3-Hydroxybutyrate Is Active Compound in Flax that Upregulates Genes Involved in DNA Methylation
Abstract
1. Introduction
2. Results
2.1. Transgenic Flax Plant Selection and Analysis
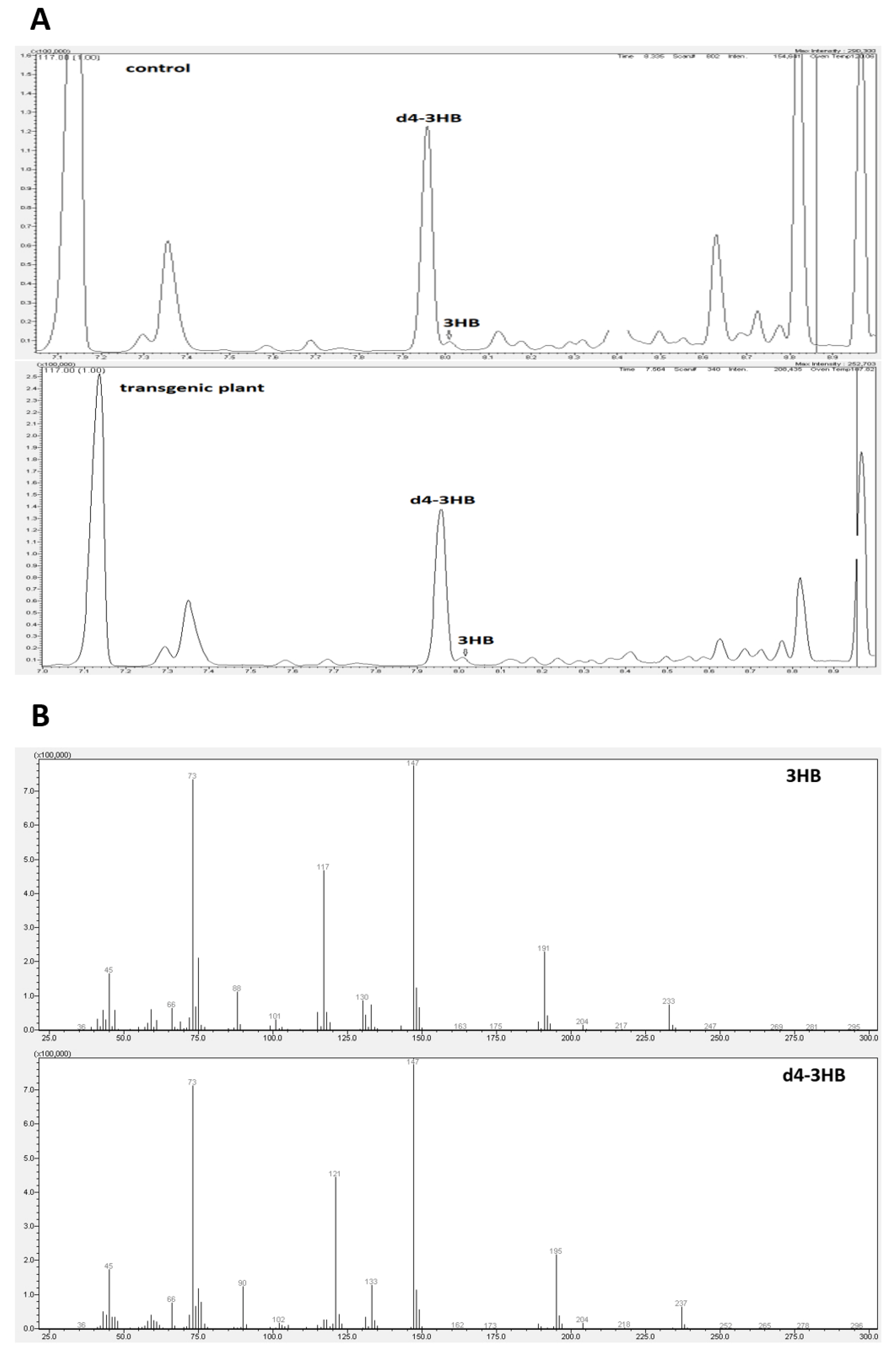
2.2. Determination of 3-Hydroxybutyrate Content in Flax
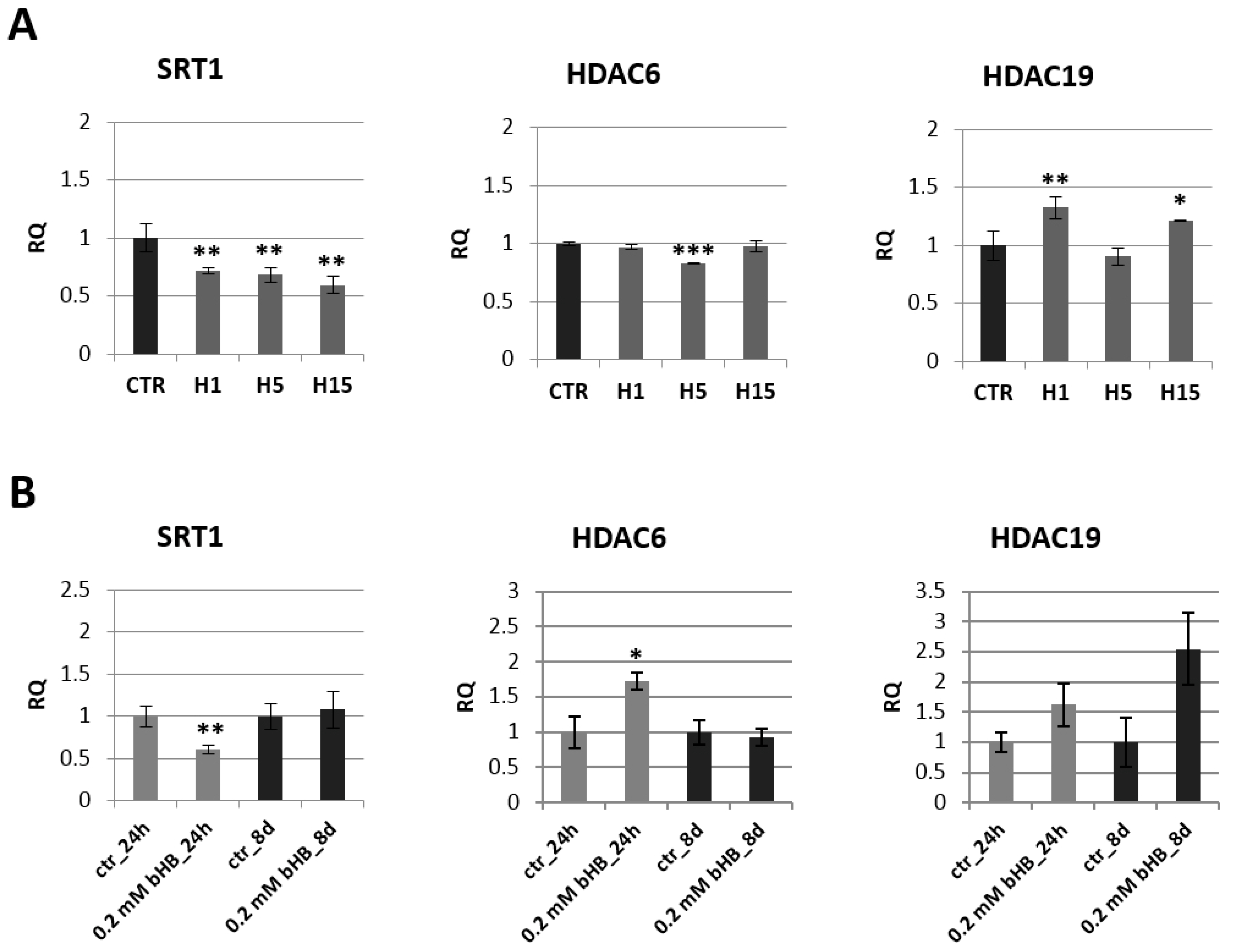
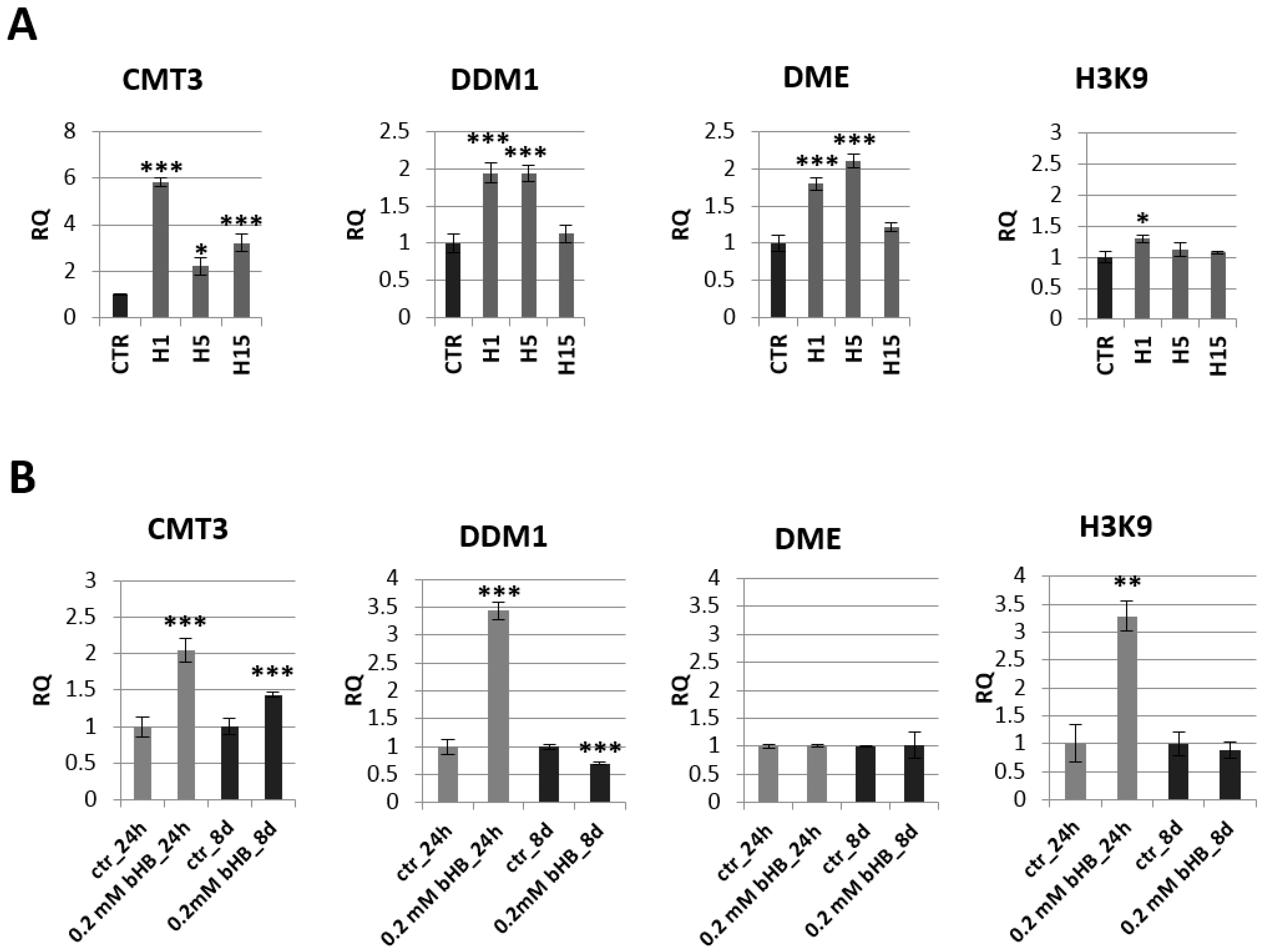
2.3. Analysis of mRNA Levels of Genes Involved in Epigenetic Modifications
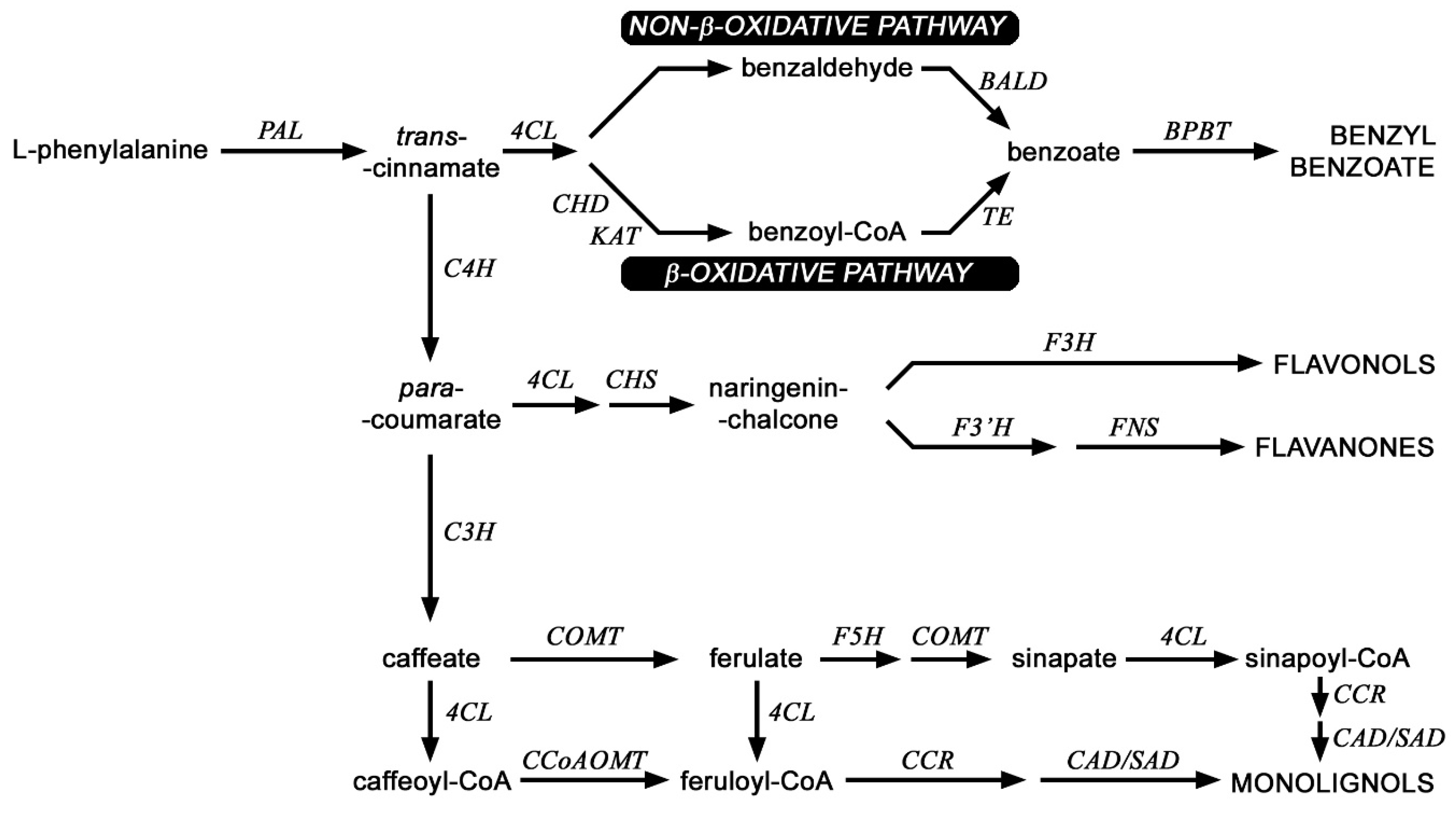
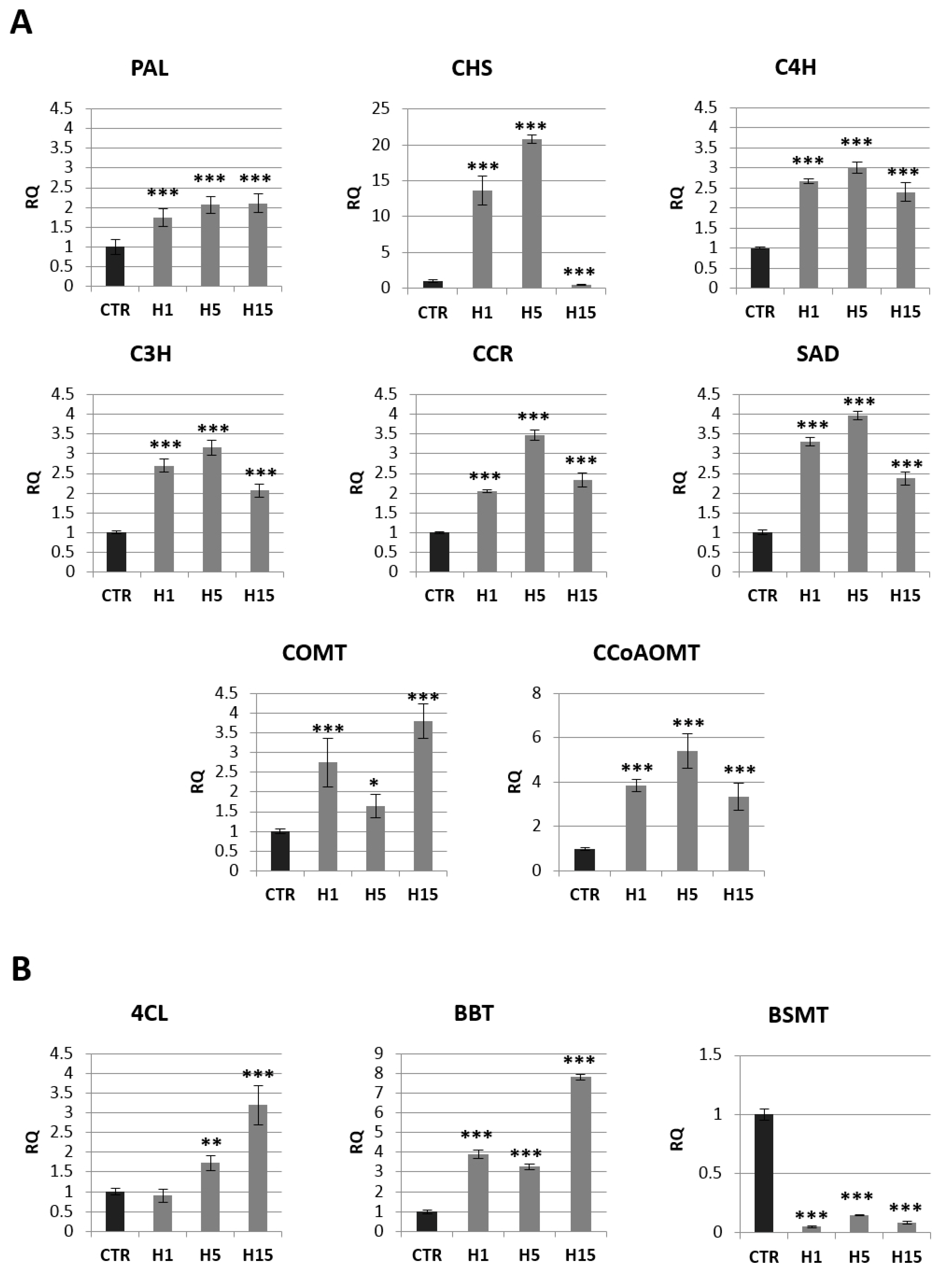
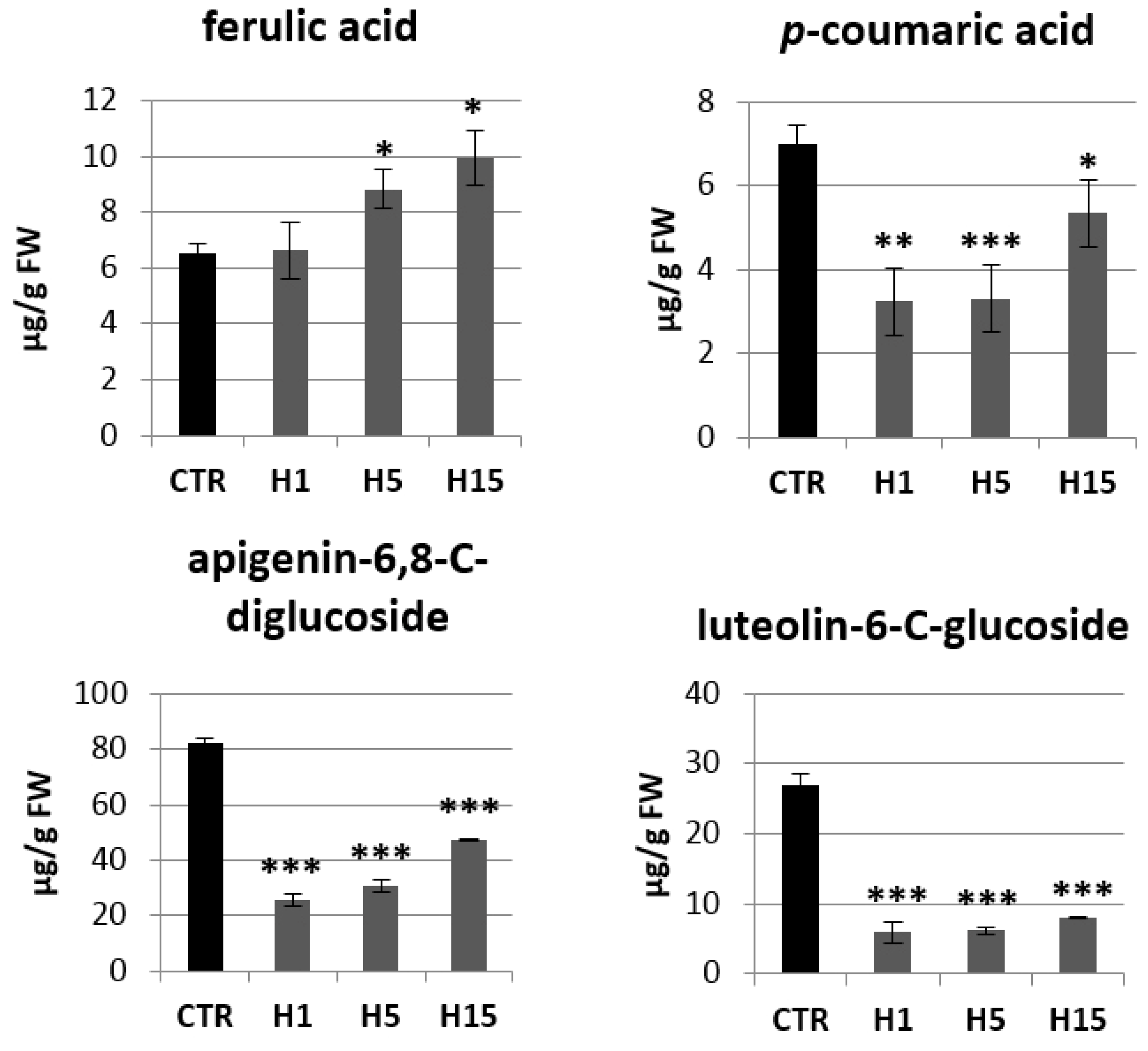
2.4. Analysis of mRNA Levels of Genes Involved in the Phenylpropanoid Pathway and the Content of Selected Metabolites of This Pathway
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Transgenic Plant Construction and Selection
4.3. Treatment of Plants with the 3-Hydroxybutyrate Standard
4.4. Treatment of Human Fibroblasts with the 3-Hydroxybutyrate Standard
4.5. Determination of 3-Hydroxybutyrate Content
4.6. Determination of mRNA Level
4.7. Determination of the Content of Phenylpropanoid Compounds
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar] [CrossRef] [PubMed]
- Gillmaier, N.; Schunder, E.; Kutzner, E.; Tlapak, H.; Rydzewski, K.; Herrmann, V.; Stammler, M.; Lasch, P.; Eisenreich, W.; Heuner, K. Growth-related Metabolism of the Carbon Storage Poly-3-hydroxybutyrate in Legionella pneumophila. J. Biol. Chem. 2016, 291, 6471–6482. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Sedlacek, P.; Koller, M.; Kucera, D.; Pernicova, I. Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: Biotechnological consequences and applications. Biotechnol. Adv. 2018, 36, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Uchino, K.; Saito, T.; Jendrossek, D. Poly(3-hydroxybutyrate) (PHB) depolymerase PhaZa1 is involved in mobilization of accumulated PHB in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2008, 74, 1058–1063. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jendrossek, D.; Handrick, R. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 2002, 56, 403–432. [Google Scholar] [CrossRef] [PubMed]
- Uchino, K.; Saito, T.; Gebauer, B.; Jendrossek, D. Isolated poly(3-hydroxybutyrate) (PHB) granules are complex bacterial organelles catalyzing formation of PHB from acetyl coenzyme A (CoA) and degradation of PHB to acetyl-CoA. J. Bacteriol. 2007, 189, 8250–8256. [Google Scholar] [CrossRef]
- Eggers, J.; Steinbuchel, A. Poly(3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl coenzyme A (CoA) via crotonyl-CoA. J. Bacteriol. 2013, 195, 3213–3223. [Google Scholar] [CrossRef]
- Emeruwa, A.C.; Hawirko, R.Z. Poly-beta-hydroxybutyrate metabolism during growth and sporulation of Clostridium botulinum. J. Bacteriol. 1973, 116, 989–993. [Google Scholar] [CrossRef]
- Maalouf, M.; Sullivan, P.G.; Davis, L.; Kim, D.Y.; Rho, J.M. Ketones inhibit mitochondrial production of reactive oxygen species production following glutamate excitotoxicity by increasing NADH oxidation. Neuroscience 2007, 145, 256–264. [Google Scholar] [CrossRef]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef]
- Sleiman, S.F.; Henry, J.; Al-Haddad, R.; El Hayek, L.; Abou Haidar, E.; Stringer, T.; Ulja, D.; Karuppagounder, S.S.; Holson, E.B.; Ratan, R.R.; et al. Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body beta-hydroxybutyrate. eLife 2016, 5. [Google Scholar] [CrossRef]
- Edwards, C.; Canfield, J.; Copes, N.; Rehan, M.; Lipps, D.; Bradshaw, P.C. D-beta-hydroxybutyrate extends lifespan in C. elegans. Aging 2014, 6, 621–644. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Han, P.; Tang, Z.; Liu, Q.; Shi, J. Sirtuin 3 mediates neuroprotection of ketones against ischemic stroke. J. Cereb. Blood Flow Metab. 2015, 35, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Marosi, K.; Kim, S.W.; Moehl, K.; Scheibye-Knudsen, M.; Cheng, A.; Cutler, R.; Camandola, S.; Mattson, M.P. 3-Hydroxybutyrate regulates energy metabolism and induces BDNF expression in cerebral cortical neurons. J. Neurochem. 2016, 139, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Svart, M.; Gormsen, L.C.; Hansen, J.; Zeidler, D.; Gejl, M.; Vang, K.; Aanerud, J.; Moeller, N. Regional cerebral effects of ketone body infusion with 3-hydroxybutyrate in humans: Reduced glucose uptake, unchanged oxygen consumption and increased blood flow by positron emission tomography. A randomized, controlled trial. PLoS ONE 2018, 13, e0190556. [Google Scholar] [CrossRef] [PubMed]
- Snell, K.; Peoples, O. PHA bioplastic: A value-added coproduct for biomass biorefineries. Biofuels Bioprod. Biorefining 2009, 3, 456–467. [Google Scholar] [CrossRef]
- Peterson, A.; Fischer, C. Conversion of Natural Products Including Cellulose to Hydrocarbons, Hydrogen and/or Other Related Compounds. U.S. Patent Application No. 2010/0228067, 1 October 2013. [Google Scholar]
- Coons, R. Industrial biotechnology. Chem. Week 2010, 172, 22–26. [Google Scholar]
- Wrobel, M.; Zebrowski, J.; Szopa, J. Polyhydroxybutyrate synthesis in transgenic flax. J. Biotechnol. 2004, 107, 41–54. [Google Scholar] [CrossRef]
- Tsuda, H.; Shiraki, M.; Inoue, E.; Saito, T. Generation of poly-beta-hydroxybutyrate from acetate in higher plants: Detection of acetoacetyl CoA reductase- and PHB synthase-activities in rice. J. Plant Physiol. 2016, 201, 9–16. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Suzuki, F.; Emerich, D.W. Enzymes of the Poly-beta-Hydroxybutyrate and Citric Acid Cycles of Rhizobium japonicum Bacteroids. Plant Physiol. 1984, 75, 1158–1162. [Google Scholar] [CrossRef]
- Wang, C.; Saldanha, M.; Sheng, X.; Shelswell, K.J.; Walsh, K.T.; Sobral, B.W.; Charles, T.C. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 2007, 153, 388–398. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Silveira Alves, L.P.; Plucani do Amaral, F.; Kim, D.; Todo Bom, M.; Pinero Gavidia, M.; Silvano Teixeira, C.; Holthman, F.; de Oliveira Pedrosa, F.; Maltempi de Souza, E.; Chubatsu, L.S.; et al. Importance of Poly-3-Hydroxybutyrate Metabolism to the Ability of Herbaspirillum seropedicae To Promote Plant Growth. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Wrobel-Kwiatkowska, M.; Zuk, M.; Szopa, J.; Dyminska, L.; Maczka, M.; Hanuza, J. Poly-3-hydroxy butyric acid interaction with the transgenic flax fibers: FT-IR and Raman spectra of the composite extracted from a GM flax. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2009, 73, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Sun, X.; Tian, X.; Li, Z.; Zhang, Y. Synthesis of acrylamide molecularly imprinted polymers immobilized on graphite oxide through surface-initiated atom transfer radical polymerization. Anal. Lett. 2013, 46, 969–981. [Google Scholar] [CrossRef]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Sepúlveda, L.; González-Morales, S.; Benavides-Mendoza, A. Benzoic acid: Biosynthesis, modification and function in plants. Rev. Mex. Cienc. Agric. 2015, 67, 1667–1678. [Google Scholar]
- Ladurner, A.G. Rheostat control of gene expression by metabolites. Mol. Cell 2006, 24, 1–11. [Google Scholar] [CrossRef]
- Taylor-Teeples, M.; Lin, L.; de Lucas, M.; Turco, G.; Toal, T.W.; Gaudinier, A.; Young, N.F.; Trabucco, G.M.; Veling, M.T.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009, 324, 1076–1080. [Google Scholar] [CrossRef]
- Takahashi, H.; McCaffery, J.M.; Irizarry, R.A.; Boeke, J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell 2006, 23, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- El-Osta, A.; Wolffe, A.P. DNA methylation and histone deacetylation in the control of gene expression: Basic biochemistry to human development and disease. Gene Expr. 2000, 9, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Coffee, B.; Zhang, F.; Warren, S.T.; Reines, D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999, 22, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhong, X.; Bernatavichute, Y.V.; Stroud, H.; Feng, S.; Caro, E.; Vashisht, A.A.; Terragni, J.; Chin, H.G.; Tu, A.; et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012, 151, 167–180. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Blevins, T.; Pontes, O.; Pikaard, C.S.; Meins, F., Jr. Heterochromatic siRNAs and DDM1 independently silence aberrant 5S rDNA transcripts in Arabidopsis. PLoS ONE 2009, 4, e5932. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Heredia, F.; Sarazin, A.; Roudier, F.; Boccara, M.; Ciaudo, C.; Cruaud, C.; Poulain, J.; Berdasco, M.; Fraga, M.F.; et al. A role for RNAi in the selective correction of DNA methylation defects. Science 2009, 323, 1600–1604. [Google Scholar] [CrossRef]
- Park, J.S.; Frost, J.M.; Park, K.; Ohr, H.; Park, G.T.; Kim, S.; Eom, H.; Lee, I.; Brooks, J.S.; Fischer, R.L.; et al. Control of DEMETER DNA demethylase gene transcription in male and female gamete companion cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 2078–2083. [Google Scholar] [CrossRef]
- Jones, P.A. Genome wide analysis of DNA methylation and nucleosome positioning. Epigenet. Chromatin 2013, 6, O37. [Google Scholar] [CrossRef][Green Version]
- Dzialo, M.; Szopa, J.; Czuj, T.; Zuk, M. Oligodeoxynucleotides can transiently up- and downregulate CHS gene expression in flax by changing DNA methylation in a sequence-specific manner. Front. Plant. Sci. 2017, 8, 755. [Google Scholar] [CrossRef] [PubMed]
- Widhalm, J.R.; Dudareva, N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant. 2015, 8, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C.; Jarvis, A.P.; Xiang, L.; Moore, B.S.; Oldham, N.J. A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid beta-oxidation. Chembiochem A Eur. J. Chem. Biol. 2001, 2, 784–786. [Google Scholar] [CrossRef]
- Xiang, L.; Moore, B.S. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium “Streptomyces maritimus”. J. Bacteriol. 2003, 185, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Bewick, A.J.; Schmitz, R.J. Gene body DNA methylation in plants. Curr. Opin. Plant. Biol. 2017, 36, 103–110. [Google Scholar] [CrossRef]
- Zuk, M.; Dzialo, M.; Richter, D.; Dyminska, L.; Matula, J.; Kotecki, A.; Hanuza, J.; Szopa, J. Chalcone Synthase (CHS) gene suppression in flax leads to changes in wall synthesis and sensing genes, cell wall chemistry and stem morphology parameters. Front. Plant. Sci. 2016, 7, 894. [Google Scholar] [CrossRef]
- Mierziak, J.; Wojtasik, W.; Kostyn, K.; Czuj, T.; Szopa, J.; Kulma, A. Crossbreeding of transgenic flax plants overproducing flavonoids and glucosyltransferase results in progeny with improved antifungal and antioxidative properties. Mol. Breed. New Strateg. Plant Improv. 2014, 34, 1917–1932. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mierziak, J.; Wojtasik, W.; Kulma, A.; Dziadas, M.; Kostyn, K.; Dymińska, L.; Hanuza, J.; Żuk, M.; Szopa, J. 3-Hydroxybutyrate Is Active Compound in Flax that Upregulates Genes Involved in DNA Methylation. Int. J. Mol. Sci. 2020, 21, 2887. https://doi.org/10.3390/ijms21082887
Mierziak J, Wojtasik W, Kulma A, Dziadas M, Kostyn K, Dymińska L, Hanuza J, Żuk M, Szopa J. 3-Hydroxybutyrate Is Active Compound in Flax that Upregulates Genes Involved in DNA Methylation. International Journal of Molecular Sciences. 2020; 21(8):2887. https://doi.org/10.3390/ijms21082887
Chicago/Turabian StyleMierziak, Justyna, Wioleta Wojtasik, Anna Kulma, Mariusz Dziadas, Kamil Kostyn, Lucyna Dymińska, Jerzy Hanuza, Magdalena Żuk, and Jan Szopa. 2020. "3-Hydroxybutyrate Is Active Compound in Flax that Upregulates Genes Involved in DNA Methylation" International Journal of Molecular Sciences 21, no. 8: 2887. https://doi.org/10.3390/ijms21082887
APA StyleMierziak, J., Wojtasik, W., Kulma, A., Dziadas, M., Kostyn, K., Dymińska, L., Hanuza, J., Żuk, M., & Szopa, J. (2020). 3-Hydroxybutyrate Is Active Compound in Flax that Upregulates Genes Involved in DNA Methylation. International Journal of Molecular Sciences, 21(8), 2887. https://doi.org/10.3390/ijms21082887





