Molecular Markers Associated with Agro-Physiological Traits under Terminal Drought Conditions in Bread Wheat
Abstract
1. Introduction
2. Results
2.1. Analysis of Variance and Heritability
2.2. GWAS Analysis in Diverse Pre-Breeding Germplasm
2.2.1. Physiological Traits
2.2.2. Plant Height and Earliness
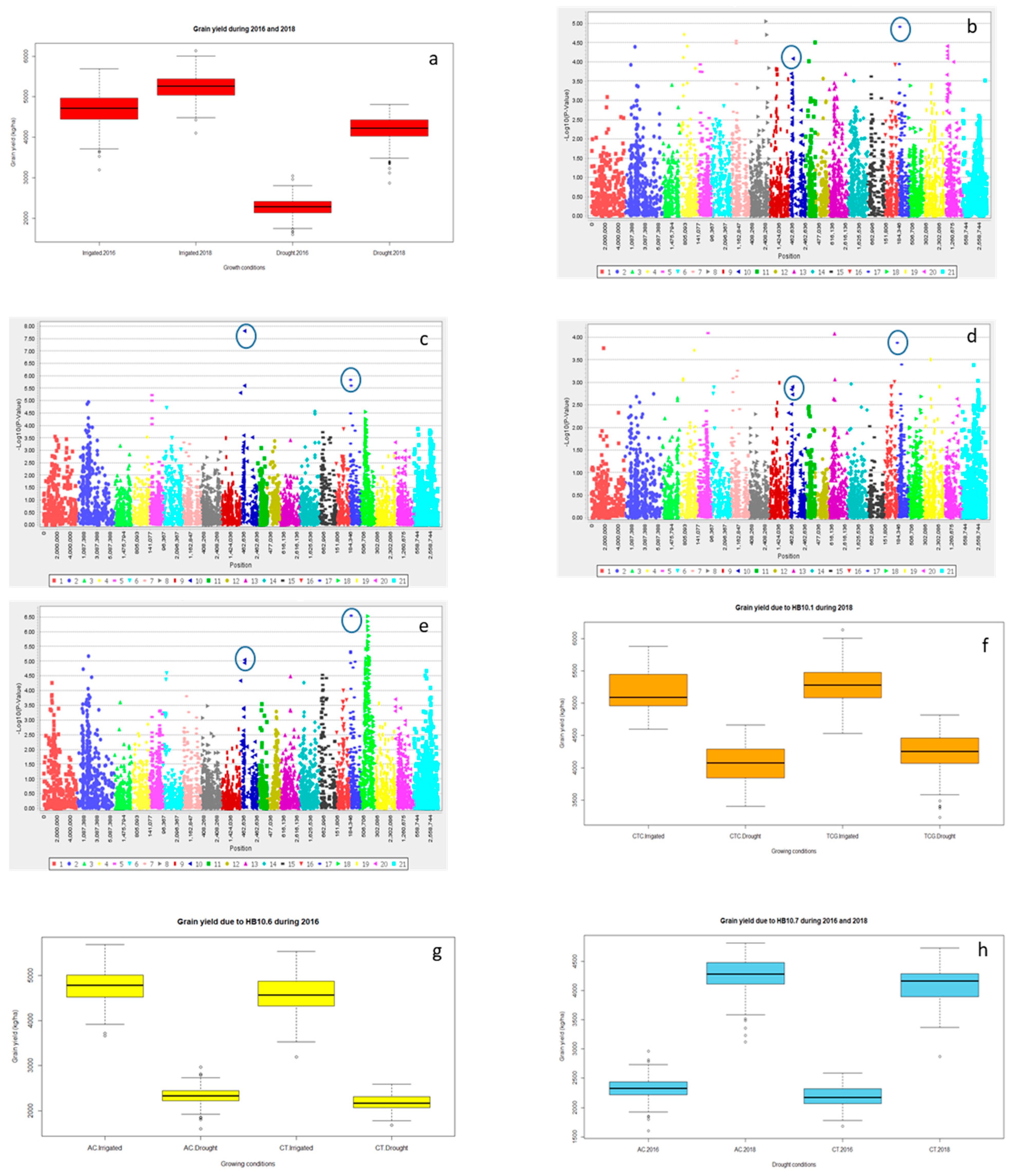
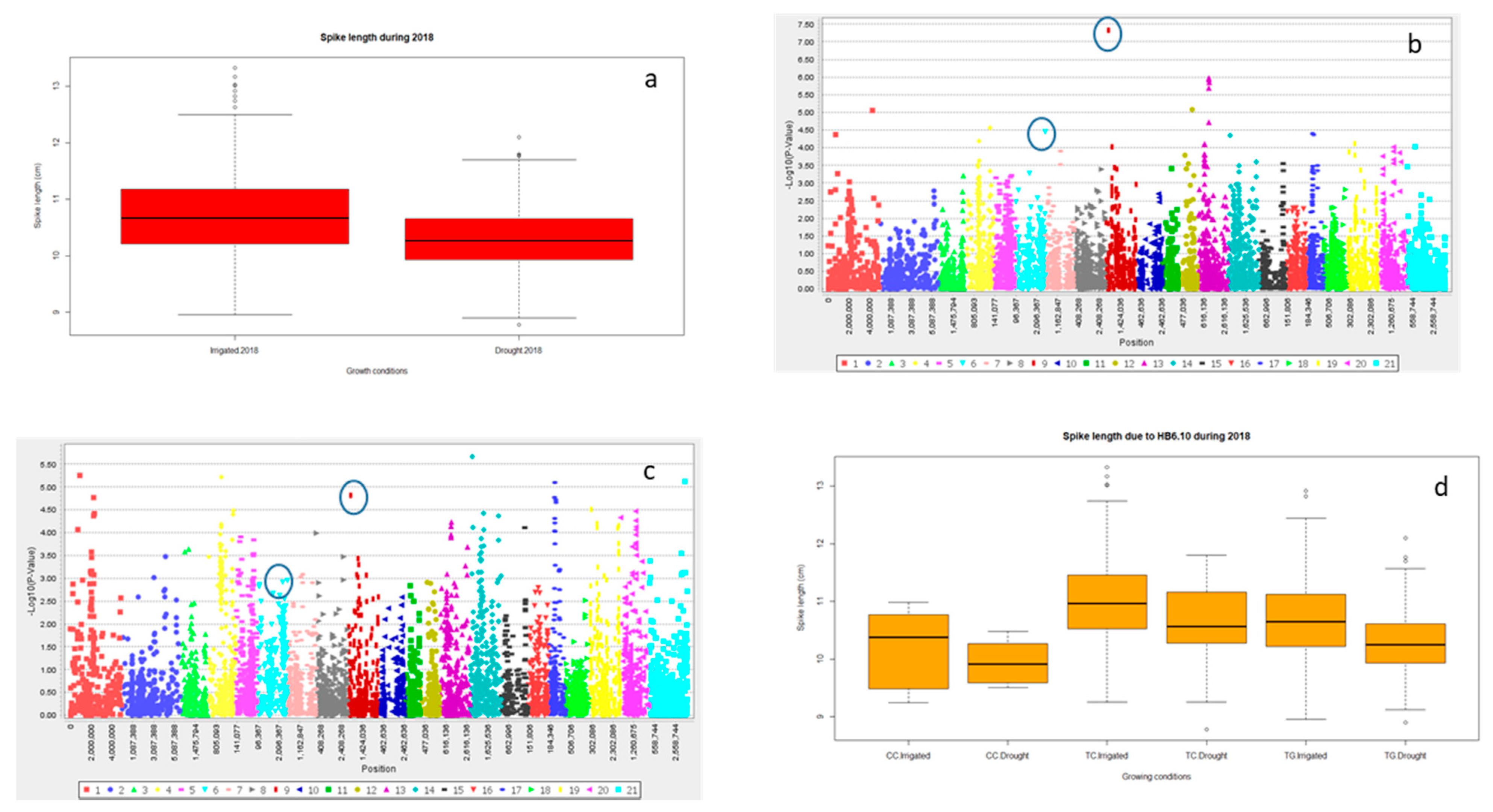
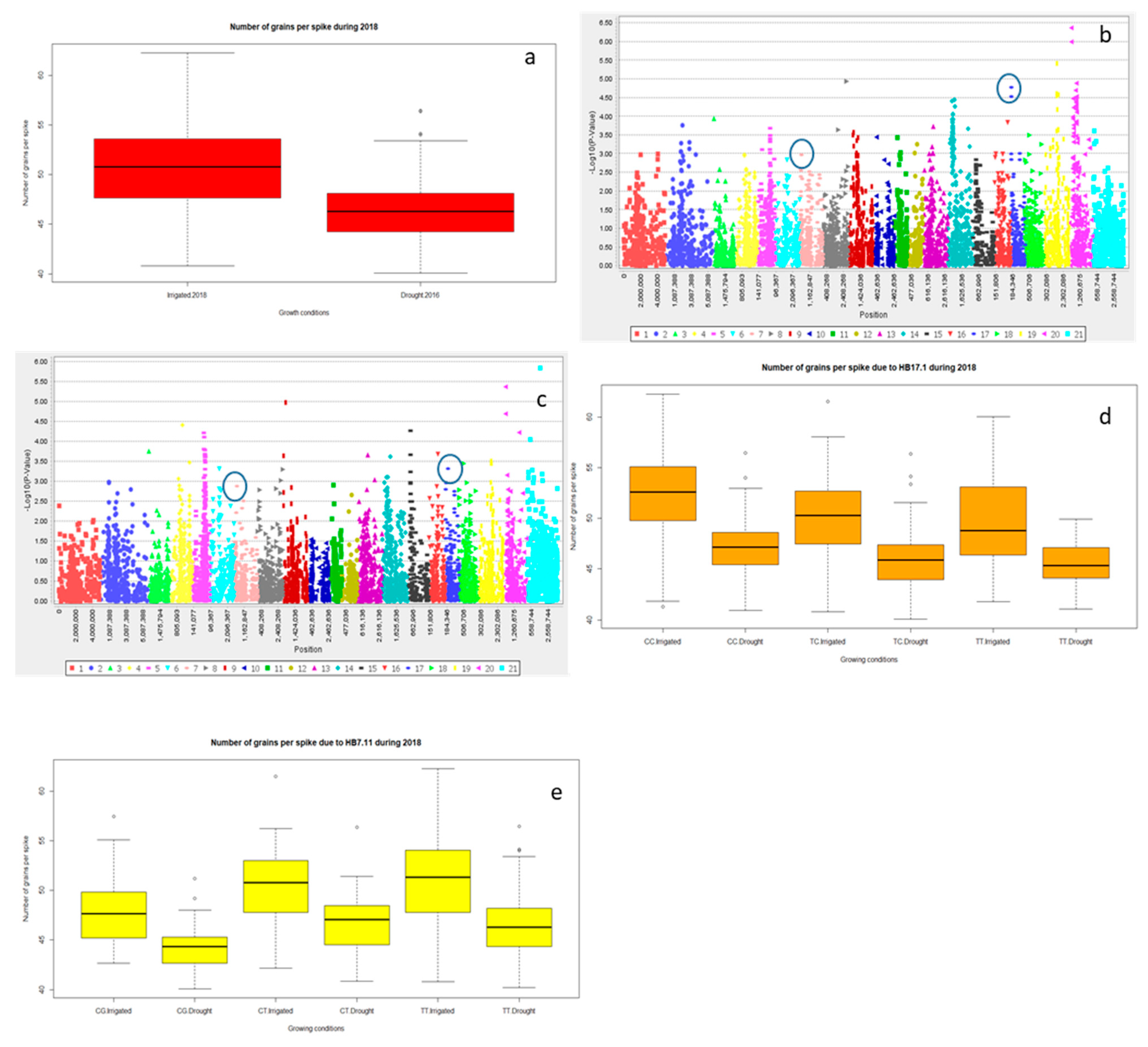
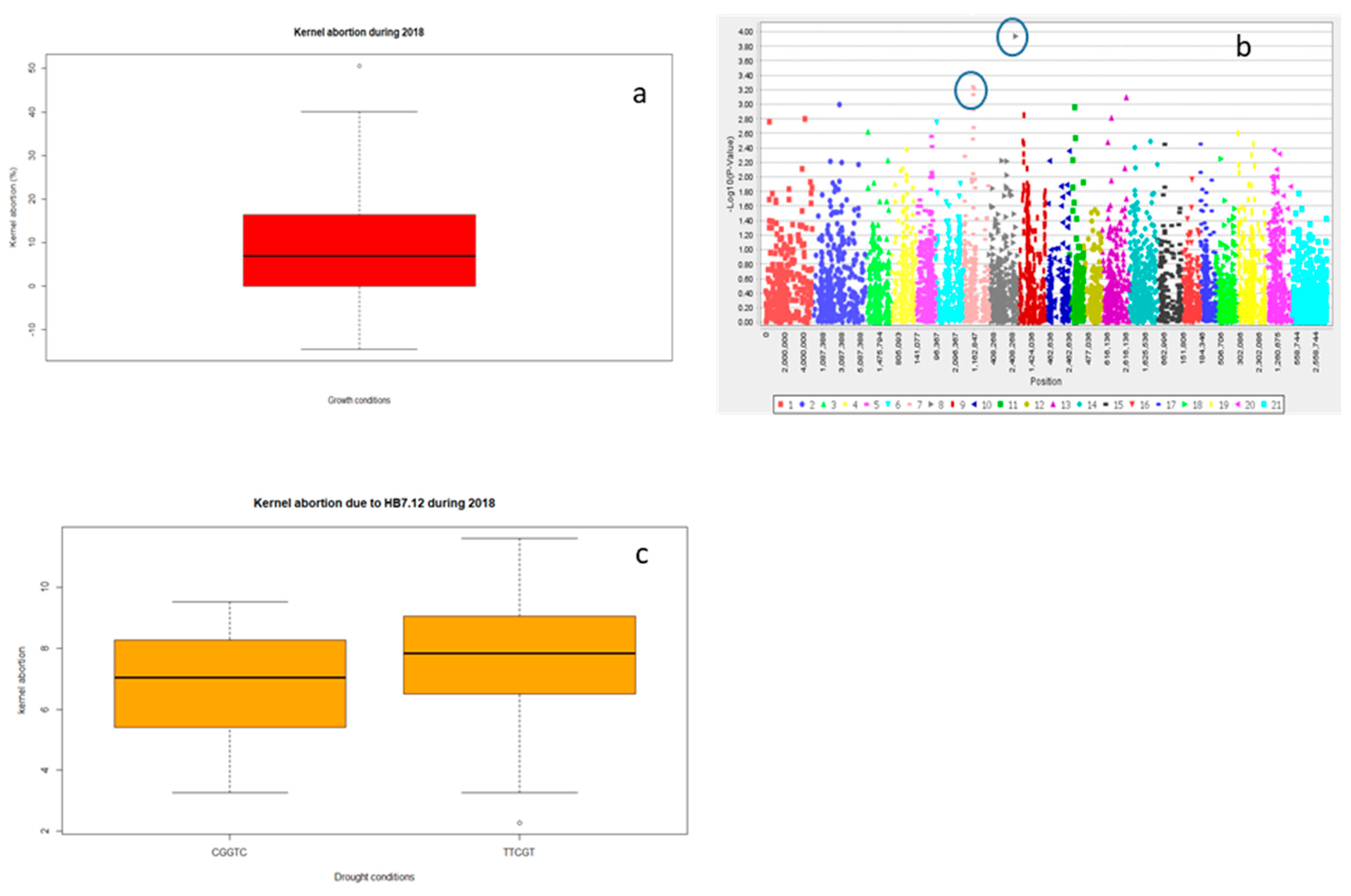
2.2.3. Grain Yield (Y) and Yield Related Traits
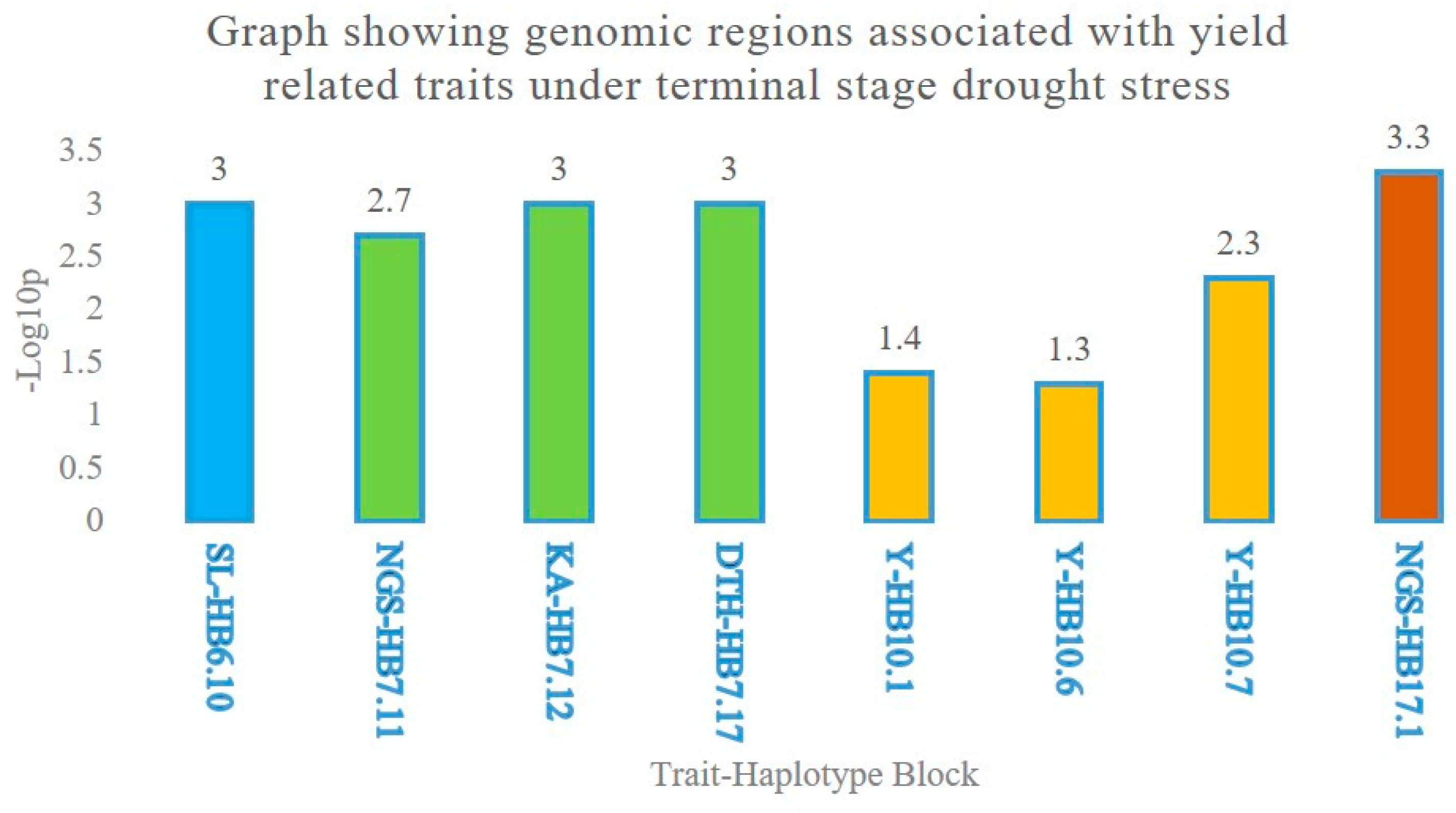
2.3. Multiple Quantitative Traits Loci (QTL’s) at the Same Chromosome
Haplotype Blocks at Same Chromosomes
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Population Structure and Linkage Disequilibrium
4.3. Experimental Setup
4.4. Phenotypic Characterization
- (a)
- Canopy temperature depression (CTD) was measured with infrared thermometer sixth sense (LT300). The measurement was adjusted 0.5 m distance above the canopy and viewing angle maintained around 45° middle area about 2 m of each plot was targeted.
- (b)
- Normalized difference in vegetative index (NDVI) was measured with the help of Green seeker variable rate application and mapping systems with portable sensor (Mod:505) for 5 s for both well-watered and drought genotypes. The average data were used with values 0–1 (no-green to maximum green).
- (c)
- Days to heading (DTH) were recorded by counting the number of days taken from sowing to 50% flowering.
- (d)
- Days to maturity (DTM) were counted by number of days taken from sowing to until all spikes were turn to their specific color of maturity.
- (e)
- Plant height (PH) was measured from 5 random plants of each replication from ground level to the end spike excluding awns
- (f)
- Spike length (SL) was calculated by taking 5 random spikes from each replication and length was measured from the start of spike to the end of spike excluding awns
- (g)
- Number of grains spike−1 (NGS) were counted by taking five random spikes from each replication and their averages were recorded
- (h)
- Thousand kernel weight (TKW) were recorded by counting thousand grains from each replication and their weight was recorded in grams
- (i)
- Kernel abortion (KA) was recorded using following formula:
- (NGS for drought plants/NGS for well-watered) × 100
- (j)
- Grain yield (Y) of each genotype was recorded after harvesting the both the rows and yield was expressed in kilograms per hectare.
4.5. Molecular Characterization
4.6. Haplotype Characterization
4.7. Genome Wide Association Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. Statistics. World Food and Agriculture—Statistical Pocketbook; FAO: Rome, Italy, 2018. [Google Scholar]
- Lobell, D.; Burke, M.; Tebaldi, C.; Mastrandrea, M.; Falcon, W.; Naylor, R. Needs for food security in 2030 region. Science 2008, 319, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Amiri, R.; Bahraminejad, S.; Jalali-Honarmand, S. Effect of terminal drought stress on grain yield and some morphological traits in 80 bread wheat genotypes. Int. J. Agric. Crop Sci. 2013, 5, 1145–1153. [Google Scholar]
- Giunta, F.; Motzo, R.; Deidda, M. Effect of drought on yield and yield components of durum wheat and triticale in a Mediterranean environment. Field Crop. Res. 1993, 33, 399–409. [Google Scholar] [CrossRef]
- Saeidi, M.; Abdoli, M. Effect of drought stress during grain filling on yield and its components, gas exchange variables, and some physiological traits of wheat cultivars. J. Agric. Sci. Technol. 2015, 17, 885–898. [Google Scholar]
- Shokat, S.; Großkinsky, D.K.; Roitsch, T.; Liu, F. Activity of leaf and spike carbohydrate-metabolic and antioxidant enzymes reveals yield performance in three spring wheat genotypes grown under well-watered and drought conditions. BMC Plant Biol. 2020. [Google Scholar] [CrossRef]
- Pingali, P.L. Green revolution: Impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. USA 2012, 109, 12302–12308. [Google Scholar] [CrossRef]
- Vikram, P.; Franco, J.; Burgueño-Ferreira, J.; Li, H.; Sehgal, D.; Saint Pierre, C.; Ortiz, C.; Sneller, C.; Tattaris, M.; Guzman, C.; et al. Unlocking the genetic diversity of Creole wheats. Sci. Rep. 2016, 6, 23092. [Google Scholar] [CrossRef]
- Moore, G. Strategic pre-breeding for wheat improvement. Nat. Plants 2015, 1, 1–3. [Google Scholar] [CrossRef]
- SeeD. Seed of Discovery. International Maize and Wheat Improvement Center (CIMMYT). 2011. Available online: http://seedsofdiscovery.org (accessed on 31 May 2018).
- Singh, S.; Vikram, P.; Sehgal, D.; Burgueño, J.; Sharma, A.; Singh, S.K.; Sansaloni, C.P.; Joynson, R.; Brabbs, T.; Ortiz, C.; et al. Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Sci. Rep. 2018, 8, 12527. [Google Scholar] [CrossRef]
- Ledesma-Ramírez, L.; Solís-Moya, E.; Iturriaga, G.; Sehgal, D.; Reyes-Valdes, M.H.; Montero-Tavera, V.; Sansaloni, C.P.; Burgueño, J.; Ortiz, C.; Aguirre-Mancilla, C.L.; et al. GWAS to identify genetic loci for resistance to yellow rust in wheat pre-breeding lines derived from diverse exotic crosses. Front. Plant Sci. 2019, 10, 1390. [Google Scholar] [CrossRef]
- Crossa, J.; Burgueño, J.; Dreisigacker, S.; Vargas, M.; Herrera-Foessel, S.A.; Lillemo, M.; Singh, R.P.; Trethowan, R.; Warburton, M.; Franco, J.; et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 2007, 177, 1889–1913. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Kobiljski, B.; Denčić, S.; Varshney, R.K.; Börner, A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.). Mol. Breed. 2011, 27, 37–58. [Google Scholar] [CrossRef]
- Bordes, J.; Ravel, C.; Jaubertie, J.P.; Duperrier, B.; Gardet, O.; Heumez, E.; Pissavy, A.L.; Charmet, G.; Le Gouis, J.; Balfourier, F. Genomic regions associated with the nitrogen limitation response revealed in a global wheat core collection. Theor. Appl. Genet. 2013, 126, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.S.; El-Basyoni, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A.; et al. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Mora, F.; Castillo, D.; Lado, B.; Matus, I.; Poland, J.; Belzile, F.; von Zitzewitz, J.; del Pozo, A. Genome-wide association mapping of agronomic traits and carbon isotope discrimination in a worldwide germplasm collection of spring wheat using SNP markers. Mol. Breed. 2015, 35, 69. [Google Scholar] [CrossRef]
- Shokat, S.; Großkinsky, D.K. Tackling salinity in sustainable agriculture—What developing countries may learn from approaches of the developed world. Sustainability 2019, 11, 4558. [Google Scholar] [CrossRef]
- Sehgal, D.; Mondal, S.; Guzman, C.; Garcia Barrios, G.; Franco, C.; Singh, R.; Dreisigacker, S. Validation of candidate gene-based markers and identification of novel loci for thousand-grain weight in spring bread wheat. Front. Plant Sci. 2019, 10, 1189. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Liu, J.; Zhang, Y.; Cao, S.; He, Z.; Rasheed, A.; Jin, H. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019, 19, 168. [Google Scholar] [CrossRef]
- Qian, L.; Hickey, L.T.; Stahl, A.; Werner, C.R.; Hayes, B.; Snowdon, R.J.; Voss-Fels, K.P. Exploring and harnessing haplotype diversity to improve yield stability in crops. Front. Plant Sci. 2017, 8, 1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, H.; Yuan, L.; Wang, Z.; Yang, J.; Zhang, J. Post-anthesis alternate wetting and moderate soil drying enhances activities of key enzymes in sucrose-to-starch conversion in inferior spikelets of rice. J. Exp. Bot. 2012, 63, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, A.J.; Hamblin, M.T.; Jannink, J.L. Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in barley. PLoS ONE 2010, 5, e14079. [Google Scholar] [CrossRef] [PubMed]
- N’Diaye, A.; Haile, J.K.; Cory, A.T.; Clarke, F.R.; Clarke, J.M.; Knox, R.E.; Pozniak, C.J. Single marker and haplotype-based association analysis of semolina and pasta colour in elite durum wheat breeding lines using a high-density consensus map. PLoS ONE 2017, 12, e0187178. [Google Scholar]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; Reif, J.C. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef]
- Zegeye, H.; Rasheed, A.; Makdis, F.; Badebo, A.; Ogbonnaya, F.C. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS ONE 2014, 9, e105593. [Google Scholar] [CrossRef]
- Sehgal, D.; Autrique, E.; Singh, R.; Ellis, M.; Singh, S.; Dreisigacker, S. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci. Rep. 2017, 7, 41578. [Google Scholar] [CrossRef]
- Somers, D.J.; Banks, T.; Depauw, R.; Fox, S.; Clarke, J.; Pozniak, C.; Mccartney, C. Genome-wide linkage disequilibrium analysis in bread wheat and durum wheat. Genome 2007, 50, 557–567. [Google Scholar] [CrossRef]
- Benson, J.; Brown-guedira, G.; Murphy, J.P.; Sneller, C. Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for Fusarium head blight resistance. Plant Genome 2012, 5, 71–80. [Google Scholar] [CrossRef]
- Chen, X.; Min, D.; Yasir, T.A.; Hu, Y. Genetic diversity, population structure and linkage disequilibrium in elite Chinese winter wheat investigated with SSR markers. PLoS ONE 2012, 7, e44510. [Google Scholar] [CrossRef]
- Shokat, S.; Azhar, F.M.; Nabi, G.; Iqbal, Q. Estimation of heritability and genetic advance for some characters related to earliness in tomato (Solanum lycopersicum L.). J. Agric. Res. 2015, 53, 351–356. [Google Scholar]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Chapman, J.-J.O.-V.; Scott, C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.M.; Kirigwi, F.M.; Fritz, A.K.; Fellers, J.P. Mapping and quantitative trait loci analysis of drought tolerance in a spring wheat population using amplified fragment length polymorphism and diversity array technology markers. Crop Sci. 2012, 52, 253–261. [Google Scholar] [CrossRef]
- Ballesta, P.; Mora, F.; Pozo, A. Del Association mapping of drought tolerance indices in wheat: QTL-rich regions on chromosome 4A. Sci. Agric. 2018, 77, e20180153. [Google Scholar] [CrossRef]
- Guo, Z.; Zhao, Y.; Röder, M.S.; Reif, J.C.; Ganal, M.W.; Chen, D.; Schnurbusch, T. Manipulation and prediction of spike morphology traits for the improvement of grain yield in wheat. Sci. Rep. 2018, 8, 14435. [Google Scholar] [CrossRef]
- Mwadzingeni, L.; Shimelis, H.; Tesfay, S.; Tsilo, T.J. Screening of bread wheat genotypes for drought tolerance using phenotypic and proline analyses. Front. Plant Sci. 2016, 7, 1276. [Google Scholar] [CrossRef]
- Chai, L.; Chen, Z.; Bian, R.; Zhai, H.; Cheng, X.; Peng, H.; Yao, Y. Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2019, 132, 1815–1831. [Google Scholar] [CrossRef]
- Zhai, H.; Feng, Z.; Li, J.; Liu, X.; Xiao, S.; Ni, Z. QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front. Plant Sci. 2016, 7, 1617. [Google Scholar] [CrossRef]
- Ji, X.; Shiran, B.; Wan, J.; Lewis, D.C.; Jenkins, C.L.D.; Condon, A.G.; Richards, R.A.; Dolferus, R. Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat. Plant Cell Environ. 2010, 33, 926–942. [Google Scholar] [CrossRef]
- Rajala, A.; Hakala, K.; Mäkelä, P.; Peltonen-Sainio, P. Drought effect on grain number and grain weight at spike and spikelet level in six-row spring barley. J. Agron. Crop Sci. 2011, 197, 103–112. [Google Scholar] [CrossRef]
- Pradhan, S.; Robbins, K.; Bai, G.; Mason, R.E.; Khan, J.; Shahi, D.; Avci, M.; Guo, J.; Hossain, M.M.; Bhatta, M.; et al. Understanding the genetic basis of spike fertility to improve grain number, harvest index, and grain yield in wheat under high temperature stress environments. Front. Plant Sci. 2019, 10, 1481. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kobayashi, F.; Joshi, G.P.; Onuki, R.; Sakai, H.; Kanamori, H.; Wu, J.; Šimková, H.; Nasuda, S.; Endo, T.R.; et al. Next-generation survey sequencing and the molecular organization of wheat chromosome 6B. DNA Res. 2014, 21, 103–114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nadolska-Orczyk, A.; Izabela, O.; Wacław, K.R. Major genes determining yield-related traits in wheat and barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhou, L.; Chen, J.; Qiu, Z.; He, Y. GainTKW: A Measurement system of thousand kernel weight based on the android platform. Agronomy 2018, 8, 178. [Google Scholar] [CrossRef]
- Duggan, B.L.; Domitruk, D.R.; Fowler, D.B. Yield component variation in winter wheat grown under drought stress. Can. J. Plant Sci. 2000, 80, 739–745. [Google Scholar] [CrossRef]
- Liu, Y.; Bowman, B.; Hu, Y.-G.; Liang, X.; Zhao, W.; Wheeler, J.; Klassen, N.; Bockelman, H.; Bonman, J.; Chen, J. Evaluation of agronomic traits and drought tolerance of winter wheat accessions from the USDA-ARS national small grains collection. Agronomy 2017, 7, 51. [Google Scholar] [CrossRef]
- Lehnert, H.; Serfling, A.; Friedt, W.; Ordon, F. Genome-wide association studies reveal genomic regions associated with the response of wheat (Triticum aestivum L.) to mycorrhizae under drought stress conditions. Front. Plant Sci. 2018, 9, 1728. [Google Scholar] [CrossRef]
- LujánBasile, S.M.; Ramírez, I.A.; Crescente, J.M.; Conde, M.B.; Demichelis, M.; Abbate, P.; Rogers, W.J.; Pontaroli, A.C.; Helguera, M.; Vanzetti, L.S. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019, 19, 553. [Google Scholar]
- Rustenholz, C.; Choulet, F.; Laugier, C.; Šafář, J.; Šimková, H.; Doležel, J.; Magni, F.; Scalabrin, S.; Cattonaro, F.; Vautrin, S.; et al. A 3,000-loci transcription map of chromosome 3B unravels the structural and functional features of gene islands in hexaploid wheat. Plant Physiol. 2011, 157, 1596–1608. [Google Scholar] [CrossRef]
- Ogihara, Y.; Kawaura, K.; Mishina, K.; Tarui, H.; Kawai, J. Functional Partitioning of the Expressed Homoeoloci Based on the Fully Collected cDNAs of Allohexaploid Wheat; Kohei Mishina Yokohama City University, Kihara Institute for Biological Research: Yokohama, Japan, 2016. [Google Scholar]
- Gabriel, S.; Schaffner, S.F.; Nguyen, H.; Moore, J.; Roy, J.; Blumenstiel, B.; Higgins, J.; DeFelice, M.; Lochner, A.; Faggart, M.; et al. The structure of haplotype blocks in the human genome. Science 2002, 296, 2225–2229. [Google Scholar] [CrossRef]
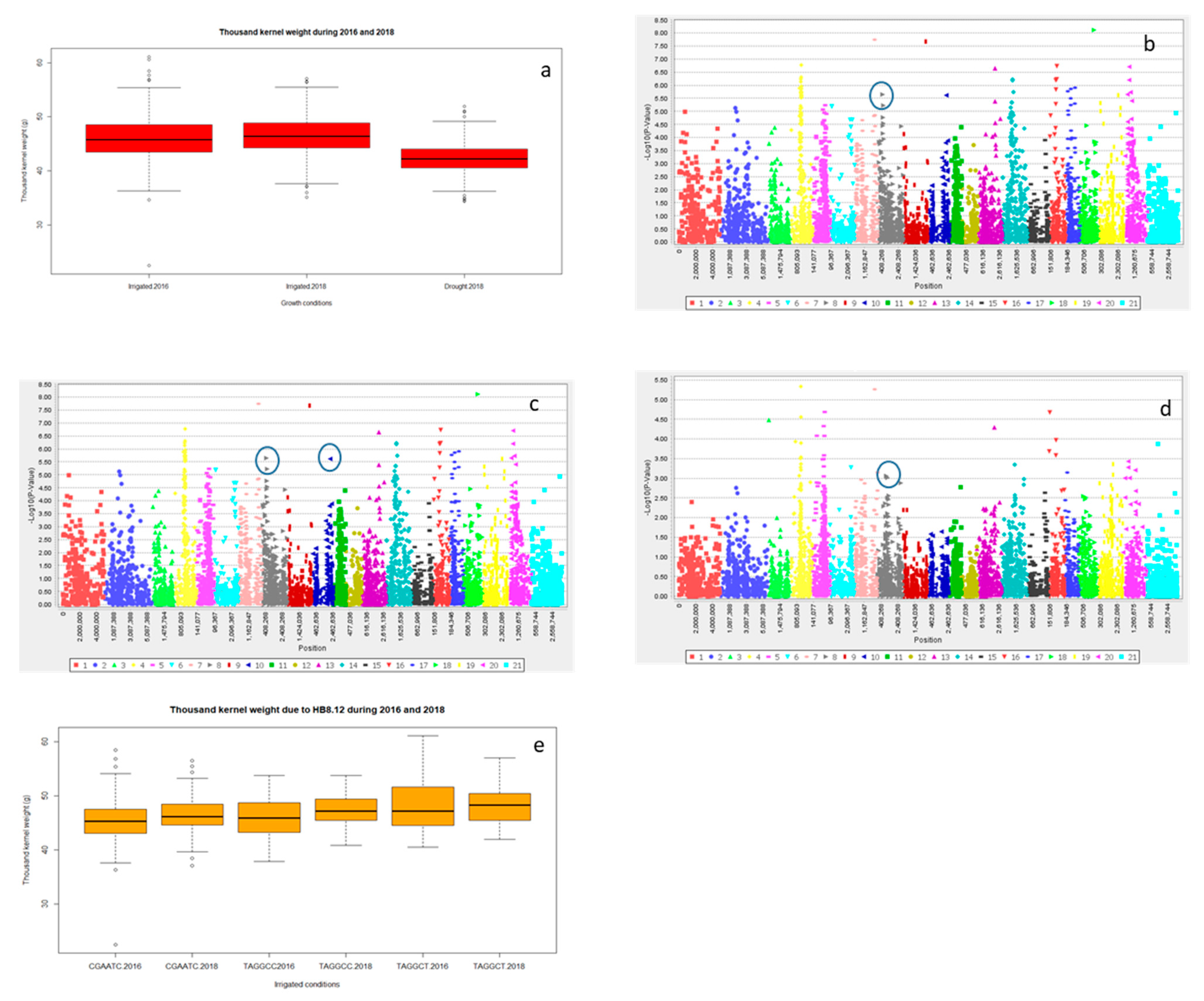
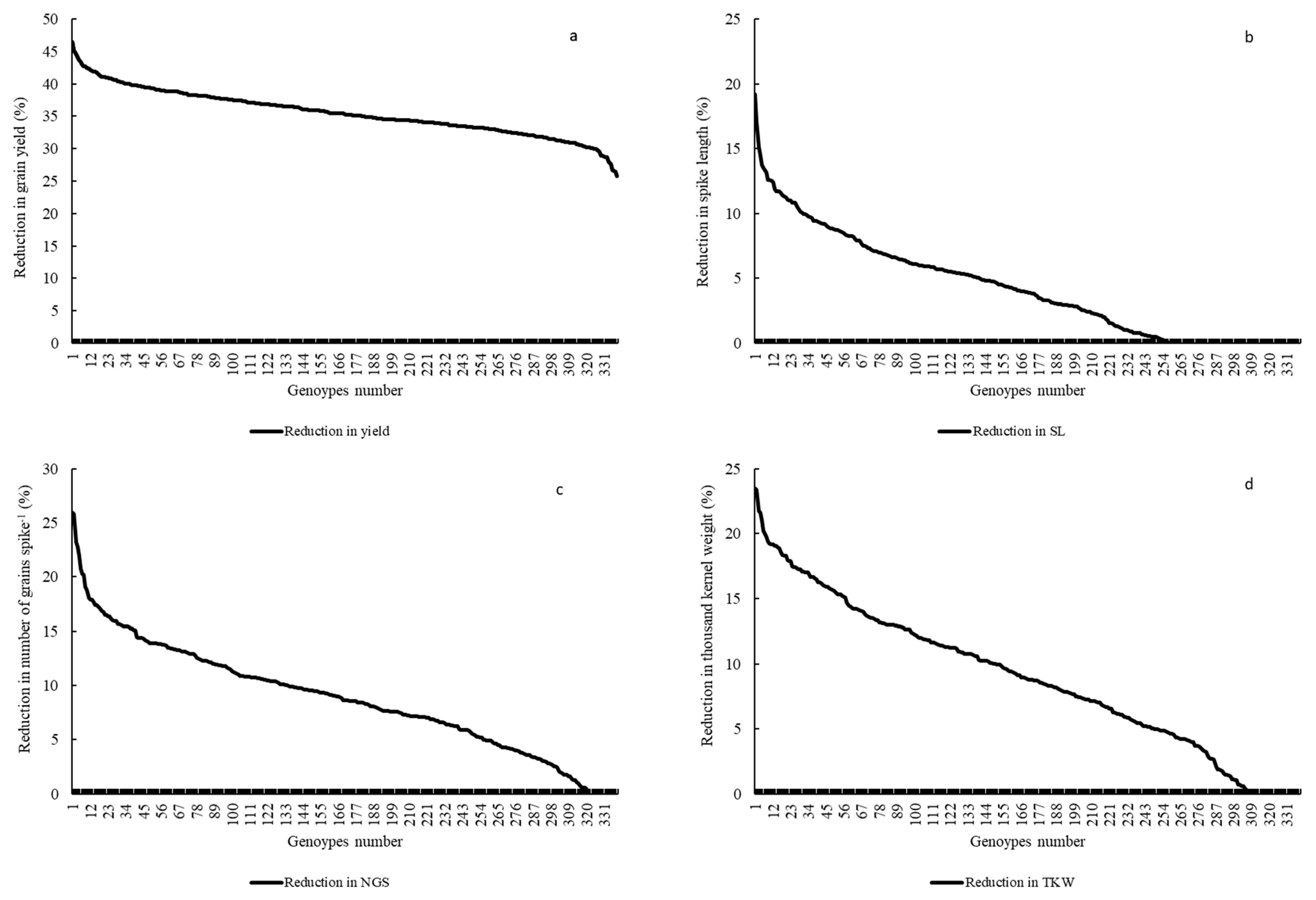
| Variables | DTH | DTM | PH | NDVI | CTD | NGS | SL | TKW | Y | KA |
|---|---|---|---|---|---|---|---|---|---|---|
| 2018-IR | 67.41 ± 0.15 | 120.7 ± 0.1 | 96.7 ± 0.18 | 0.76 ± 0.0002 | 24.15 ± 0.004 | 50.82 ± 0.17 | 10.71 ± 0.03 | 46.44 ± 0.1 | 5242.59 ± 11.9 | |
| 2016-IR | 77.09 ± 0.02 | 119.99 ± 0.1 | 99.7 ± 0.2 | 45.96 ± 0.2 | 4698.03 ± 15.3 | |||||
| 2018-DR | 64.84 ± 0.01 | 104.77 ± 0.2 | 87.33 ± 0.13 | 0.65 ± 0.001 | 22.87 ± 0.006 | 46.24 ± 0.11 | 10.32 ± 0.02 | 42.30 ± 0.1 | 4203.02 ± 12.4 | 9.31 ± 0.4 |
| 2016-DR | 76.08 ± 0.12 | 110.31 ± 0.1 | 81.95 ± 0.19 | 2277.12 ± 8.38 | ||||||
| Genotypes | ns | <0.001 | <0.001 | ns | ns | ns | <0.001 | <0.001 | <0.001 | |
| Year | <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| Treatment | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| GenotypesxYear | ns | <0.001 | <0.001 | <0.05 | ||||||
| GenotypesxTreatment | ns | ns | ns | <0.05 | ns | ns | ns | <0.001 | ns | |
| YearxTreatment | <0.001 | <0.001 | <0.001 | <0.001 | ||||||
| GenotypesxYearxTreatment | ns | ns | ns | ns | ||||||
| Heritability broad sense (hbs) Irrigated | 0.77 | 0.85 | 0.81 | 0.25 | 0.13 | 0.72 | 0.80 | 0.84 | 0.61 | |
| Heritability broad sense (hbs) Drought | 0.08 | 0.82 | 0.71 | 0.68 | 0.22 | 0.6 | 0.72 | 0.67 | 0.65 | 0.17 |
| Year | Treatment | Trait | Marker | Chromosome | Log10 | p-Value | R2 (%) |
|---|---|---|---|---|---|---|---|
| 2018 | Irrigated | SL | 1,216,938 | 3D | 5.46 | 3.43 × 10−6 | 8.2 |
| Drought | 1,216,938 | 3D | 2.39 | 0.0041 | 2.8 | ||
| 2018 | Irrigated | SL | 1,113,306 | 2D | 3.32 | 4.76 × 10−4 | 4.8 |
| Drought | 1,046,144 | 2D | 1.98 | 0.01047 | 2.1 | ||
| 2018 | Irrigated | NGS | 3,064,504 | 6B | 2.46 | 0.00346 | 2.9 |
| Drought | 2,256,902 | 6B | 2.54 | 0.00287 | 2.9 | ||
| 2018 | Irrigated | NGS | 1,079,850 | 3A | 2.12 | 0.00761 | 2.3 |
| Drought | 1,190,017 | 3A | 2.27 | 0.00532 | 2.6 | ||
| 2018 | Drought | KA | 4,910,194 | 3B | 2.78 | 0.00167 | 3.4 |
| Drought | KA | 1,076,524 | 3A | 2.71 | 0.0019 | 3.1 | |
| 2016 | Irrigated | TKW | 2,260,800 | 3B | 2.04 | 0.0096 | 2.5 |
| 2018 | Irrigated | 3,064,863 | 3B | 3.02 | 9.61 × 10−4 | 3.7 | |
| Drought | 988,433 | 3B | 3.08 | 8.36 × 10−4 | 3.9 | ||
| 2018 | Drought | 1,664,941 | 4A | 3.06 | 8.65 × 10−4 | 4.0 | |
| 2016 | Irrigated | Y | 993,866 | 6B | 1.99 | 0.01024 | 2.1 |
| Drought | 1,219,677 | 6B | 2.47 | 0.0034 | 2.9 | ||
| 2018 | Irrigated | 3,064,504 | 6B | 3.28 | 5.22 × 10−4 | 4.1 | |
| Drought | 1,219,677 | 6B | 4.57 | 2.70 × 10−5 | 6.1 | ||
| 2016 | Irrigated | Y | 1,101,137 | 4A | 2.66 | 0.0022 | 3.6 |
| Drought | 1,041,102 | 4A | 3.40 | 3.94 × 10−4 | 5.0 | ||
| 2018 | Irrigated | 1,091,070 | 4A | 2.64 | 0.0023 | 3.2 | |
| Drought | 1,041,102 | 4A | 2.30 | 0.00502 | 2.4 |
| Traits | HB | Clone ID | Position (cM) | Significance | R2 (%) | Chromosome |
|---|---|---|---|---|---|---|
| Y | HB10.1 | 1,054,658 | 11.64 | * | 6.9 | 4A |
| 1,718,021 | 10.76 | 4A | ||||
| 2,248,753 | 13.13 | 4A | ||||
| HB10.6 | 1,130,105 | 28.98 | * | 3.0 | 4A | |
| 1,217,569 | 28.84 | 4A | ||||
| HB10.7 | 3,024,473 | 31.63 | ** | 5.9 | 4A | |
| 979,934 | 32.41 | 4A | ||||
| TKW | HB8.12 | 1,082,315 | 49.77 | ** | 8.4 | 3B |
| 1,082,914 | 50.69 | 3B | ||||
| 997,119 | 50.81 | 3B | ||||
| 1,110,218 | 51.27 | 3B | ||||
| 1,010,250 | 50.17 | 3B | ||||
| 1,130,393 | 53.18 | 3B | ||||
| KA | HB7.12 | 1,038,112 | 61.93 | *** | 10.1 | 3A |
| 1,052,021 | 61.93 | 3A | ||||
| 2,278,246 | 60.45 | 3A | ||||
| 1,250,769 | 60.37 | 3A | ||||
| 1,040,728 | 61.93 | 3A | ||||
| SL | HB6.10 | 3,029,487 | 142.74 | **** | 6.7 | 2D |
| 1,123,029 | 147.6 | 2D | ||||
| DTH | HB7.17 | 1,150,200 | 77.13 | *** | 5.5 | 3A |
| 3,024,325 | 76.17 | 3A | ||||
| DTM | HB2.5 | 3,064,540 | 98.51 | ** | 6.0 | 1B |
| 1,093,781 | 99.06 | 1B | ||||
| NGS | HB17.1 | 3,222,182 | 2.64 | **** | 7.4 | 6B |
| 1,050,615 | 6.34 | 6B | ||||
| NGS | HB7.11 | 990,692 | 56.45 | ** | 5.9 | 3A |
| 3,023,149 | 56.5 | 3A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shokat, S.; Sehgal, D.; Vikram, P.; Liu, F.; Singh, S. Molecular Markers Associated with Agro-Physiological Traits under Terminal Drought Conditions in Bread Wheat. Int. J. Mol. Sci. 2020, 21, 3156. https://doi.org/10.3390/ijms21093156
Shokat S, Sehgal D, Vikram P, Liu F, Singh S. Molecular Markers Associated with Agro-Physiological Traits under Terminal Drought Conditions in Bread Wheat. International Journal of Molecular Sciences. 2020; 21(9):3156. https://doi.org/10.3390/ijms21093156
Chicago/Turabian StyleShokat, Sajid, Deepmala Sehgal, Prashant Vikram, Fulai Liu, and Sukhwinder Singh. 2020. "Molecular Markers Associated with Agro-Physiological Traits under Terminal Drought Conditions in Bread Wheat" International Journal of Molecular Sciences 21, no. 9: 3156. https://doi.org/10.3390/ijms21093156
APA StyleShokat, S., Sehgal, D., Vikram, P., Liu, F., & Singh, S. (2020). Molecular Markers Associated with Agro-Physiological Traits under Terminal Drought Conditions in Bread Wheat. International Journal of Molecular Sciences, 21(9), 3156. https://doi.org/10.3390/ijms21093156







