang, Y. Ethylene biosynthesis and signaling is required for rice immune response and basal resistance against Magnaporthe oryzae infection. Mol. Plant Microbe Interact. 2016, 29, 831–843. [Google Scholar] [CrossRef]
- Yu, L.; Liu, Y.; Xu, F. Comparative transcriptome analysis reveals significant differences in the regulation of gene expression between hydrogen cyanide- and ethylene-treated Arabidopsis thaliana. BMC Plant Biol. 2019, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Liu, L.; Yin, Y.; Huang, M.; Chen, Y.; Xu, X.; Wu, P.; Li, M.; Wu, G.; Jiang, H.; et al. Heterogeneity in the expression and subcellular localization of POLYOL/MONOSACCHARIDE TRANSPORTER genes in Lotus japonicus. PLoS ONE 2017, 12, e0185269. [Google Scholar] [CrossRef] [PubMed]
- Pires, M.V.; Pereira Junior, A.A.; Medeiros, D.B.; Daloso, D.M.; Pham, P.A.; Barros, K.A.; Engqvist, M.K.; Florian, A.; Krahnert, I.; Maurino, V.G.; et al. The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant Cell Environ. 2016, 39, 1304–1319. [Google Scholar] [CrossRef] [PubMed]
- Fabregas, N.; Fernie, A.R. The metabolic response to drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Melandri, G.; AbdElgawad, H.; Riewe, D.; Hageman, J.A.; Asard, H.; Beemster, G.T.S.; Kadam, N.; Jagadish, K.; Altmann, T.; Ruyter-Spira, C.; et al. Biomarkers for grain yield stability in rice under drought stress. J. Exp. Bot. 2020, 71, 669–683. [Google Scholar] [CrossRef]
- Yoshida, S.; Forn, D.A.; Cock, J.H.; Gomez, K.A. Laboratory Manual for Physiological Studies of Rice; International Rice Research Institute: Los Banos, Philippines, 1976. [Google Scholar]
- Dethloff, F.; Erban, A.; Orf, I.; Alpers, J.; Fehrle, I.; Beine-Golovchuk, O.; Schmidt, S.; Schwachtje, J.; Kopka, J. Profiling methods to identify cold-regulated primary metabolites using gas chromatography coupled to mass spectrometry. Methods Mol. Biol. 2014, 1166, 171–197. [Google Scholar] [CrossRef] [PubMed]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography—mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmuller, E.; Dormann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef]
- Hummel, J.; Strehmel, N.; Selbig, J.; Walther, D.; Kopka, J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 2010, 6, 322–333. [Google Scholar] [CrossRef]
- Lisec, J.; Romisch-Margl, L.; Nikoloski, Z.; Piepho, H.P.; Giavalisco, P.; Selbig, J.; Gierl, A.; Willmitzer, L. Corn hybrids display lower metabolite variability and complex metabolite inheritance patterns. Plant J. 2011, 68, 326–336. [Google Scholar] [CrossRef]
- Komsta, L. Outliers: Tests for Outliers. R Package Version 0.14. 2011. Available online: https://cran.r-project.org/web/packages/outliers/outliers.pdf (accessed on 16 August 2019).
- Gibon, Y.; Blaesing, O.E.; Hannemann, J.; Carillo, P.; Höhne, M.; Hendriks, J.H.M.; Palacios, N.; Cross, J.; Selbig, J.; Stitt, M. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 2004, 16, 3304–3325. [Google Scholar] [CrossRef] [PubMed]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods—a Bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- RCore, T. R: A Language and Environment for Statistical Computing, 3.4.2; R foundation for statistical computing: Vienna, Austria, 2017. [Google Scholar]
- RStudio, T. RStudio: Integrated Development for R; RStudio: Boston, MA, USA, 2016. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Murrell, P. R Graphics; Chapman and Hall/CRC: Boca Raton, FL, USA, 2005. [Google Scholar]
- Auguie, B. Gridextra: Miscellaneous Functions for “Grid” Graphics. R Package Version 2.3. Available online: http://CRAN.R-project.org/package=gridExtra (accessed on 16 August 2019).
- Wickham, H. Reshaping Data with the reshape Package J. Stat. Softw. 2007, 21, 1–20. [Google Scholar]
| Experiment 1 | Experiment 2 | |||
|---|---|---|---|---|
| Season | Wet | Dry | ||
| Conditions | Control | HNT | Control | HNT |
| Cultivars | CT9993-5-10-1M | |||
| IR123 | ||||
| IR62266-42-6-2 | ||||
| IR64 | ||||
| IR72 | ||||
| M202 | ||||
| Moroberekan | ||||
| Taipei309 | ||||
| Tday (°C) | 27.7 | 26.1 | ||
| Tnight (°C) | 22.2 | 27.6 | 22.2 | 27.8 |
| RH (%) | 98.3 | 89.4 | 96.4 | 80.9 |
| Sampling time | Panicle at 50% flowering | |||
| Samples | Flag leaves, panicles | |||
| Parameter | WS | WS | WS | DS | DS | DS | Both Seasons | ||
|---|---|---|---|---|---|---|---|---|---|
| G | T | GxT | G | T | GxT | T | S | TxS | |
| Plant Height (cm) | *** | ns | ** | *** | ns. | * | ns | ns | ns |
| Biomass g/m2 | *** | * | ns | *** | ns | ns | ns | * | ns |
| Straw (g) | *** | ns | * | *** | ns | ns | ns | ns | ns |
| Rachis (g) | *** | ns | ns | *** | ns | ns | ns | * | ns |
| Tiller No | *** | ns | ns | *** | ns | ns | ns | ** | ns |
| Panicle No | *** | ns | ns | *** | ns | ns | ns | ** | ns |
| Panicle/m2 | *** | ns | ns | *** | ns | ns | ns | ** | ns |
| Spikelet/m2 | *** | * | ns | *** | ns | ns | ns | *** | ns |
| Spikelets/Panicle | *** | ** | ns | *** | ns | ns | ns | *** | ns |
| Seed set (%) | *** | ns | ns | *** | ns | ns | ** | * | ns |
| Grain yield (g/m2) | *** | *** | ns | *** | ns | ns | ns | ** | ns |
| 1000 grain weight (g) | *** | *** | *** | *** | *** | ns | ns | *** | ns |
| Harvest Index | *** | *** | ns | *** | ** | ns | ns | *** | ns |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schaarschmidt, S.; Lawas, L.M.F.; Glaubitz, U.; Li, X.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Hincha, D.K.; Zuther, E. Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field. Int. J. Mol. Sci. 2020, 21, 3187. https://doi.org/10.3390/ijms21093187
Schaarschmidt S, Lawas LMF, Glaubitz U, Li X, Erban A, Kopka J, Jagadish SVK, Hincha DK, Zuther E. Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field. International Journal of Molecular Sciences. 2020; 21(9):3187. https://doi.org/10.3390/ijms21093187
Chicago/Turabian StyleSchaarschmidt, Stephanie, Lovely Mae F. Lawas, Ulrike Glaubitz, Xia Li, Alexander Erban, Joachim Kopka, S. V. Krishna Jagadish, Dirk K. Hincha, and Ellen Zuther. 2020. "Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field" International Journal of Molecular Sciences 21, no. 9: 3187. https://doi.org/10.3390/ijms21093187
APA StyleSchaarschmidt, S., Lawas, L. M. F., Glaubitz, U., Li, X., Erban, A., Kopka, J., Jagadish, S. V. K., Hincha, D. K., & Zuther, E. (2020). Season Affects Yield and Metabolic Profiles of Rice (Oryza sativa) under High Night Temperature Stress in the Field. International Journal of Molecular Sciences, 21(9), 3187. https://doi.org/10.3390/ijms21093187






