Regulation of Translation in the Protozoan Parasite Leishmania
Abstract
1. Introduction
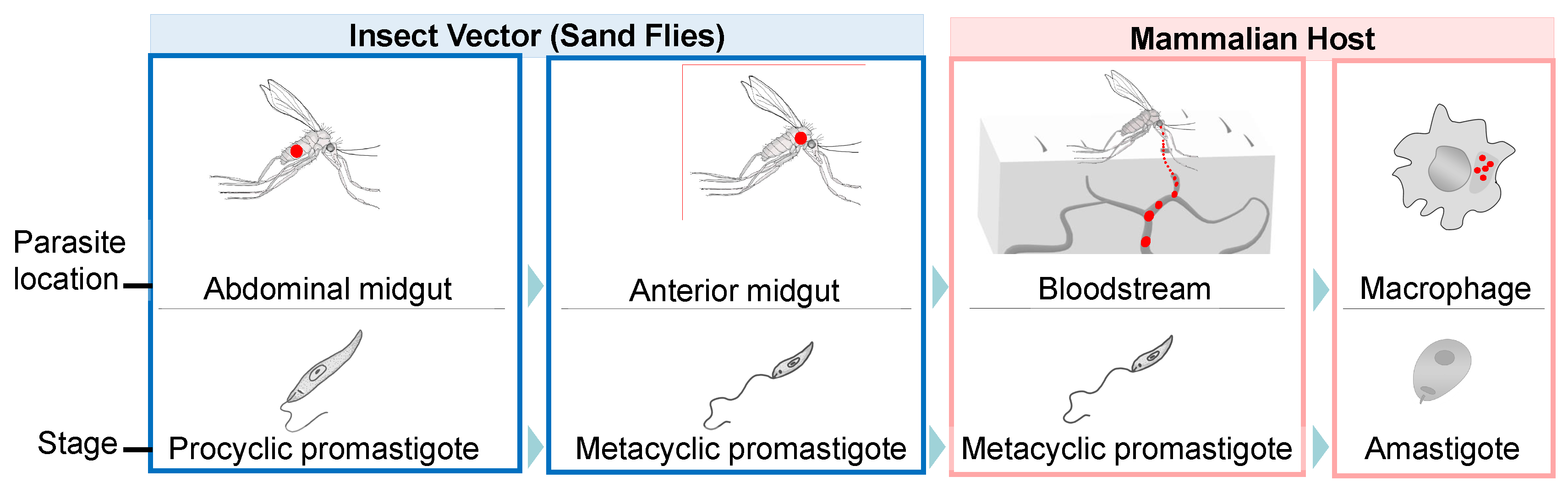
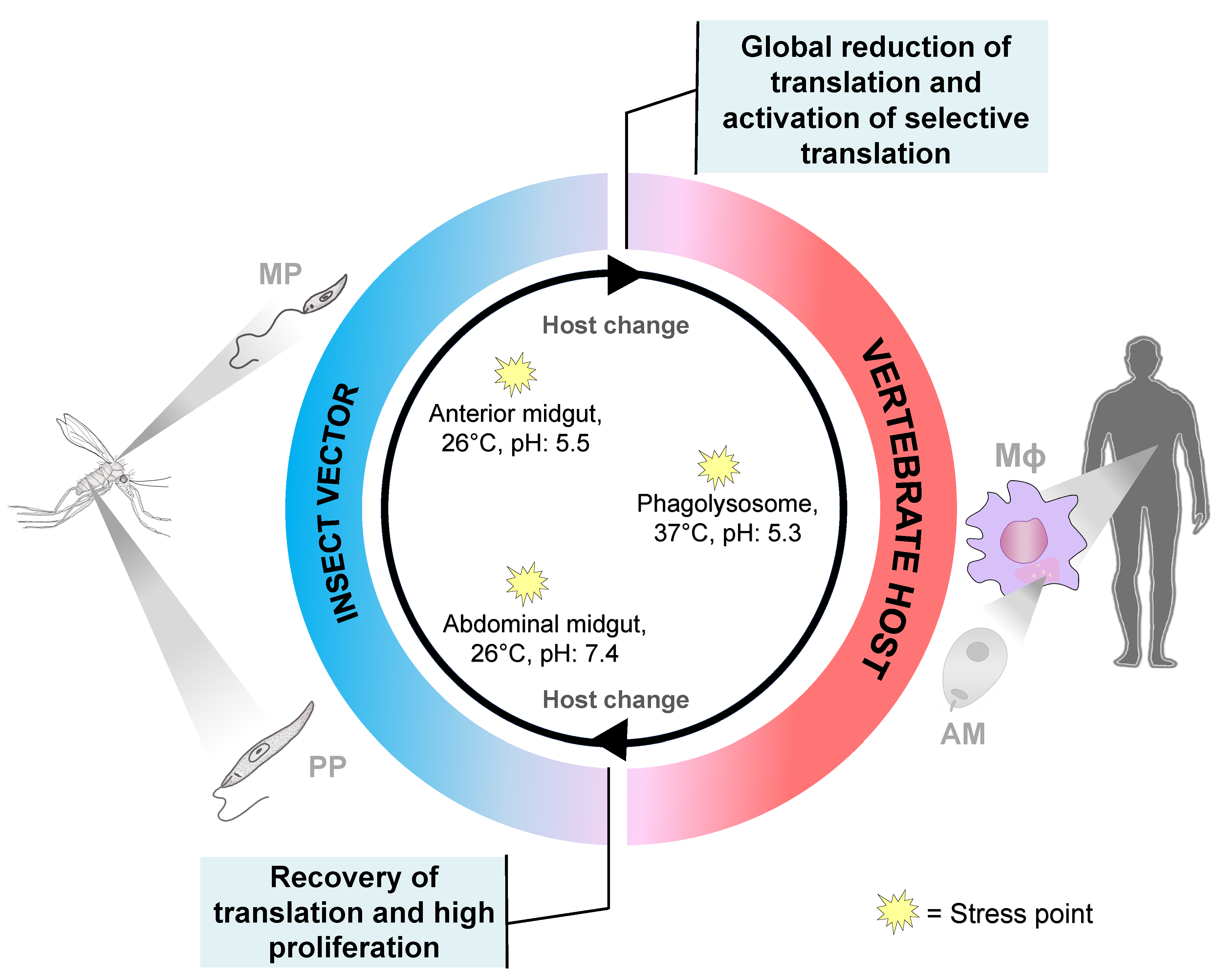
2. Translational Control during Leishmania Differentiation
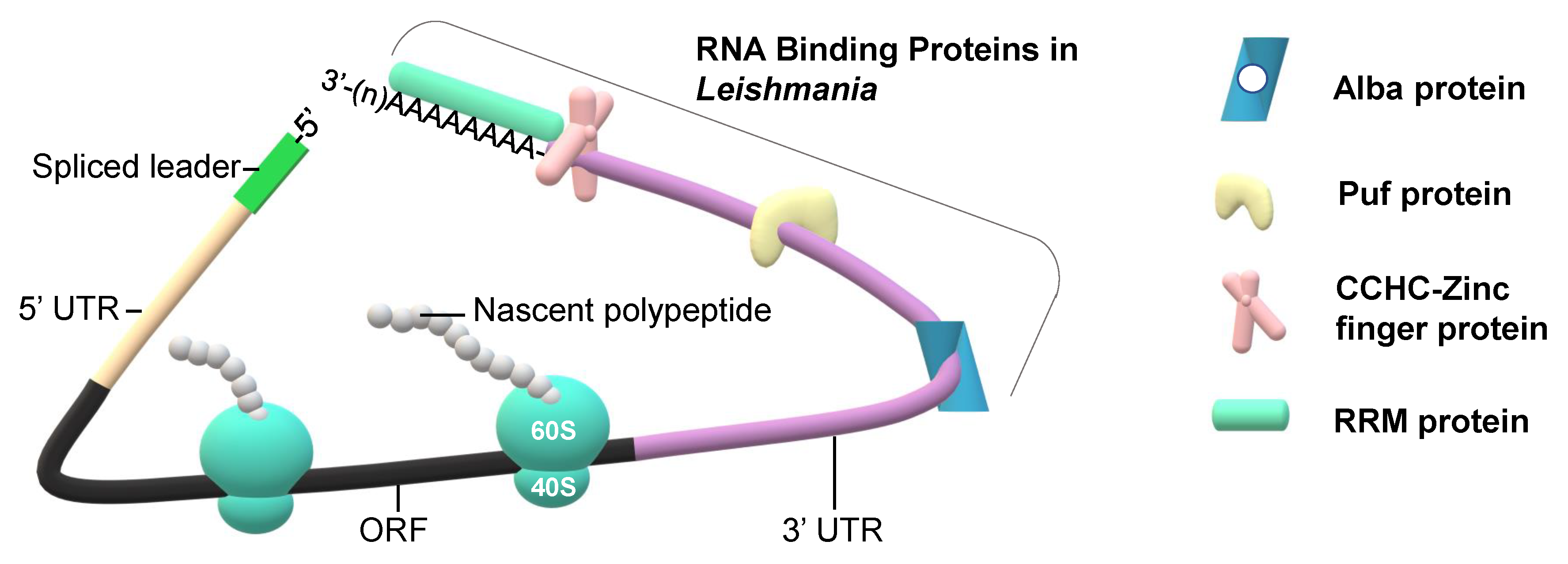
3. RNA Binding Proteins and Their Role in Regulation of Translation and Differentiation of Parasites
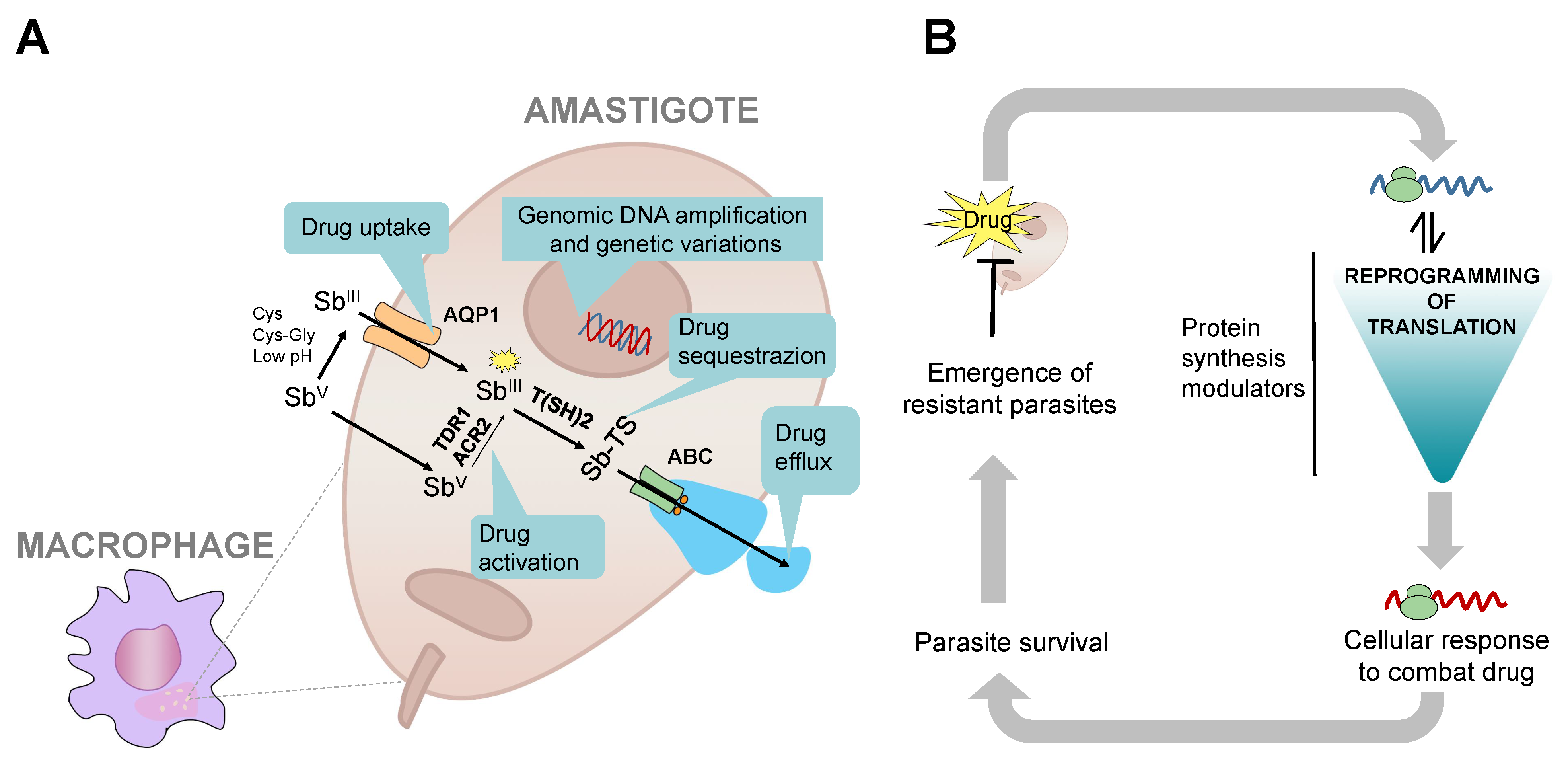
4. Drug Resistance and Translation
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| eIF2α | alpha-subunit of eukaryotic initiation factor 2 |
| RBPs | RNA binding proteins |
| Puf | Pumilio and Fem-3 binding factor |
| Alba | acetylation lowers binding affinity |
| RRM | RNA-recognition motif |
| PABP | Poly(A)-binding protein |
| CCCH | Cys3His Zinc finger |
| ZFP1 | zinc finger protein 1 |
| ZFP2 | zinc finger protein 2 |
| UTR | untranslated region |
| CDPK1 | calcium-dependent protein kinase 1 |
| TCA | tricarboxylic acid cycle |
References
- Georgiadou, S.P.; Makaritsis, K.P.; Dalekos, G.N. Leishmaniasis revisited: Current aspects on epidemiology, diagnosis and treatment. J. Transl. Int. Med. 2015, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Akhoundi, M.; Kuhls, K.; Cannet, A.; Votypka, J.; Marty, P.; Delaunay, P.; Sereno, D. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Negl. Trop. Dis. 2016, 10, e0004349. [Google Scholar] [CrossRef] [PubMed]
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; WHO Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef]
- Murray, H.W.; Berman, J.D.; Davies, C.R.; Saravia, N.G. Advances in leishmaniasis. Lancet 2005, 366, 1561–1577. [Google Scholar] [CrossRef]
- Leishmaniasis; World Health Organization, 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 10 March 2020).
- Capela, R.; Moreira, R.; Lopes, F. An Overview of Drug Resistance in Protozoal Diseases. Int. J. Mol. Sci. 2019, 20, 5748. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.; More, D.K.; Singh, M.K.; Singh, V.P.; Sharma, S.; Makharia, A.; Kumar, P.C.; Murray, H.W. Failure of pentavalent antimony in visceral leishmaniasis in India: Report from the center of the Indian epidemic. Clin. Infect. Dis. 2000, 31, 1104–1107. [Google Scholar] [CrossRef]
- Santos, D.O.; Coutinho, C.E.; Madeira, M.F.; Bottino, C.G.; Vieira, R.T.; Nascimento, S.B.; Bernardino, A.; Bourguignon, S.C.; Corte-Real, S.; Pinho, R.T.; et al. Leishmaniasis treatment--a challenge that remains: A review. Parasitol. Res. 2008, 103, 1–10. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Leprohon, P.; Bigot, S.; Padmanabhan, P.K.; Mukherjee, A.; Roy, G.; Gingras, H.; Mestdagh, A.; Papadopoulou, B.; Ouellette, M. Coupling chemical mutagenesis to next generation sequencing for the identification of drug resistance mutations in Leishmania. Nat. Commun. 2019, 10, 5627. [Google Scholar] [CrossRef]
- De Gaudenzi, J.G.; Noe, G.; Campo, V.A.; Frasch, A.C.; Cassola, A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011, 51, 31–46. [Google Scholar] [CrossRef]
- Palenchar, J.B.; Bellofatto, V. Gene transcription in trypanosomes. Mol. Biochem. Parasitol. 2006, 146, 135–141. [Google Scholar] [CrossRef]
- Liang, X.H.; Haritan, A.; Uliel, S.; Michaeli, S. Trans and cis splicing in trypanosomatids: Mechanism, factors, and regulation. Eukaryot. Cell 2003, 2, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Zilberstein, D.; Shapira, M. The role of pH and temperature in the development of Leishmania parasites. Annu. Rev. Microbiol. 1994, 48, 449–470. [Google Scholar] [CrossRef] [PubMed]
- Minia, I.; Merce, C.; Terrao, M.; Clayton, C. Translation Regulation and RNA Granule Formation after Heat Shock of Procyclic Form Trypanosoma brucei: Many Heat-Induced mRNAs Are also Increased during Differentiation to Mammalian-Infective Forms. PLoS Negl. Trop Dis. 2016, 10, e0004982. [Google Scholar] [CrossRef] [PubMed]
- Zilka, A.; Garlapati, S.; Dahan, E.; Yaolsky, V.; Shapira, M. Developmental regulation of heat shock protein 83 in Leishmania. 3′ processing and mRNA stability control transcript abundance, and translation id directed by a determinant in the 3’-untranslated region. J. Biol. Chem. 2001, 276, 47922–47929. [Google Scholar] [CrossRef]
- Shapira, M.; Zilka, A.; Garlapati, S.; Dahan, E.; Dahan, I.; Yavesky, V. Post transcriptional control of gene expression in Leishmania. Med. Microbiol. Immunol. 2001, 190, 23–26. [Google Scholar] [CrossRef]
- Cloutier, S.; Laverdiere, M.; Chou, M.N.; Boilard, N.; Chow, C.; Papadopoulou, B. Translational control through eIF2alpha phosphorylation during the Leishmania differentiation process. PLoS ONE 2012, 7, e35085. [Google Scholar] [CrossRef]
- Kloehn, J.; Saunders, E.C.; O’Callaghan, S.; Dagley, M.J.; McConville, M.J. Characterization of metabolically quiescent Leishmania parasites in murine lesions using heavy water labeling. PLoS Pathog. 2015, 11, e1004683. [Google Scholar] [CrossRef]
- Serafim, T.D.; Figueiredo, A.B.; Costa, P.A.; Marques-da-Silva, E.A.; Goncalves, R.; de Moura, S.A.; Gontijo, N.F.; da Silva, S.M.; Michalick, M.S.; Meyer-Fernandes, J.R.; et al. Leishmania metacyclogenesis is promoted in the absence of purines. PLoS Negl. Trop. Dis. 2012, 6, e1833. [Google Scholar] [CrossRef]
- Shrivastava, R.; Drory-Retwitzer, M.; Shapira, M. Nutritional stress targets LeishIF4E-3 to storage granules that contain RNA and ribosome components in Leishmania. PLoS Negl. Trop. Dis. 2019, 13, e0007237. [Google Scholar] [CrossRef]
- Zinoviev, A.; Manor, S.; Shapira, M. Nutritional stress affects an atypical cap-binding protein in Leishmania. RNA Biol. 2012, 9, 1450–1460. [Google Scholar] [CrossRef]
- Zinoviev, A.; Shapira, M. Evolutionary conservation and diversification of the translation initiation apparatus in trypanosomatids. Comp. Funct. Genomics. 2012, 2012, 813718. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, R.; Tupperwar, N.; Drory-Retwitzer, M.; Shapira, M. Deletion of a Single LeishIF4E-3 Allele by the CRISPR-Cas9 System Alters Cell Morphology and Infectivity of Leishmania. mSphere 2019, 4, e00450-19. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, P.J.; Alonso, A.; Gomez, M.J.; Sanchez-Gorostiaga, A.; Moreno-Paz, M.; Gonzalez-Pastor, E.; Torano, A.; Parro, V.; Larraga, V. Temperature increase prevails over acidification in gene expression modulation of amastigote differentiation in Leishmania infantum. BMC Genom. 2010, 11, 31. [Google Scholar] [CrossRef]
- De Pablos, L.M.; Ferreira, T.R.; Walrad, P.B. Developmental differentiation in Leishmania lifecycle progression: Post-transcriptional control conducts the orchestra. Curr. Opin. Microbiol. 2016, 34, 82–89. [Google Scholar] [CrossRef]
- Holcik, M.; Sonenberg, N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005, 6, 318–327. [Google Scholar] [CrossRef]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Wek, R.C.; Jiang, H.Y.; Anthony, T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006, 34, 7–11. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Tikhonova, E.B.; Grozdanov, P.N.; Huffman, J.C.; Baca, K.R.; Karamyshev, A.; Denison, R.B.; MacDonald, C.C.; Zhang, K.; Karamyshev, A.L. Polysome Profiling in Leishmania, Human Cells and Mouse Testis. J. Vis. Exp. 2018. [Google Scholar] [CrossRef]
- Zinoviev, A.; Leger, M.; Wagner, G.; Shapira, M. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 2011, 39, 8404–8415. [Google Scholar] [CrossRef]
- Yoffe, Y.; Zuberek, J.; Lerer, A.; Lewdorowicz, M.; Stepinski, J.; Altmann, M.; Darzynkiewicz, E.; Shapira, M. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell 2006, 5, 1969–1979. [Google Scholar] [CrossRef]
- Freire, E.R.; Malvezzi, A.M.; Vashisht, A.A.; Zuberek, J.; Saada, E.A.; Langousis, G.; Nascimento, J.D.; Moura, D.; Darzynkiewicz, E.; Hill, K.; et al. Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot. Cell 2014, 13, 896–908. [Google Scholar] [CrossRef] [PubMed]
- Freire, E.R.; Vashisht, A.A.; Malvezzi, A.M.; Zuberek, J.; Langousis, G.; Saada, E.A.; Nascimento Jde, F.; Stepinski, J.; Darzynkiewicz, E.; Hill, K.; et al. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA 2014, 20, 1272–1286. [Google Scholar] [CrossRef] [PubMed]
- Freire, E.R.; Moura, D.M.N.; Bezerra, M.J.R.; Xavier, C.C.; Morais-Sobral, M.C.; Vashisht, A.A.; Rezende, A.M.; Wohlschlegel, J.A.; Sturm, N.R.; de Melo Neto, O.P.; et al. Trypanosoma brucei EIF4E2 cap-binding protein binds a homolog of the histone-mRNA stem-loop-binding protein. Curr. Genet. 2018, 64, 821–839. [Google Scholar] [CrossRef]
- Freire, E.R.; Sturm, N.R.; Campbell, D.A.; de Melo Neto, O.P. The Role of Cytoplasmic mRNA Cap-Binding Protein Complexes in Trypanosoma brucei and Other Trypanosomatids. Pathogens 2017, 6, 55. [Google Scholar] [CrossRef]
- Kramer, S.; Carrington, M. Trans-acting proteins regulating mRNA maturation, stability and translation in trypanosomatids. Trends Parasitol. 2011, 27, 23–30. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nandan, D.; Thomas, S.A.; Nguyen, A.; Moon, K.M.; Foster, L.J.; Reiner, N.E. Comprehensive Identification of mRNA-Binding Proteins of Leishmania donovani by Interactome Capture. PLoS ONE 2017, 12, e0170068. [Google Scholar] [CrossRef] [PubMed]
- Alsford, S.; Turner, D.J.; Obado, S.O.; Sanchez-Flores, A.; Glover, L.; Berriman, M.; Hertz-Fowler, C.; Horn, D. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome. Res. 2011, 21, 915–924. [Google Scholar] [CrossRef]
- Clayton, C. The regulation of trypanosome gene expression by RNA-binding proteins. PLoS Pathog. 2013, 9, e1003680. [Google Scholar] [CrossRef]
- Kolev, N.G.; Ullu, E.; Tschudi, C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell Microbiol. 2014, 16, 482–489. [Google Scholar] [CrossRef]
- Wurst, M.; Seliger, B.; Jha, B.A.; Klein, C.; Queiroz, R.; Clayton, C. Expression of the RNA recognition motif protein RBP10 promotes a bloodstream-form transcript pattern in Trypanosoma brucei. Mol. Microbiol. 2012, 83, 1048–1063. [Google Scholar] [CrossRef]
- Urbaniak, M.D.; Guther, M.L.; Ferguson, M.A. Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLoS ONE 2012, 7, e36619. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Ramey-Butler, K.; Cross, G.A.; Ullu, E.; Tschudi, C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science 2012, 338, 1352–1353. [Google Scholar] [CrossRef] [PubMed]
- Minvielle-Sebastia, L.; Preker, P.J.; Wiederkehr, T.; Strahm, Y.; Keller, W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. USA 1997, 94, 7897–7902. [Google Scholar] [CrossRef] [PubMed]
- Amrani, N.; Minet, M.; Le Gouar, M.; Lacroute, F.; Wyers, F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol. Cell. Biol. 1997, 17, 3694–3701. [Google Scholar] [CrossRef]
- Mangus, D.A.; Amrani, N.; Jacobson, A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998, 18, 7383–7396. [Google Scholar] [CrossRef]
- De Melo Neto, O.P.; da Costa Lima, T.D.C.; Merlo, K.C.; Romao, T.P.; Rocha, P.O.; Assis, L.A.; Nascimento, L.M.; Xavier, C.C.; Rezende, A.M.; Reis, C.R.S.; et al. Phosphorylation and interactions associated with the control of the Leishmania Poly-A Binding Protein 1 (PABP1) function during translation initiation. RNA Biol. 2018, 15, 739–755. [Google Scholar] [CrossRef]
- Da Costa Lima, T.D.; Moura, D.M.; Reis, C.R.; Vasconcelos, J.R.; Ellis, L.; Carrington, M.; Figueiredo, R.C.; de Melo Neto, O.P. Functional characterization of three leishmania poly(a) binding protein homologues with distinct binding properties to RNA and protein partners. Eukaryot. Cell 2010, 9, 1484–1494. [Google Scholar] [CrossRef]
- Kramer, S.; Bannerman-Chukualim, B.; Ellis, L.; Boulden, E.A.; Kelly, S.; Field, M.C.; Carrington, M. Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS ONE 2013, 8, e54004. [Google Scholar] [CrossRef]
- Bates, E.J.; Knuepfer, E.; Smith, D.F. Poly(A)-binding protein I of Leishmania: Functional analysis and localisation in trypanosomatid parasites. Nucleic Acids Res. 2000, 28, 1211–1220. [Google Scholar] [CrossRef][Green Version]
- Lueong, S.; Merce, C.; Fischer, B.; Hoheisel, J.D.; Erben, E.D. Gene expression regulatory networks in Trypanosoma brucei: Insights into the role of the mRNA-binding proteome. Mol. Microbiol. 2016, 100, 457–471. [Google Scholar] [CrossRef]
- Klein, C.; Terrao, M.; Inchaustegui Gil, D.; Clayton, C. Polysomes of Trypanosoma brucei: Association with Initiation Factors and RNA-Binding Proteins. PLoS ONE 2015, 10, e0135973. [Google Scholar] [CrossRef]
- Hendriks, E.F.; Robinson, D.R.; Hinkins, M.; Matthews, K.R. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 2001, 20, 6700–6711. [Google Scholar] [CrossRef]
- Hendriks, E.F.; Matthews, K.R. Disruption of the developmental programme of Trypanosoma brucei by genetic ablation of TbZFP1, a differentiation-enriched CCCH protein. Mol. Microbiol. 2005, 57, 706–716. [Google Scholar] [CrossRef]
- Ling, A.S.; Trotter, J.R.; Hendriks, E.F. A zinc finger protein, TbZC3H20, stabilizes two developmentally regulated mRNAs in trypanosomes. J. Biol. Chem. 2011, 286, 20152–20162. [Google Scholar] [CrossRef]
- Droll, D.; Minia, I.; Fadda, A.; Singh, A.; Stewart, M.; Queiroz, R.; Clayton, C. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013, 9, e1003286. [Google Scholar] [CrossRef] [PubMed]
- Delhi, P.; Queiroz, R.; Inchaustegui, D.; Carrington, M.; Clayton, C. Is there a classical nonsense-mediated decay pathway in trypanosomes? PLoS ONE 2011, 6, e25112. [Google Scholar] [CrossRef] [PubMed]
- Kramer, S.; Kimblin, N.C.; Carrington, M. Genome-wide in silico screen for CCCH-type zinc finger proteins of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major. BMC Genom. 2010, 11, 283. [Google Scholar] [CrossRef] [PubMed]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef]
- Spassov, D.S.; Jurecic, R. The PUF family of RNA-binding proteins: Does evolutionarily conserved structure equal conserved function? IUBMB Life 2003, 55, 359–366. [Google Scholar] [CrossRef]
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Zamore, P.D.; Williamson, J.R.; Lehmann, R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 1997, 3, 1421–1433. [Google Scholar] [PubMed]
- Wharton, R.P.; Sonoda, J.; Lee, T.; Patterson, M.; Murata, Y. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1998, 1, 863–872. [Google Scholar] [CrossRef]
- Gu, W.; Deng, Y.; Zenklusen, D.; Singer, R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004, 18, 1452–1465. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.S., Jr.; Houshmandi, S.S.; Lopez Leban, F.; Olivas, W.M. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 2004, 10, 1625–1636. [Google Scholar] [CrossRef]
- Ulbricht, R.J.; Olivas, W.M. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA 2008, 14, 246–262. [Google Scholar] [CrossRef]
- Caro, F.; Bercovich, N.; Atorrasagasti, C.; Levin, M.J.; Vazquez, M.P. Trypanosoma cruzi: Analysis of the complete PUF RNA-binding protein family. Exp. Parasitol. 2006, 113, 112–124. [Google Scholar] [CrossRef]
- Luu, V.D.; Brems, S.; Hoheisel, J.D.; Burchmore, R.; Guilbride, D.L.; Clayton, C. Functional analysis of Trypanosoma brucei PUF1. Mol. Biochem. Parasitol. 2006, 150, 340–349. [Google Scholar] [CrossRef]
- Kemp, D.J.; Coppel, R.L.; Anders, R.F. Repetitive proteins and genes of malaria. Annu. Rev. Microbiol. 1987, 41, 181–208. [Google Scholar] [CrossRef]
- Folgueira, C.; Martinez-Bonet, M.; Requena, J.M. The Leishmania infantum PUF proteins are targets of the humoral response during visceral leishmaniasis. BMC Res. Notes 2010, 3, 13. [Google Scholar] [CrossRef]
- Clayton, C.; Shapira, M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007, 156, 93–101. [Google Scholar] [CrossRef]
- Fernandez-Moya, S.M.; Estevez, A.M. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip. Rev. RNA 2010, 1, 34–46. [Google Scholar] [CrossRef]
- Dallagiovanna, B.; Correa, A.; Probst, C.M.; Holetz, F.; Smircich, P.; de Aguiar, A.M.; Mansur, F.; da Silva, C.V.; Mortara, R.A.; Garat, B.; et al. Functional genomic characterization of mRNAs associated with TcPUF6, a pumilio-like protein from Trypanosoma cruzi. J. Biol. Chem. 2008, 283, 8266–8273. [Google Scholar] [CrossRef]
- Dupe, A.; Dumas, C.; Papadopoulou, B. An Alba-domain protein contributes to the stage-regulated stability of amastin transcripts in Leishmania. Mol. Microbiol. 2014, 91, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Mani, J.; Guttinger, A.; Schimanski, B.; Heller, M.; Acosta-Serrano, A.; Pescher, P.; Spath, G.; Roditi, I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS ONE 2011, 6, e22463. [Google Scholar] [CrossRef] [PubMed]
- Subota, I.; Rotureau, B.; Blisnick, T.; Ngwabyt, S.; Durand-Dubief, M.; Engstler, M.; Bastin, P. ALBA proteins are stage regulated during trypanosome development in the tsetse fly and participate in differentiation. Mol. Biol. Cell 2011, 22, 4205–4219. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, K.S.; Galucio, J.M.P.; Leonardo, E.S.; Cardoso, G.; Leal, E.; Conde, G.; Lameira, J. Structural and evolutionary analysis of Leishmania Alba proteins. Mol. Biochem. Parasitol. 2017, 217, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Dupe, A.; Dumas, C.; Papadopoulou, B. Differential Subcellular Localization of Leishmania Alba-Domain Proteins throughout the Parasite Development. PLoS ONE 2015, 10, e0137243. [Google Scholar] [CrossRef]
- Hendrickx, S.; Caljon, G.; Maes, L. Need for sustainable approaches in antileishmanial drug discovery. Parasitol. Res. 2019, 118, 2743–2752. [Google Scholar] [CrossRef]
- Rojas, R.; Valderrama, L.; Valderrama, M.; Varona, M.X.; Ouellette, M.; Saravia, N.G. Resistance to antimony and treatment failure in human Leishmania (Viannia) infection. J. Infect. Dis. 2006, 193, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, J.; Ramirez, L.; Adaui, V.; Zimic, M.; Tulliano, G.; Miranda-Verastegui, C.; Lazo, M.; Loayza-Muro, R.; De Doncker, S.; Maurer, A.; et al. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J. Infect. Dis. 2007, 195, 1846–1851. [Google Scholar] [CrossRef]
- Purkait, B.; Kumar, A.; Nandi, N.; Sardar, A.H.; Das, S.; Kumar, S.; Pandey, K.; Ravidas, V.; Kumar, M.; De, T.; et al. Mechanism of amphotericin B resistance in clinical isolates of Leishmania donovani. Antimicrob. Agents Chemother. 2012, 56, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Mishra, J.; Gupta, A.K.; Singh, A.; Shankar, P.; Singh, S. Laboratory confirmed miltefosine resistant cases of visceral leishmaniasis from India. Parasites Vectors 2017, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Jeddi, F.; Mary, C.; Aoun, K.; Harrat, Z.; Bouratbine, A.; Faraut, F.; Benikhlef, R.; Pomares, C.; Pratlong, F.; Marty, P.; et al. Heterogeneity of molecular resistance patterns in antimony-resistant field isolates of Leishmania species from the western Mediterranean area. Antimicrob. Agents Chemother. 2014, 58, 4866–4874. [Google Scholar] [CrossRef] [PubMed]
- Coelho, A.C.; Beverley, S.M.; Cotrim, P.C. Functional genetic identification of PRP1, an ABC transporter superfamily member conferring pentamidine resistance in Leishmania major. Mol. Biochem. Parasitol. 2003, 130, 83–90. [Google Scholar] [CrossRef]
- Coelho, A.C.; Messier, N.; Ouellette, M.; Cotrim, P.C. Role of the ABC transporter PRP1 (ABCC7) in pentamidine resistance in Leishmania amastigotes. Antimicrob. Agents Chemother. 2007, 51, 3030–3032. [Google Scholar] [CrossRef]
- Mondelaers, A.; Hendrickx, S.; Van Bockstal, L.; Maes, L.; Caljon, G. Miltefosine-resistant Leishmania infantum strains with an impaired MT/ROS3 transporter complex retain amphotericin B susceptibility. J. Antimicrob. Chemother. 2018, 73, 392–394. [Google Scholar] [CrossRef]
- De Moura, T.R.; Santos, M.L.; Braz, J.M.; Santos, L.F.; Aragao, M.T.; de Oliveira, F.A.; Santos, P.L.; da Silva, A.M.; de Jesus, A.R.; de Almeida, R.P. Cross-resistance of Leishmania infantum isolates to nitric oxide from patients refractory to antimony treatment, and greater tolerance to antileishmanial responses by macrophages. Parasitol. Res. 2016, 115, 713–721. [Google Scholar] [CrossRef]
- Perry, M.R.; Wyllie, S.; Raab, A.; Feldmann, J.; Fairlamb, A.H. Chronic exposure to arsenic in drinking water can lead to resistance to antimonial drugs in a mouse model of visceral leishmaniasis. Proc. Natl. Acad. Sci. USA 2013, 110, 19932–19937. [Google Scholar] [CrossRef]
- Hendrickx, S.; Mondelaers, A.; Eberhardt, E.; Delputte, P.; Cos, P.; Maes, L. In Vivo Selection of Paromomycin and Miltefosine Resistance in Leishmania donovani and L. infantum in a Syrian Hamster Model. Antimicrob. Agents Chemother. 2015, 59, 4714–4718. [Google Scholar] [CrossRef]
- Marquis, N.; Gourbal, B.; Rosen, B.P.; Mukhopadhyay, R.; Ouellette, M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol. Microbiol. 2005, 57, 1690–1699. [Google Scholar] [CrossRef]
- Perea, A.; Manzano, J.I.; Castanys, S.; Gamarro, F. The LABCG2 Transporter from the Protozoan Parasite Leishmania Is Involved in Antimony Resistance. Antimicrob. Agents Chemother. 2016, 60, 3489–3496. [Google Scholar] [CrossRef] [PubMed]
- Laffitte, M.N.; Leprohon, P.; Papadopoulou, B.; Ouellette, M. Plasticity of the Leishmania genome leading to gene copy number variations and drug resistance. F1000Research 2016, 5, 2350. [Google Scholar] [CrossRef] [PubMed]
- Patino, L.H.; Imamura, H.; Cruz-Saavedra, L.; Pavia, P.; Muskus, C.; Mendez, C.; Dujardin, J.C.; Ramirez, J.D. Major changes in chromosomal somy, gene expression and gene dosage driven by Sb(III) in Leishmania braziliensis and Leishmania panamensis. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Monte-Neto, R.; Laffitte, M.C.; Leprohon, P.; Reis, P.; Frezard, F.; Ouellette, M. Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony resistant Leishmania (Viannia) guyanensis. PLoS Negl. Trop. Dis. 2015, 9, e0003476. [Google Scholar] [CrossRef]
- Rastrojo, A.; Garcia-Hernandez, R.; Vargas, P.; Camacho, E.; Corvo, L.; Imamura, H.; Dujardin, J.C.; Castanys, S.; Aguado, B.; Gamarro, F.; et al. Genomic and transcriptomic alterations in Leishmania donovani lines experimentally resistant to antileishmanial drugs. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 246–264. [Google Scholar] [CrossRef]
- Patino, L.H.; Muskus, C.; Ramirez, J.D. Transcriptional responses of Leishmania (Leishmania) amazonensis in the presence of trivalent sodium stibogluconate. Parasites Vectors 2019, 12, 348. [Google Scholar] [CrossRef]
- Brotherton, M.C.; Bourassa, S.; Leprohon, P.; Legare, D.; Poirier, G.G.; Droit, A.; Ouellette, M. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS ONE 2013, 8, e81899. [Google Scholar] [CrossRef]
- Das, S.; Shah, P.; Baharia, R.K.; Tandon, R.; Khare, P.; Sundar, S.; Sahasrabuddhe, A.A.; Siddiqi, M.I.; Dube, A. Over-expression of 60s ribosomal L23a is associated with cellular proliferation in SAG resistant clinical isolates of Leishmania donovani. PLoS Negl. Trop. Dis. 2013, 7, e2527. [Google Scholar] [CrossRef]
- Rugani, J.N.; Gontijo, C.M.F.; Frézard, F.; Soares, R.P.; Monte-Neto, R.L.d. Antimony resistance in Leishmania (Viannia) braziliensis clinical isolates from atypical lesions associates with increased ARM56/ARM58 transcripts and reduced drug uptake. Mem. Inst. Oswaldo Cruz. 2019, 114, e190111. [Google Scholar] [CrossRef]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef]
- Gong, C.; Tsoi, H.; Mok, K.C.; Cheung, J.; Man, E.P.S.; Fujino, K.; Wong, A.; Lam, E.W.F.; Khoo, U.S. Phosphorylation independent eIF4E translational reprogramming of selective mRNAs determines tamoxifen resistance in breast cancer. Oncogene 2020. [Google Scholar] [CrossRef] [PubMed]
- Falletta, P.; Sanchez-Del-Campo, L.; Chauhan, J.; Effern, M.; Kenyon, A.; Kershaw, C.J.; Siddaway, R.; Lisle, R.; Freter, R.; Daniels, M.J.; et al. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 2017, 31, 18–33. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Ruggero, D. A wobbly road to drug resistance in melanoma: tRNA-modifying enzymes in translation reprogramming. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- El-Naggar, A.M.; Sorensen, P.H. Translational control of aberrant stress responses as a hallmark of cancer. J. Pathol. 2018, 244, 650–666. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karamysheva, Z.N.; Gutierrez Guarnizo, S.A.; Karamyshev, A.L. Regulation of Translation in the Protozoan Parasite Leishmania. Int. J. Mol. Sci. 2020, 21, 2981. https://doi.org/10.3390/ijms21082981
Karamysheva ZN, Gutierrez Guarnizo SA, Karamyshev AL. Regulation of Translation in the Protozoan Parasite Leishmania. International Journal of Molecular Sciences. 2020; 21(8):2981. https://doi.org/10.3390/ijms21082981
Chicago/Turabian StyleKaramysheva, Zemfira N., Sneider Alexander Gutierrez Guarnizo, and Andrey L. Karamyshev. 2020. "Regulation of Translation in the Protozoan Parasite Leishmania" International Journal of Molecular Sciences 21, no. 8: 2981. https://doi.org/10.3390/ijms21082981
APA StyleKaramysheva, Z. N., Gutierrez Guarnizo, S. A., & Karamyshev, A. L. (2020). Regulation of Translation in the Protozoan Parasite Leishmania. International Journal of Molecular Sciences, 21(8), 2981. https://doi.org/10.3390/ijms21082981




