Potential Implications of Interactions between Fe and S on Cereal Fe Biofortification
Abstract
1. Introduction
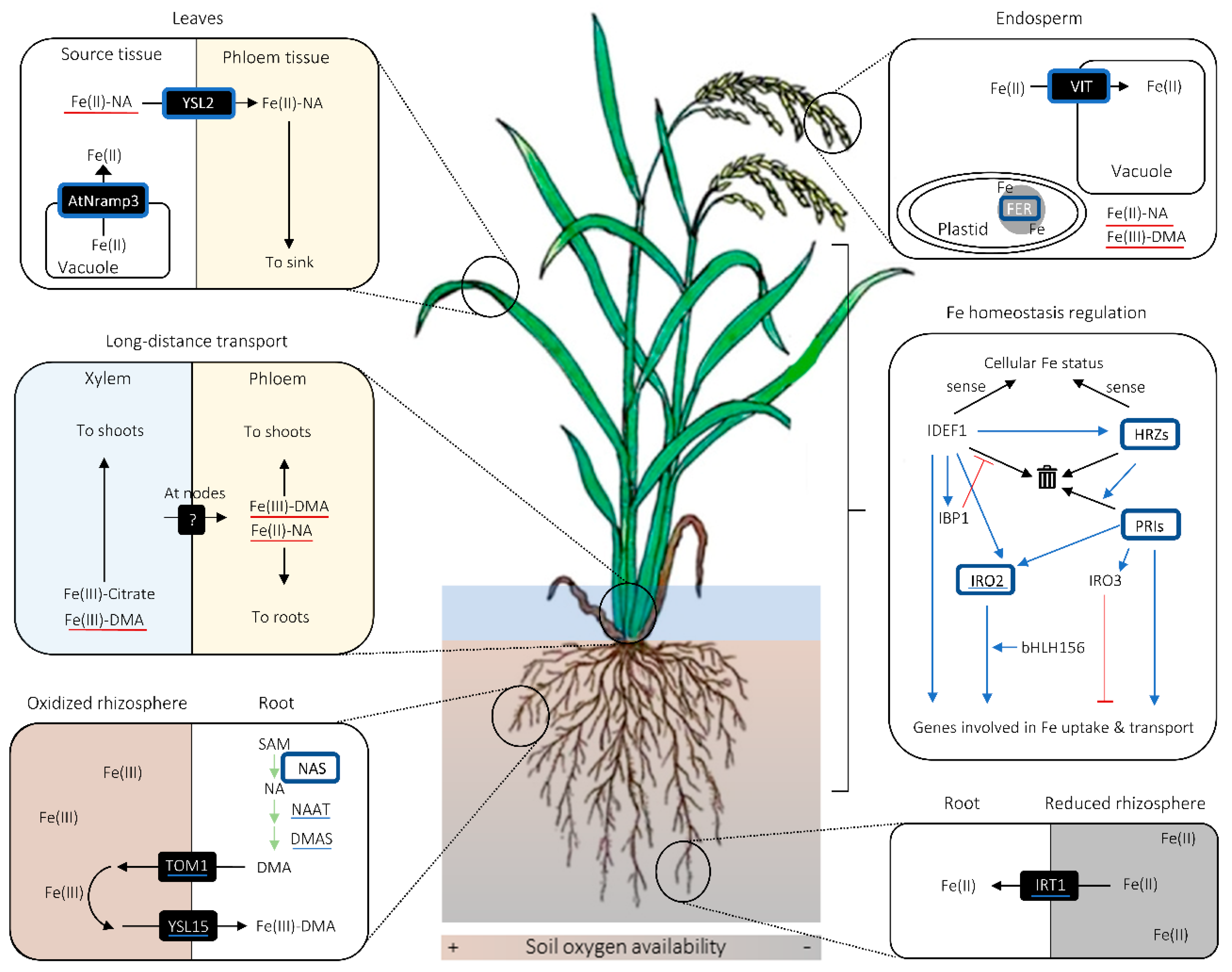
2. Fe Uptake and Transport in Cereals
2.1. Fe Uptake System and Phytosiderophore Synthesis
2.2. Potential Diversity in the Manner of Fe Uptake and Radial Fe Transport in Roots
2.3. Fe Mobilization for Long-Distance and Intercellular Transport
2.4. Fe Intracellular Homeostasis
2.5. Fe Homeostasis Regulation
3. Biotechnological Strategies for Cereal Fe Biofortification
3.1. Target Fe Increase in Biofortification
3.2. Fe Biofortification Strategies Involving NAS Overexpression
3.3. Fe Biofortification Strategies Involving Endosperm-Specific FER or VIT Expression
3.4. Combinatory Strategies Lead to Further Increase in Grain Fe Concentration
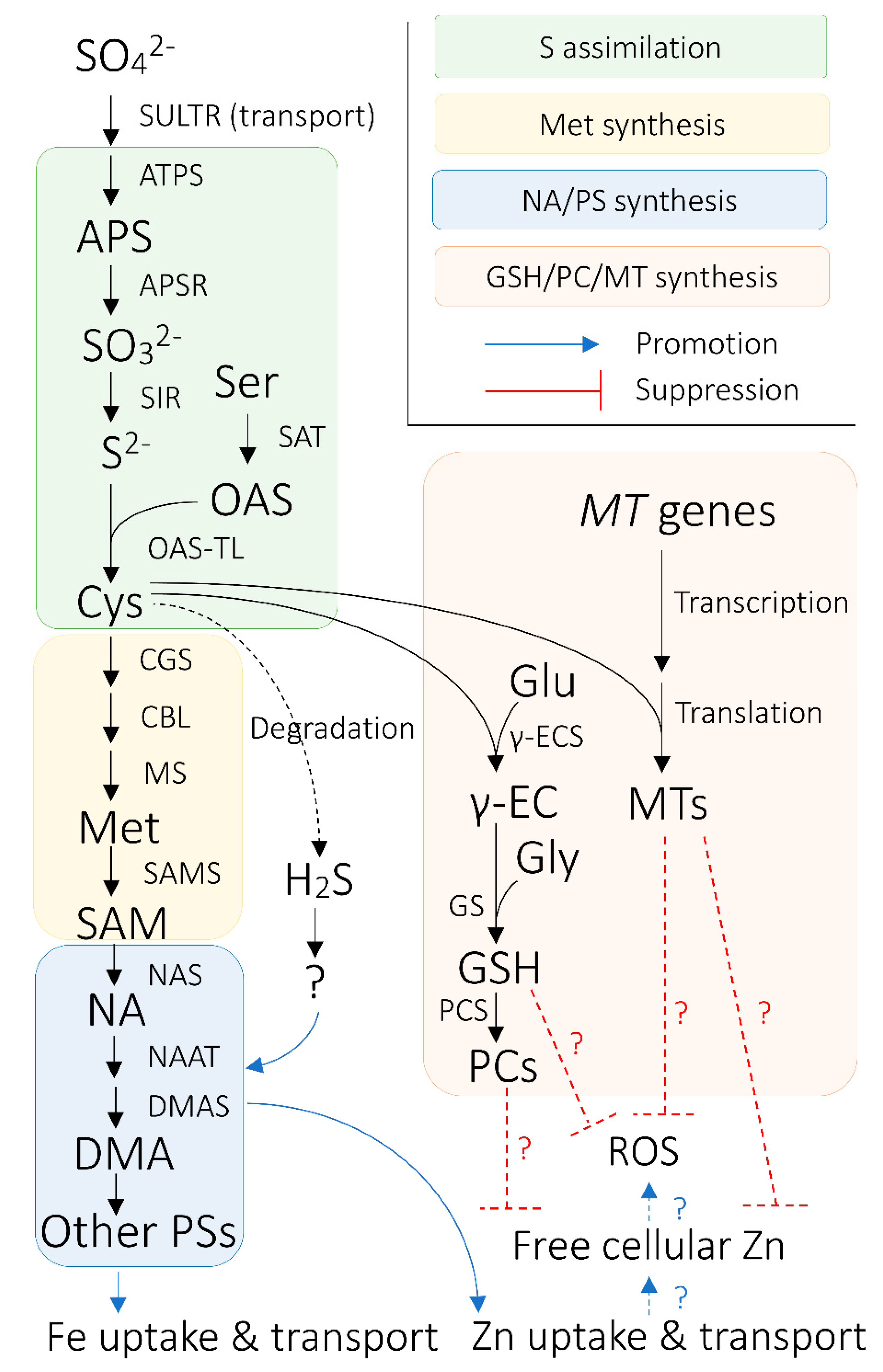
4. Uptake, Transport, and Metabolism of S and Its Biological Functions Related to Metal Homeostasis in Plants
4.1. Sulfate Uptake
4.2. Sulfate Long-Distance Transport Via Xylem
4.3. S Assimilation and Synthesis of S-Containing Biomolecules Related to Metal Homeostasis
4.4. S Intracellular Homeostasis and Long-Distance Transport Via Phloem
5. Aspects of Fe and S Interaction with Particular Implications for Cereal Fe Biofortification
5.1. Effect of Crop S Status on Fe Acquisition and Translocation
5.2. H2S as a Potential Regulator of Crop Fe Homeostasis
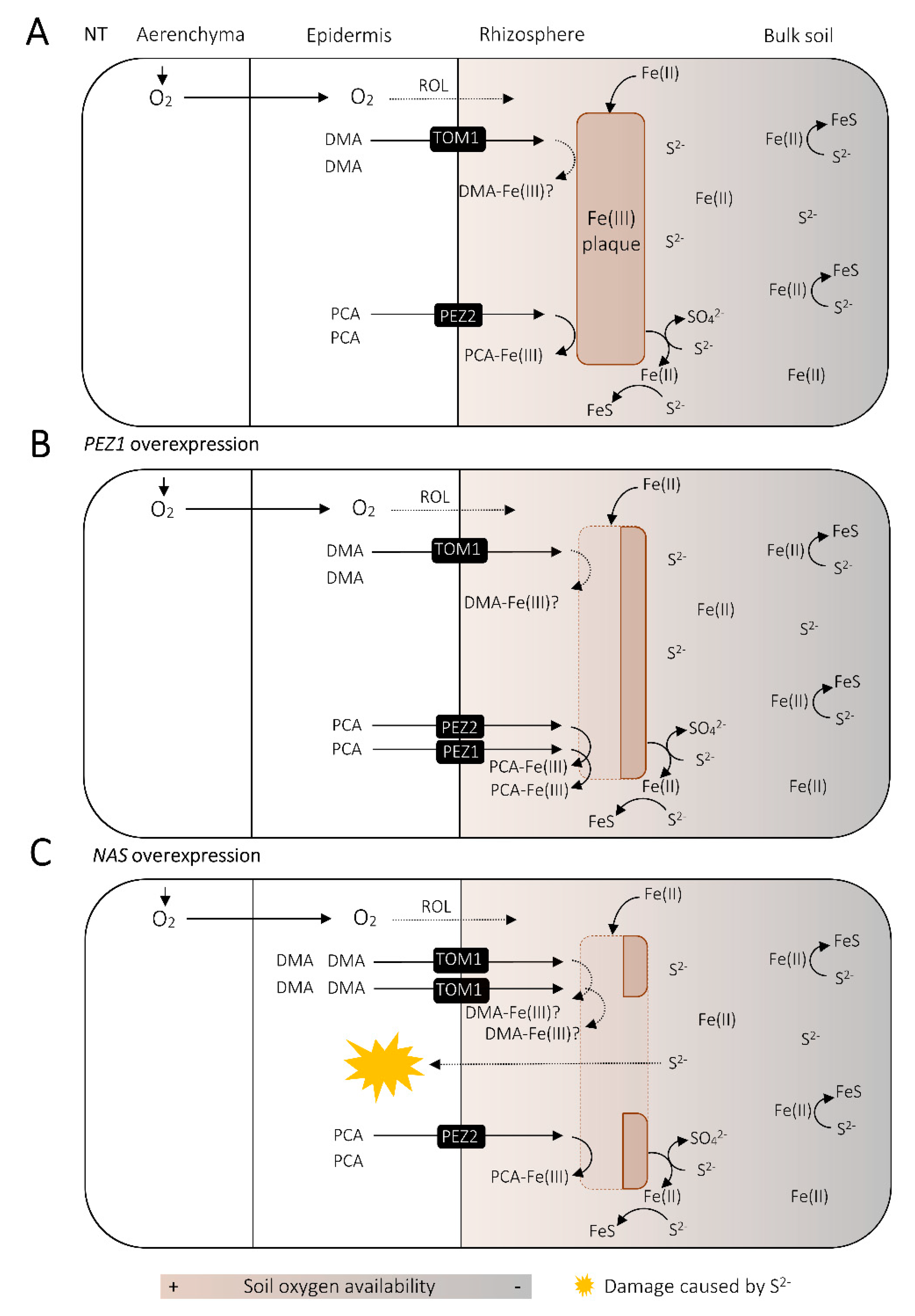
5.3. Fe as a Buffer for Sulfide Damage in the Rice Rhizosphere
5.4. Roles of S-Containing Biomolecules in Response to Concomitant Increase in Heavy Metal Uptake by Fe-Biofortified Crops
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McLean, E.; Cogswell, M.; Egli, I.; Wojdyla, D.; De Benoist, B. Worldwide prevalence of anaemia, WHO Vitamin and Mineral Nutrition Information System, 1993–2005. Public Health Nutr. 2009, 12, 444–454. [Google Scholar] [CrossRef]
- WHO. The Global Prevalence of Anemia in 2011; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Graham, R.D.; Welch, R.M.; Bouis, H.E. Addressing micronutrient malnutrition through enhancing the nutritional quality of staple foods: Principles, perspectives and knowledge gaps. Adv. Agron. 2001, 70, 77–142. [Google Scholar]
- Bouis, H.E.; Welch, R.M. Biofortification—A sustainable agricultural strategy for reducing micronutrient malnutrition in the Global South. Crop Sci. 2010, 50 (Suppl. 1), S20–S32. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J. Iron biofortification of staple crops: Lessons and challenges in plant genetics. Plant Cell Physiol. 2019, 60, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Aung, M.S.; Nishizawa, N.K. Iron biofortification of rice using different transgenic approaches. Rice 2013, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Slamet-Loedin, I.H.; Johnson-Beebout, S.E.; Impa, S.; Tsakirpaloglou, N. Enriching rice with Zn and Fe while minimizing Cd risk. Front. Plant Sci. 2015, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, Y.; Slamet-Loedin, I.H. Genetic biofortification to enrich rice and wheat grain iron: From genes to product. Front. Plant Sci. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Kawakami, Y.; Bhullar, N.K. Molecular processes in iron and zinc homeostasis and their modulation for biofortification in rice. J. Integr. Plant Biol. 2018, 60, 1181–1198. [Google Scholar] [CrossRef]
- Moreno-Moyano, L.T.; Bonneau, J.P.; Sánchez-Palacios, J.T.; Tohme, J.; Johnson, A.A.T. Association of increased grain iron and zinc concentrations with agro-morphological traits of biofortified rice. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Wang, M.; Gruissem, W.; Bhullar, N.K. Nicotianamine synthase overexpression positively modulates iron homeostasis-related genes in high iron rice. Front. Plant Sci. 2013, 4, 1–15. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Nozoye, T.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Exploiting new tools for iron bio-fortification of rice. Biotechnol. Adv. 2013, 31, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Connorton, J.M.; Balk, J.; Rodríguez-Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Römheld, V.; Marschner, H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 1986, 80, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Takagi, S. Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Sci. Plant Nutr. 1976, 22, 423–433. [Google Scholar] [CrossRef]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Takahashi, M.; Sato, Y.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N.K. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011, 286, 5446–5454. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Ma, J.F.; Yamaji, N.; Ueno, D.; Nomoto, K.; Iwashita, T. A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 2006, 46, 563–572. [Google Scholar] [CrossRef]
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.F.; Walker, E.L. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 2001, 409, 346–349. [Google Scholar] [CrossRef]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef]
- Lee, S.; Chiecko, J.C.; Kim, S.A.; Walker, E.L.; Lee, Y.; Guerinot, M.L.; An, G. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009, 150, 786–800. [Google Scholar] [CrossRef]
- Mori, S.; Nishizawa, N. Methionine as a dominant precursor of phytosiderophores in Graminaceae plants. Plant Cell Physiol. 1987, 28, 1081–1092. [Google Scholar]
- Shojima, S.; Nishizawa, N.-K.; Fushiya, S.; Nozoe, S.; Irifune, T.; Mori, S. Biosynthesis of phytosiderophores. Plant Physiol. 1990, 93, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Shinada, T.; Matsuda, C.; Nomoto, K. Biosynthesis of phytosiderophores, mugineic acids, associated with methionine cycling. J. Biol. Chem. 1995, 270, 16549–16554. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, N.; Mori, S. The particular vesicle appearing in barley root cells and its relation to mugineic acid secretion. J. Plant Nutr. 1987, 10, 1013–1020. [Google Scholar] [CrossRef]
- Negishi, T.; Nakanishi, H.; Yazaki, J.; Kishimoto, N.; Fujii, F.; Shimbo, K.; Yamamoto, K.; Sakata, K.; Sasaki, T.; Kikuchi, S.; et al. cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J. 2002, 30, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, D.; Higuchi, K.; Sakamoto, T.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol. 2003, 132, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Nozoye, T.; Nagasaka, S.; Bashir, K.; Takahashi, M.; Kobayashi, T.; Nakanishi, H.; Nishizawa, N.K. Nicotianamine synthase 2 localizes to the vesicles of iron-deficient rice roots, and its mutation in the YXXφ or LL motif causes the disruption of vesicle formation or movement in rice. Plant J. 2014, 77, 246–260. [Google Scholar] [CrossRef]
- Nozoye, T.; Tsunoda, K.; Nagasaka, S.; Bashir, K.; Takahashi, M.; Kobayashi, T.; Nakanishi, H.; Nishizawa, N.K. Rice nicotianamine synthase localizes to particular vesicles for proper function. Plant Signal. Behav. 2014, 9, e28660. [Google Scholar] [CrossRef]
- Zheng, L.; Fujii, M.; Yamaji, N.; Sasaki, A.; Yamane, M.; Sakurai, I.; Sato, K.; Ma, J.F. Isolation and characterization of a barley yellow stripe-like gene, HvYSL5. Plant Cell Physiol. 2011, 52, 765–774. [Google Scholar] [CrossRef]
- Higuchi, K.; Suzuki, K.; Nakanishi, H.; Yamaguchi, H.; Nishizawa, N.-K.; Mori, S. Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 1999, 119, 471–480. [Google Scholar] [CrossRef]
- Takahashi, M.; Yamaguchi, H.; Nakanishi, H.; Shioiri, T.; Nishizawa, N.K.; Mori, S. Cloning two genes for nicotianamine aminotransferase, a critical enzyme in iron acquisition (Strategy II) in graminaceous plants. Plant Physiol. 1999, 121, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Inoue, H.; Nagasaka, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 2006, 281, 32395–32402. [Google Scholar] [CrossRef] [PubMed]
- Kawai, S.; Takagi, S.I.; Sato, Y. Mugineic acid-family phytosiderophores in root-secretions of barley, corn and sorghum varieties. J. Plant Nutr. 1988, 11, 633–642. [Google Scholar] [CrossRef]
- Ma, J.F.; Taketa, S.; Chang, Y.-C.; Takeda, K.; Matsumoto, H. Biosynthesis of phytosiderophores in several Triticeae species with different genomes. J. Exp. Bot. 1999, 50, 723–726. [Google Scholar] [CrossRef]
- Nakanishi, H.; Yamaguchi, H.; Sasakuma, T.; Nishizawa, N.K.; Mori, S. Two dioxygenase genes, Ids3 and Ids2, from Hordeum vulgare are involved in the biosynthesis of mugineic acid family phytosiderophores. Plant Mol. Biol. 2000, 44, 199–207. [Google Scholar] [CrossRef] [PubMed]
- von Wirén, N.; Khodr, H.; Hider, R.C. Hydroxylated phytosiderophore species possess an enhanced chelate stability and affinity for Iron(III). Plant Physiol. 2002, 124, 1149–1158. [Google Scholar] [CrossRef][Green Version]
- Ueno, D.; Rombolà, A.D.; Iwashita, T.; Nomoto, K.; Ma, J.F. Identification of two novel phytosiderophores secreted by perennial grasses. New Phytol. 2007, 174, 304–310. [Google Scholar] [CrossRef]
- Nozoye, T.; Aung, M.S.; Masuda, H.; Nakanishi, H.; Nishizawa, N.K. Bioenergy grass [Erianthus ravennae (L.) Beauv.] secretes two members of mugineic acid family phytosiderophores which involved in their tolerance to Fe deficiency. Soil Sci. Plant Nutr. 2017, 63, 543–552. [Google Scholar] [CrossRef]
- Takagi, S.-I. Production of phytosiderophores. In Iron Chelation in Plants and Soil Microorganisms; Barton, L., Hemming, B.C., Eds.; Academic Press: San Diego, CA, USA, 1993; pp. 111–131. [Google Scholar]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M.; et al. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Wairich, A.; de Oliveira, B.H.N.; Arend, E.B.; Duarte, G.L.; Ponte, L.R.; Sperotto, R.A.; Ricachenevsky, F.K.; Fett, J.P. The Combined Strategy for iron uptake is not exclusive to domesticated rice (Oryza sativa). Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Wang, P.; Yamaji, N.; Inoue, K.; Mochida, K.; Ma, J.F. Plastic transport systems of rice for mineral elements in response to diverse soil environmental changes. New Phytol. 2020, 226, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Zaharieva, T.; Römheld, V. Specific Fe2+ uptake system in strategy I plants inducible under Fe deficiency. J. Plant Nutr. 2000, 23, 1733–1744. [Google Scholar] [CrossRef]
- Bughio, N.; Yamaguchi, H.; Nishizawa, N.K.; Nakanishi, H.; Mori, S. Cloning an iron-regulated metal transporter from rice. J. Exp. Bot. 2002, 53, 1677–1682. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gao, T.; Liu, Y.; Liu, J.; Li, F.; Chen, Z.; Li, Y.; Lv, Y.; Song, Z.; Reinfelder, J.R.; et al. Isotopic fingerprints indicate distinct strategies of Fe uptake in rice. Chem. Geol. 2019, 524, 323–328. [Google Scholar] [CrossRef]
- Enstone, D.E.; Peterson, C.A.; Ma, F. Root endodermis and exodermis: Structure, function, and responses to the environment. J. Plant Growth Regul. 2002, 21, 335–351. [Google Scholar] [CrossRef]
- Perumalla, C.J.; Peterson, C.A.; Enstone, D.E. A survey of angiosperm species to detect hypodermal Casparian bands. I. Roots with a uniseriate hypodermis and epidermis. Bot. J. Linn. Soc. 1990, 103, 93–112. [Google Scholar] [CrossRef]
- Peterson, C.A.; Emanuel, M.E.; Wilson, C. Identification of a Casparian band in the hypodermis of onion and corn roots. Can. J. Bot. 1982, 60, 1529–1535. [Google Scholar] [CrossRef]
- Clark, L.H.; Harris, W.H. Observations on the root anatomy of rice (Oryza sativa L.). Am. J. Bot. 1981, 68, 154–161. [Google Scholar] [CrossRef]
- Tylová, E.; Pecková, E.; Blascheová, Z.; Soukup, A. Casparian bands and suberin lamellae in exodermis of lateral roots: An important trait of roots system response to abiotic stress factors. Ann. Bot. 2017, 120, 71–85. [Google Scholar] [CrossRef]
- Kreszies, T.; Eggels, S.; Kreszies, V.; Osthoff, A.; Shellakkutti, N.; Baldauf, J.A.; Zeisler-Diehl, V.V.; Hochholdinger, F.; Ranathunge, K.; Schreiber, L. Seminal roots of wild and cultivated barley differentially respond to osmotic stress in gene expression, suberization, and hydraulic conductivity. Plant Cell Environ. 2020, 43, 344–357. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Ma, J.F. Further characterization of ferric—Phytosiderophore transporters ZmYS1 and HvYS1 in maize and barley. J. Exp. Bot. 2009, 60, 3513–3520. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Murata, J.; Murata, Y. A novel barley yellow stripe 1-like transporter (HvYSL2) localized to the root endodermis transports metal-phytosiderophore complexes. Plant Cell Physiol. 2011, 52, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.B.; Armstrong, W. Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1999, 1, 274–287. [Google Scholar] [CrossRef]
- Kawai, M.; Samarajeewa, P.K.; Barrero, R.A.; Nishiguchi, M.; Uchimiya, H. Cellular dissection of the degradation pattern of cortical cell death during aerenchyma formation of rice roots. Planta 1998, 204, 277–287. [Google Scholar] [CrossRef]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Transporters involved in mineral nutrient uptake in rice. J. Exp. Bot. 2016, 67, 3645–3653. [Google Scholar] [CrossRef]
- Che, J.; Yamaji, N.; Ma, J.F. Efficient and flexible uptake system for mineral elements in plants. New Phytol. 2018, 219, 513–517. [Google Scholar] [CrossRef]
- Ogo, Y.; Kakei, Y.; Itai, R.N.; Kobayashi, T.; Nakanishi, H.; Takahashi, H.; Nakazono, M.; Nishizawa, N.K. Spatial transcriptomes of iron-deficient and cadmium-stressed rice. New Phytol. 2014, 201, 781–794. [Google Scholar] [CrossRef]
- Hell, R.; Stephan, U.W. Iron uptake, trafficking and homeostasis in plants. Planta 2003, 216, 541–551. [Google Scholar] [CrossRef]
- von Wirén, N.; Klair, S.; Bansal, S.; Briat, J.-F.; Khodr, H.; Shioiri, T.; Leigh, R.A.; Hider, R.C. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in Plants. Plant Physiol. 1999, 119, 1107–1114. [Google Scholar] [CrossRef]
- Ariga, T.; Hazama, K.; Yanagisawa, S.; Yoneyama, T. Chemical forms of iron in xylem sap from graminaceous and non-graminaceous plants. Soil Sci. Plant Nutr. 2014, 60, 460–469. [Google Scholar] [CrossRef]
- Kakei, Y.; Yamaguchi, I.; Kobayashi, T.; Takahashi, M.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. A highly sensitive, quick and simple quantification method for nicotianamine and 2′-deoxymugineic acid from minimum samples using LC/ESI-TOF-MS achieves functional analysis of these components in plants. Plant Cell Physiol. 2009, 50, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Rellán-Álvarez, R.; Abadía, J.; Álvarez-Fernández, A. Formation of metal-nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange. A study by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2008, 22, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Ueno, D.; Mitani, N.; Ma, J.F. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009, 149, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, R.; Kato, M.; Nagata, S.; Yanagisawa, S.; Yoneyama, T. Identification of Zn-nicotianamine and Fe-2’-deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.). Plant Cell Physiol. 2012, 53, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Kamei, S.; Kawai, S. Effect of iron deficiency on the chemical composition of the xylem sap of barley. Soil Sci. Plant Nutr. 2001, 47, 643–649. [Google Scholar] [CrossRef]
- Mori, S.; Nishizawa, N.; Hayashi, H.; Chino, M.; Yoshimura, E.; Ishihara, J. Why are young rice plants highly susceptible to iron deficiency? Plant Soil 1991, 130, 143–156. [Google Scholar] [CrossRef]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N.K. The phytosiderophore efflux transporter TOM2 is involved in metal transport in Rice. J. Biol. Chem. 2015, 290, 27688–27699. [Google Scholar] [CrossRef]
- Nozoye, T.; von Wirén, N.; Sato, Y.; Higashiyama, T.; Nakanishi, H.; Nishizawa, N.K. Characterization of the nicotianamine exporter ENA1 in rice. Front. Plant Sci. 2019, 10, 502. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Aoyama, T.; Kobayashi, T.; Takahashi, M.; Nagasaka, S.; Usuda, K.; Kakei, Y.; Ishimaru, Y.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL18 is a rice iron(III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol. Biol. 2009, 70, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, T.; Nakanishi, H.; Uchida, H.; Watanabe, S.; Matsuhashi, S.; Mori, S.; Nishizawa, N.K. 52Fe translocation in barley as monitored by a positron-emitting tracer imaging system (PETIS): Evidence for the direct translocation of Fe from roots to young leaves via phloem. Plant Cell Physiol. 2009, 50, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Ma, J.F. The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 2014, 19, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Ma, J.F. Node-controlled allocation of mineral elements in Poaceae. Curr. Opin. Plant Biol. 2017, 39, 18–24. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Ishikawa, S.; Abe, T.; Baba, K.; Arao, T.; Terada, Y. Role of the node in controlling traffic of cadmium, zinc, and manganese in rice. J. Exp. Bot. 2012, 63, 2729–2737. [Google Scholar] [CrossRef]
- Moore, K.L.; Chen, Y.; van de Meene, A.M.L.; Hughes, L.; Liu, W.; Geraki, T.; Mosselmans, F.; Mcgrath, S.P.; Grovenor, C.; Zhao, F.J. Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 2014, 201, 104–115. [Google Scholar] [CrossRef]
- Yamaji, N.; Ma, J.F. Bioimaging of multiple elements by high-resolution LA-ICP-MS reveals altered distribution of mineral elements in the nodes of rice mutants. Plant J. 2019, 99, 1254–1263. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Ma, J.F. OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J. Exp. Bot. 2016, 67, 5485–5494. [Google Scholar] [CrossRef]
- Bienfait, H.F.; van den Briel, W.; Mesland-Mul, N.T. Free space iron pools in roots: Generation and mobilization. Plant Physiol. 1985, 78, 596–600. [Google Scholar] [CrossRef]
- Zhang, F.; Römheld, V.; Marschner, H. Role of the root apoplasm for iron acquisition by wheat plants. Plant Physiol. 1991, 97, 1302–1305. [Google Scholar] [CrossRef]
- Shi, R.; Melzer, M.; Zheng, S.; Benke, A.; Stich, B.; von Wirén, N. Iron retention in root hemicelluloses causes genotypic variability in the tolerance to iron deficiency-induced chlorosis in maize. Front. Plant Sci. 2018, 9, 557. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Kakei, Y.; Shimo, H.; Bashir, K.; Sato, Y.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N.K. A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem. 2011, 286, 24649–24655. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Ishimaru, Y.; Shimo, H.; Kakei, Y.; Senoura, T.; Takahashi, R.; Sato, Y.; Sato, Y.; Uozumi, N.; Nakanishi, H.; et al. Rice phenolics efflux transporter 2 (PEZ2) plays an important role in solubilizing apoplasmic iron. Soil Sci. Plant Nutr. 2011, 57, 803–812. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Clemens, S.; Williams, L.E.; Krämer, U.; Borg, S.; Schjørring, J.K.; Sanders, D. Zinc biofortification of cereals: Problems and solutions. Trends Plant Sci. 2008, 13, 464–473. [Google Scholar] [CrossRef]
- Zee, S.; O’brien, T. A special type of tracheary element associated with “xylem discontinuity” in the floral axis of wheat. Aust. J. Biol. Sci. 1970, 23, 783. [Google Scholar] [CrossRef]
- Stomph, T.J.; Jiang, W.; Struik, P.C. Zinc biofortification of cereals: Rice differs from wheat and barley. Trends Plant Sci. 2009, 14, 123–124. [Google Scholar] [CrossRef]
- Zee, S.-Y. Vascular tissue and transfer cell distribution in the rice spikelet. Aust. J. Biol. Sci. 1972, 25, 411. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Li, D.; Liu, C.M. Rice caryopsis development I: Dynamic changes in different cell layers. J. Integr. Plant Biol. 2016, 58, 772–785. [Google Scholar] [CrossRef]
- Beasley, J.T.; Bonneau, J.P.; Sánchez-Palacios, J.T.; Moreno-Moyano, L.T.; Callahan, D.L.; Tako, E.; Glahn, R.P.; Lombi, E.; Johnson, A.A.T. Metabolic engineering of bread wheat improves grain iron concentration and bioavailability. Plant Biotechnol. J. 2019, 17, 1514–1526. [Google Scholar] [CrossRef]
- Kyriacou, B.; Moore, K.L.; Paterson, D.; De Jonge, M.D.; Howard, D.L.; Stangoulis, J.; Tester, M.; Lombi, E.; Johnson, A.A.T. Localization of iron in rice grain using synchrotron X-ray fluorescence microscopy and high resolution secondary ion mass spectrometry. J. Cereal Sci. 2014, 59, 173–180. [Google Scholar] [CrossRef]
- Johnson, A.A.T.; Kyriacou, B.; Callahan, D.L.; Carruthers, L.; Stangoulis, J.; Lombi, E.; Tester, M. Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS ONE 2011, 6, e24476. [Google Scholar] [CrossRef] [PubMed]
- Senoura, T.; Sakashita, E.; Kobayashi, T.; Takahashi, M.; Aung, M.S.; Masuda, H.; Nakanishi, H.; Nishizawa, N.K. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 2017, 95, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Thomine, S.; Vert, G. Iron transport in plants: Better be safe than sorry. Curr. Opin. Plant Biol. 2013, 16, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Nouet, C.; Motte, P.; Hanikenne, M. Chloroplastic and mitochondrial metal homeostasis. Trends Plant Sci. 2011, 16, 395–404. [Google Scholar] [CrossRef]
- Vigani, G.; Solti, Á.; Thomine, S.; Philippar, K. Essential and detrimental—An update on intracellular iron trafficking and homeostasis. Plant Cell Physiol. 2019, 60, 1420–1439. [Google Scholar] [CrossRef]
- Bashir, K.; Ishimaru, Y.; Shimo, H.; Nagasaka, S.; Fujimoto, M.; Takanashi, H.; Tsutsumi, N.; An, G.; Nakanishi, H.; Nishizawa, N.K. The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2011, 2, 322. [Google Scholar] [CrossRef]
- Han, J.H.; Song, X.F.; Li, P.; Yang, H.J.; Yin, L.P. Maize ZmFDR3 localized in chloroplasts is involved in iron transport. Sci. China, Ser. C Life Sci. 2009, 52, 864–871. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zhang, X.; Zhang, Q.; Pan, X.-X.; Yan, L.-C.; Ma, X.-J.; Zhao, W.-Z.; Qi, X.-T.; Yin, L.-P. Zea mays Fe deficiency-related 4 (ZmFDR4) functions as an iron transporter in the plastids of monocots. Plant J. 2017, 90, 147–163. [Google Scholar] [CrossRef]
- Fobis-Loisy, I.; Loridon, K.; Lobreaux, S.; Lebrun, M.; Briat, J.-F. Structure and differential expression of two maize ferritin genes in response to iron and abscisic acid. Eur. J. Biochem. 1995, 231, 609–619. [Google Scholar] [CrossRef]
- Stein, R.J.; Ricachenevsky, F.K.; Fett, J.P. Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2). Plant Sci. 2009, 177, 563–569. [Google Scholar] [CrossRef]
- Borg, S.; Brinch-Pedersen, H.; Tauris, B.; Madsen, L.H.; Darbani, B.; Noeparvar, S.; Holm, P.B. Wheat ferritins: Improving the iron content of the wheat grain. J. Cereal Sci. 2012, 56, 204–213. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.H.; Yi, H.Y.; Gong, J.M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Connorton, J.M.; Jones, E.R.; Rodríguez-Ramiro, I.; Fairweather-Tait, S.; Uauy, C.; Balk, J. Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 2017, 174, 2434–2444. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Yokosho, K.; Yamaji, N.; Ma, J.F. A vacuolar phytosiderophore transporter alters iron and zinc accumulation in polished rice grains. Plant Physiol. 2019, 181, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, L.; Kong, Q.; Shou, H. A vacuolar membrane ferric-chelate reductase, OsFRO1, alleviates Fe toxicity in rice (Oryza sativa L.). Front. Plant Sci. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Suzuki, M.; Inoue, H.; Itai, R.N.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J. Exp. Bot. 2005, 56, 1305–1316. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, N.; Zhao, F.; Song, X.; Yin, Z.; Huang, R.; Zhang, C. Changes in the transcriptomic profiles of maize roots in response to iron-deficiency stress. Plant Mol. Biol. 2014, 85, 349–363. [Google Scholar] [CrossRef]
- Wang, M.; Kawakami, Y.; Bhullar, N.K. Molecular Analysis of Iron Deficiency Response in Hexaploid Wheat. Front. Sustain. Food Syst. 2019, 3, 67. [Google Scholar]
- Nagasaka, S.; Takahashi, M.; Nakanishi-Itai, R.; Bashir, K.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Time course analysis of gene expression over 24 hours in Fe-deficient barley roots. Plant Mol. Biol. 2009, 69, 621–631. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron sensors and signals in response to iron deficiency. Plant Sci. 2014, 224, 36–43. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ogo, Y.; Itai, R.N.; Nakanishi, H.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 19150–19155. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Itai, R.N.; Ogo, Y.; Kakei, Y.; Nakanishi, H.; Takahashi, M.; Nishizawa, N.K. The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J. 2009, 60, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Itai, R.N.; Aung, M.S.; Senoura, T.; Nakanishi, H.; Nishizawa, N.K. The rice transcription factor IDEF1 directly binds to iron and other divalent metals for sensing cellular iron status. Plant J. 2012, 69, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nakayama, Y.; Itai, R.N.; Nakanishi, H.; Yoshihara, T.; Mori, S.; Nishizawa, N.K. Identification of novel cis-acting elements, IDE1 and IDE2, of the barley IDS2 gene promoter conferring iron-deficiency-inducible, root-specific expression in heterogeneous tobacco plants. Plant J. 2003, 36, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Nakanishi Itai, R.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K.; Kobayashi, T. The Bowman-Birk trypsin inhibitor IBP1 interacts with and prevents degradation of IDEF1 in rice. Plant Mol. Biol. Report. 2014, 32, 841–851. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ogo, Y.; Aung, M.S.; Nozoye, T.; Itai, R.N.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. The spatial expression and regulation of transcription factors IDEF1 and IDEF2. Ann. Bot. 2010, 105, 1109–1117. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Nakanishi, H.; Inoue, H.; Kobayashi, T.; Suzuki, M.; Takahashi, M.; Mori, S.; Nishizawa, N.K. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006, 57, 2867–2878. [Google Scholar] [CrossRef]
- Ogo, Y.; Nakanishi Itai, R.; Nakanishi, H.; Kobayashi, T.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007, 51, 366–377. [Google Scholar] [CrossRef]
- Wang, S.; Li, L.; Ying, Y.; Wang, J.; Shao, J.F.; Yamaji, N.; Whelan, J.; Ma, J.F.; Shou, H. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice. New Phytol. 2019, 225, 1247–1260. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Yao, X.; Liang, G.; Yu, D. POSITIVE REGULATOR OF IRON HOMEOSTASIS1, OSPRI1, facilitates Iron homeostasis. Plant Physiol. 2017, 175, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Y.; Pu, M.; Xu, P.; Liang, G.; Yu, D. Oryza sativa POSITIVE REGULATOR OF IRON DEFICIENCY RESPONSE 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant. Cell Environ. 2020, 43, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ozu, A.; Kobayashi, S.; An, G.; Seong, J. OsbHLH058 and OsbHLH059 transcription factors positively regulate iron deficiency responses in rice. Plant Mol. Biol. 2019, 101, 471–486. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Ying, Y.; Wang, L.; Wang, F.; Whelan, J.; Shou, H. Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 2010, 10, 1–9. [Google Scholar] [CrossRef]
- Ogo, Y.; Kobayashi, T.; Nakanishi Itai, R.; Nakanishi, H.; Kakei, Y.; Takahashi, M.; Toki, S.; Mori, S.; Nishizawa, N.K. A novel NAC transcription factor, IDEF2, that recognizes the Iron Deficiency-responsive Element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 2008, 283, 13407–13417. [Google Scholar] [CrossRef]
- Wang, L.; Ying, Y.; Narsai, R.; Ye, L.; Zheng, L.; Tian, J.; Whelan, J.; Shou, H. Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant. Cell Environ. 2013, 36, 224–236. [Google Scholar] [CrossRef]
- Grillet, L.; Lan, P.; Li, W.; Mokkapati, G.; Schmidt, W. IRON MAN is a ubiquitous family of peptides that control iron transport in plants. Nat. Plants 2018, 4, 953–963. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef]
- Distelfeld, A.; Pearce, S.P.; Avni, R.; Scherer, B.; Uauy, C.; Piston, F.; Slade, A.; Zhao, R.; Dubcovsky, J. Divergent functions of orthologous NAC transcription factors in wheat and rice. Plant Mol. Biol. 2012, 78, 515–524. [Google Scholar] [CrossRef]
- Kobayashi, T.; Itai, R.N.; Senoura, T.; Oikawa, T.; Ishimaru, Y.; Ueda, M.; Nakanishi, H.; Nishizawa, N.K. Jasmonate signaling is activated in the very early stages of iron deficiency responses in rice roots. Plant Mol. Biol. 2016, 91, 533–547. [Google Scholar] [CrossRef]
- Shen, C.; Yue, R.; Sun, T.; Zhang, L.; Yang, Y.; Wang, H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 2015, 231, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Curie, C.; Briat, J.-F. Iron transport and signaling in plants. Annu. Rev. Plant Biol. 2003, 54, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, F.; Shou, H.; Huang, F.; Zheng, L.; He, F.; Li, J.; Zhao, F.-J.; Ueno, D.; Ma, J.F.; et al. Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 2007, 145, 1647–1657. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Cheng, Z.; Ai, C.; Jiang, X.; Bei, X.; Zheng, Y.; Raymond, P.; Welch, R.M.; Miller, D.D.; Lei, X.G.; et al. Nicotianamine, a novel enhancer of rice iron bioavailability to humans. PLoS ONE 2010, 5, 1–7. [Google Scholar] [CrossRef]
- Lee, S.; Jeon, U.S.; Lee, S.J.; Kim, Y.-K.; Persson, D.P.; Husted, S.; Schjorring, J.K.; Kakei, Y.; Masuda, H.; Nishizawa, N.K.; et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. USA 2009, 106, 22014–22019. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.S.; Jeon, U.S.; Kim, Y.K.; Schjoerring, J.K.; An, G. Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 2012, 33, 269–275. [Google Scholar] [CrossRef]
- Banakar, R.; Alvarez Fernandez, A.; Díaz-Benito, P.; Abadia, J.; Capell, T.; Christou, P. Phytosiderophores determine thresholds for iron and zinc accumulation in biofortified rice endosperm while inhibiting the accumulation of cadmium. J. Exp. Bot. 2017, 68, 4983–4995. [Google Scholar] [CrossRef]
- Singh, S.P.; Keller, B.; Gruissem, W.; Bhullar, N.K. Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor. Appl. Genet. 2017, 130, 283–292. [Google Scholar] [CrossRef]
- Masuda, H.; Usuda, K.; Kobayashi, T.; Ishimaru, Y.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2009, 2, 155–166. [Google Scholar] [CrossRef]
- Beasley, J.T.; Hart, J.J.; Tako, E.; Glahn, R.P.; Johnson, A.A.T. Investigation of nicotianamine and 2′ deoxymugineic acid as enhancers of iron bioavailability in Caco-2 cells. Nutrients 2019, 11, 1502. [Google Scholar] [CrossRef]
- Goto, F.; Yoshihara, T.; Shigemoto, N.; Toki, S.; Takaiwa, F. Iron fortification of rice seed by the soybean ferritin gene. Nat. Biotechnol. 1999, 17, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Lucca, P.; Hurrell, R.; Potrykus, I. Approaches to improving the bioavailability and level of iron in rice seeds. J. Sci. Food Agric. 2001, 81, 828–834. [Google Scholar] [CrossRef]
- Vasconcelos, M.; Datta, K.; Oliva, N.; Khalekuzzaman, M.; Torrizo, L.; Krishnan, S.; Oliveira, M.; Goto, F.; Datta, S.K. Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci. 2003, 164, 371–378. [Google Scholar] [CrossRef]
- Oliva, N.; Chadha-Mohanty, P.; Poletti, S.; Abrigo, E.; Atienza, G.; Torrizo, L.; Garcia, R.; Dueñas, C.; Poncio, M.A.; Balindong, J.; et al. Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol. Breed. 2014, 33, 23–37. [Google Scholar] [CrossRef]
- Davila-Hicks, P.; Theil, E.C.; Lönnerdal, B. Iron in ferritin or in salts (ferrous sulfate) is equally bioavailable in nonanemic women. Am. J. Clin. Nutr. 2004, 80, 936–940. [Google Scholar] [CrossRef]
- Neal, A.L.; Geraki, K.; Borg, S.; Quinn, P.; Mosselmans, J.F.; Brinch-Pedersen, H.; Shewry, P.R. Iron and zinc complexation in wild-type and ferritin-expressing wheat grain: Implications for mineral transport into developing grain. J. Biol. Inorg. Chem. 2013, 18, 557–570. [Google Scholar] [CrossRef]
- Masuda, H.; Ishimaru, Y.; Aung, M.S.; Kobayashi, T.; Kakei, Y.; Takahashi, M.; Higuchi, K.; Nakanishi, H.; Nishizawa, N.K. Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2012, 2, 543. [Google Scholar] [CrossRef]
- Wu, T.-Y.; Gruissem, W.; Bhullar, N.K. Targeting intracellular transport combined with efficient uptake and storage significantly increases grain iron and zinc levels in rice. Plant Biotechnol. J. 2018, 17, 9–20. [Google Scholar] [CrossRef]
- Trijatmiko, K.R.; Dueñas, C.; Tsakirpaloglou, N.; Torrizo, L.; Arines, F.M.; Adeva, C.; Balindong, J.; Oliva, N.; Sapasap, M.V.; Borrero, J.; et al. Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Saito, K. Sulfur assimilatory metabolism. The long and smelling Road. Plant Physiol. 2004, 136, 2443–2450. [Google Scholar] [CrossRef]
- Kopriva, S. Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 2006, 97, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Lara, L.O.; Medrano-Macías, J.; Pérez-Labrada, F.; Rivas-Martínez, E.N.; García-Enciso, E.L.; González-Morales, S.; Juárez-Maldonado, A.; Rincón-Sánchez, F.; Benavides-Mendoza, A. From elemental sulfur to hydrogen sulfide in agricultural soils and plants. Molecules 2019, 24, 2282. [Google Scholar] [CrossRef] [PubMed]
- Pierzynski, G.M.; Sims, J.T.; Vance, G.F. Soils and Environmental Quality; CRC Press: Boca Raton, FL, USA, 2005; ISBN 9780203496367. [Google Scholar]
- Ponnamperuma, F.N. The Cchemistry of submerged soils. Adv. Agron. 1972, 24, 29–96. [Google Scholar]
- Achtnich, C.; Bak, F.; Conrad, R. Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 1995, 19, 65–72. [Google Scholar] [CrossRef]
- Wind, T.; Conrad, R. Sulfur compounds, potential turnover of sulfate and thiosulfate, and numbers of sulfate-reducing bacteria in planted and unplanted paddy soil. FEMS Microbiol. Ecol. 1995, 18, 257–266. [Google Scholar] [CrossRef]
- Stubner, S.; Wind, T.; Conrad, R. Sulfur oxidation in rice field soil: Activity, enumeration, isolation and characterization of thiosulfate-oxidizing bacteria. Syst. Appl. Microbiol. 1998, 21, 569–578. [Google Scholar] [CrossRef]
- Hu, Z.; Hanekalus, S.; Cao, Z.; Schnug, E. Chemical behavior of soil sulphur in the rhizosphere and its ecological effects. In Proceedings of the 1st Sino-German Workshop on Aspects of Sulfur Nutrition of Plants; 23–27 May 2004 Shenyang, China; De Kok, L.J., Schnug, E., Eds.; FAL Agricultural Research: Braunschweig, Germany, 2005. [Google Scholar]
- Takahashi, H. Regulation of sulfate transport and assimilation in plants. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 281, pp. 129–159. [Google Scholar]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef]
- Takahashi, H.; Buchner, P.; Yoshimoto, N.; Hawkesford, M.J.; Shiu, S.H. Evolutionary relationships and functional diversity of plant sulfate transporters. Front. Plant Sci. 2012, 2, 1–9. [Google Scholar] [CrossRef]
- Takahashi, H.; Watanabe-Takahashi, A.; Smith, F.W.; Blake-Kalff, M.; Hawkesford, M.J.; Saito, K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000, 23, 171–182. [Google Scholar] [CrossRef]
- Shibagaki, N.; Rose, A.; McDermott, J.P.; Fujiwara, T.; Hayashi, H.; Yoneyama, T.; Davies, J.P. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 2002, 29, 475–486. [Google Scholar] [CrossRef]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Trivedi, P.K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genom. 2011, 11, 259–273. [Google Scholar] [CrossRef]
- Godwin, R.M.; Rae, A.L.; Carroll, B.J.; Smith, F.W. Cloning and characterization of two genes encoding sulfate transporters from rice (Oryza sativa L.). Plant Soil 2003, 257, 113–123. [Google Scholar] [CrossRef]
- Smith, F.W.; Hawkesford, M.J.; Ealing, P.M.; Clarkson, D.T.; Vanden Berg, P.J.; Belcher, A.R.; Warrilow, A.G.S. Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J. 1997, 12, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Rae, A.L.; Smith, F.W. Localisation of expression of a high-affinity sulfate transporter in barley roots. Planta 2002, 215, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Vidmar, J.J.; Schjoerring, J.K.; Touraine, B.; Glass, A.D.M. Regulation of the hvst1 gene encoding a high-affinity sulfate transporter from Hordeum vulgare. Plant Mol. Biol. 1999, 40, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Bolchi, A.; Petrucco, S.; Tenca, P.L.; Foroni, C.; Ottonello, S. Coordinate modulation of maize sulfate permease and ATP sulfurylase mRNAs in response to variations in sulfur nutritional status: Stereospecific down-regulation by L-cysteine. Plant Mol. Biol. 1999, 39, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, L.; Parmar, S.; Bouranis, D.L.; Howarth, J.R.; Hawkesford, M.J. Coordinated expression of sulfate uptake and components of the sulfate assimilatory pathway in maize. Plant Biol. 2004, 6, 408–414. [Google Scholar] [CrossRef]
- Buchner, P.; Prosser, I.M.; Hawkesford, M.J. Phylogeny and expression of paralogous and orthologous sulphate transporter genes in diploid and hexaploid wheats. Genome 2004, 47, 526–534. [Google Scholar] [CrossRef]
- Bell, R.W. Sulfur and the Production of Rice in Wetland and Dryland Ecosystems. In Sulfur: A Missing Link between Soils, Crops, and Nutrition; Jez, J., Ed.; American Society of Agronomy, Crop Science Society of America and Soil Science Society of America: Madison, WI, USA, 2008; pp. 197–218. ISBN 978-0-89118-186-6. [Google Scholar]
- Rausch, T.; Wachter, A. Sulfur metabolism: A versatile platform for launching defence operations. Trends Plant Sci. 2005, 10, 503–509. [Google Scholar] [CrossRef]
- Kataoka, T.; Hayashi, N.; Yamaya, T.; Takahashi, H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004, 136, 4198–4204. [Google Scholar] [CrossRef]
- Moura, J.J.G.; Bursakov, S.A.; Gavel, O.; Moura, I. Sulfate activation. Encycl. Catal. 2002. [Google Scholar] [CrossRef]
- Leustek, T.; Murillo, M.; Cervantes, M. Cloning of a cDNA encoding ATP sulfurylase from Arabidopsis thaliana by functional expression in Saccharomyces cerevisiae. Plant Physiol. 1994, 105, 897–902. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Setya, A.; Murillo, M.; Leustek, T. Sulfate reduction in higher plants: Molecular evidence for a novel 5′-adenylylsulfate reductase. Proc. Natl. Acad. Sci. USA 1996, 93, 13383–13388. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, M.; Akashi, T.; Hase, T. Plant sulfite reductase: Molecular structure, catalytic function and interaction with ferredoxin. J. Inorg. Biochem. 2000, 82, 27–32. [Google Scholar] [CrossRef]
- Brühl, A.; Haverkamp, T.; Gisselmann, G.; Schwenn, J.D. A cDNA clone from Arabidopsis thaliana encoding plastidic gerredoxin: Sulfite reductase. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1996, 1295, 119–124. [Google Scholar] [CrossRef]
- Saito, K.; Miura, N.; Yamazaki, M.; Hirano, H.; Murakoshi, I. Molecular cloning and bacterial expression of cDNA encoding a plant cysteine synthase. Proc. Natl. Acad. Sci. USA 1992, 89, 8078–8082. [Google Scholar] [CrossRef]
- Hell, R.; Bork, C.; Bogdanova, N.; Frolov, I.; Hauschild, R. Isolation and characterization of two cDNAs encoding for compartment specific isoforms of O-acetylserine (thiol) lyase from Arabidopsis thaliana. FEBS Lett. 1994, 351, 257–262. [Google Scholar] [CrossRef]
- Noji, M.; Inoue, K.; Kimura, N.; Gouda, A.; Saito, K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. J. Biol. Chem. 1998, 273, 32739–32745. [Google Scholar] [CrossRef]
- Hell, R.; Wirtz, M. Metabolism of cysteine in plants and phototrophic bacteria. In Sulfur Metabolism in Phototrophic Organisms; Hell, R., Dahl, C., Knaff, D.B., Leustek, T., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 59–91. [Google Scholar]
- Lu, Y. Assembly and transfer of iron–sulfur clusters in the plastid. Front. Plant Sci. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Balk, J.; Pilon, M. Ancient and essential: The assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011, 16, 218–226. [Google Scholar] [CrossRef]
- Na, G.N.; Salt, D.E. The role of sulfur assimilation and sulfur-containing compounds in trace element homeostasis in plants. Environ. Exp. Bot. 2011, 72, 18–25. [Google Scholar] [CrossRef]
- Ravanel, S.; Gakiere, B.; Job, D.; Douce, R. The specific features of methionine biosynthesis and metabolism in plants. Proc. Natl. Acad. Sci. USA 1998, 95, 7805–7812. [Google Scholar] [CrossRef]
- Ravanel, S.; Block, M.A.; Rippert, P.; Jabrin, S.; Curien, G.; Rébeillé, F.; Douce, R. Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 2004, 279, 22548–22557. [Google Scholar] [CrossRef]
- Roje, S. S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 2006, 67, 1686–1698. [Google Scholar] [CrossRef]
- Takahashi, M.; Terada, Y.; Nakai, I.; Nakanishi, H.; Yoshimura, E.; Mori, S.; Nishizawa, N.K. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 2003, 15, 1263–1280. [Google Scholar] [CrossRef]
- Marschner, H. Functions of mineral nutrients: Macronutrients. In Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; pp. 229–312. [Google Scholar]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Wang, F.; Chen, F.; Cai, Y.; Zhang, G.; Wu, F. Modulation of exogenous glutathione in ultrastructure and photosynthetic performance against Cd stress in the two barley genotypes differing in Cd tolerance. Biol. Trace Elem. Res. 2011, 144, 1275–1288. [Google Scholar] [CrossRef]
- Chen, F.; Wang, F.; Wu, F.; Mao, W.; Zhang, G.; Zhou, M. Modulation of exogenous glutathione in antioxidant defense system against Cd stress in the two barley genotypes differing in Cd tolerance. Plant Physiol. Biochem. 2010, 48, 663–672. [Google Scholar] [CrossRef]
- Yamazaki, S.; Ueda, Y.; Mukai, A.; Ochiai, K.; Matoh, T. Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance. Plant Direct 2018, 2, e00034. [Google Scholar] [CrossRef]
- Hell, R.; Bergmann, L. λ-Glutamylcysteine synthetase in higher plants: Catalytic properties and subcellular localization. Planta 1990, 180, 603–612. [Google Scholar] [CrossRef]
- Wang, C.L.; Oliver, D.J. Cloning of the cDNA and genomic clones for glutathione synthetase from Arabidopsis thaliana and complementation of a gsh2 mutant in fission yeast. Plant Mol. Biol. 1996, 31, 1093–1104. [Google Scholar] [CrossRef]
- Cobbett, C.; Goldsbrough, P. Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 2002, 53, 159–182. [Google Scholar] [CrossRef]
- Vatamaniuk, O.K.; Mari, S.; Lu, Y.-P.; Rea, P.A. AtPCS1, a phytochelatin synthase from Arabidopsis: Isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. USA 1999, 96, 7110–7115. [Google Scholar] [CrossRef]
- Clemens, S.; Kim, E.J.; Neumann, D.; Schroeder, J.I. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999, 18, 3325–3333. [Google Scholar] [CrossRef]
- Song, W.Y.; Mendoza-Cózatl, D.G.; Lee, Y.; Schroeder, J.I.; Ahn, S.N.; Lee, H.S.; Wicker, T.; Martinoia, E. Phytochelatin-metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant Cell Environ. 2014, 37, 1192–1201. [Google Scholar] [CrossRef]
- Uraguchi, S.; Tanaka, N.; Hofmann, C.; Abiko, K.; Ohkama-Ohtsu, N.; Weber, M.; Kamiya, T.; Sone, Y.; Nakamura, R.; Takanezawa, Y.; et al. Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains. Plant Cell Physiol. 2017, 58, 1730–1742. [Google Scholar] [CrossRef]
- Hasan, M.K.; Cheng, Y.; Kanwar, M.K.; Chu, X.Y.; Ahammed, G.J.; Qi, Z.Y. Responses of plant proteins to heavy metal stress—A review. Front. Plant Sci. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Hegelund, J.N.; Schiller, M.; Kichey, T.; Hansen, T.H.; Pedas, P.; Husted, S.; Schjoerring, J.K. Barley metallothioneins: MT3 and MT4 are localized in the grain aleurone layer and show Differential zinc binding. Plant Physiol. 2012, 159, 1125–1137. [Google Scholar] [CrossRef]
- Mekawy, A.M.M.; Assaha, D.V.M.; Munehiro, R.; Kohnishi, E.; Nagaoka, T.; Ueda, A.; Saneoka, H. Characterization of type 3 metallothionein-like gene (OsMT-3a) from rice, revealed its ability to confer tolerance to salinity and heavy metal stresses. Environ. Exp. Bot. 2018, 147, 157–166. [Google Scholar] [CrossRef]
- Nezhad, R.M.; Shahpiri, A.; Mirlohi, A. Discrimination between two rice metallothionein isoforms belonging to type 1 and type 4 in metal-binding ability. Biotechnol. Appl. Biochem. 2013, 60, 275–282. [Google Scholar] [CrossRef]
- Lane, B.; Kajioka, R.; Kennedy, T. The wheat-germ E c protein is a zinc-containing metallothionein. Biochem. Cell Biol. 1987, 65, 1001–1005. [Google Scholar] [CrossRef]
- Yu, L.H.; Umeda, M.; Liu, J.Y.; Zhao, N.M.; Uchimiya, H. A novel MT gene of rice plants is strongly expressed in the node portion of the stem. Gene 1998, 206, 29–35. [Google Scholar] [CrossRef]
- Zhang, H.; Lv, S.; Xu, H.; Hou, D.; Li, Y.; Wang, F. H2O2 is involved in the metallothionein-mediated rice tolerance to copper and cadmium toxicity. Int. J. Mol. Sci. 2017, 18, 2083. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, Y.; Li, Y.; Ling, H.-Q.; Chu, C. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol. Biol. 2009, 70, 219–229. [Google Scholar] [CrossRef]
- Li, Z.G. Analysis of some enzymes activities of hydrogen sulfide metabolism in plants. In Methods in Enzymology; Cadenas, E., Packer, L., Eds.; Academic Press: San Diego, CA, USA, 2015; Volume 555, pp. 253–269. [Google Scholar]
- Li, Z.G.; Min, X.; Zhou, Z.H. Hydrogen sulfide: A signal molecule in plant cross-adaptation. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Cao, M.J.; Wang, Z.; Wirtz, M.; Hell, R.; Oliver, D.J.; Xiang, C. Bin SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 2013, 73, 607–616. [Google Scholar] [CrossRef]
- Kataoka, T.; Watanabe-Takahashi, A.; Hayashi, N.; Ohnishi, M.; Mimura, T.; Buchner, P.; Hawkesford, M.J.; Yamaya, T.; Takahashi, H. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 2004, 16, 2693–2704. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Kopriva, S. Transporters in plant sulfur metabolism. Front. Plant Sci. 2014, 5, 1–16. [Google Scholar] [CrossRef]
- Herschbach, C.; Rennenberg, H. Significance of phloem-translocated organic sulfur compounds for the regulation of sulfur nutrition. In Progress in Botany; Esser, K., Lüttge, U., Kadereit, J.W., Beyschlag, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 62, pp. 177–193. [Google Scholar]
- Bourgis, F.; Roje, S.; Nuccio, M.L.; Fisher, D.B.; Tarczynski, M.C.; Li, C.; Herschbach, C.; Rennenberg, H.; Pimenta, M.J.; Shen, T.L.; et al. S-methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. Plant Cell 1999, 11, 1485–1497. [Google Scholar] [CrossRef]
- Kuzuhara, Y.; Isobe, A.; Awazuhara, M.; Fujiwara, T.; Hayashi, H. Glutathione levels in phloem sap of rice plants under sulfur deficient conditions. Soil Sci. Plant Nutr. 2000, 46, 265–270. [Google Scholar] [CrossRef]
- Cagnac, O.; Bourbouloux, A.; Chakrabarty, D.; Zhang, M.Y.; Delrot, S. AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiol. 2004, 135, 1378–1387. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Chino, M. Nitrate and other anions in the rice phloem sap. Plant Cell Physiol. 1985, 26, 325–330. [Google Scholar]
- Yoshimoto, N.; Inoue, E.; Saito, K.; Yamaya, T.; Takahashi, H. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 2003, 131, 1511–1517. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, C.; Dai, C.; Ge, Y. Sufficient sulfur supply promotes seedling growth, alleviates oxidation stress, and regulates iron uptake and translocation in rice. Biol. Plant. 2015, 59, 788–792. [Google Scholar] [CrossRef]
- Astolfi, S.; Cesco, S.; Zuchi, S.; Neumann, G.; Roemheld, V. Sulfur starvation reduces phytosiderophores release by iron-deficient barley plants. Soil Sci. Plant Nutr. 2006, 52, 43–48. [Google Scholar] [CrossRef]
- Zuchi, S.; Cesco, S.; Astolfi, S. High S supply improves Fe accumulation in durum wheat plants grown under Fe limitation. Environ. Exp. Bot. 2012, 77, 25–32. [Google Scholar] [CrossRef]
- Astolfi, S.; Zuchi, S.; Passera, C.; Cesco, S. Does the sulfur assimilation pathway play a role in the response to Fe deficiency in maize (Zea mays L.) plants. J. Plant Nutr. 2003, 26, 2111–2121. [Google Scholar] [CrossRef]
- Ciaffi, M.; Paolacci, A.R.; Celletti, S.; Catarcione, G.; Kopriva, S.; Astolfi, S. Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J. Exp. Bot. 2013, 64, 1663–1675. [Google Scholar] [CrossRef]
- Wu, Z.; Naveed, S.; Zhang, C.; Ge, Y. Adequate supply of sulfur simultaneously enhances iron uptake and reduces cadmium accumulation in rice grown in hydroponic culture. Environ. Pollut. 2020, 114327. [Google Scholar] [CrossRef]
- Wu, C.Y.H.; Lu, J.; Hu, Z.Y. Influence of sulfur supply on the iron accumulation in rice plants. Commun. Soil Sci. Plant Anal. 2014, 45, 1149–1161. [Google Scholar] [CrossRef]
- Kuwajima, K.; Kawai, S. Relationship between sulfur metabolism and biosynthesis of phytosiderophores in barley roots. In Plant Nutrition for Sustainable Food Production and Environment; Springer: Dordrecht, The Netherlands, 1997; pp. 285–286. [Google Scholar]
- Inoue, H.; Higuchi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 2003, 36, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, K.; Takahashi, M.; Nakanishi, H.; Kawasaki, S.; Nishizawa, N.K.; Mori, S. Analysis of transgenic rice containing barley nicotianamine synthase gene. Soil Sci. Plant Nutr. 2001, 47, 315–322. [Google Scholar] [CrossRef]
- Takahashi, M.; Nakanishi, H.; Kawasaki, S.; Nishizawa, N.K.; Mori, S. Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat. Biotechnol. 2001, 19, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Banakar, R.; Fernandez, A.A.; Zhu, C.; Abadia, J.; Capell, T.; Christou, P. The ratio of phytosiderophores nicotianamine to deoxymugenic acid controls metal homeostasis in rice. Planta 2019, 250, 1339–1354. [Google Scholar] [CrossRef]
- Díaz-Benito, P.; Banakar, R.; Rodríguez-Menéndez, S.; Capell, T.; Pereiro, R.; Christou, P.; Abadía, J.; Fernández, B.; Álvarez-Fernández, A. Iron and zinc in the embryo and endosperm of rice (Oryza sativa L.) seeds in contrasting 2′-deoxymugineic acid/nicotianamine scenarios. Front. Plant Sci. 2018, 9, 1190. [Google Scholar] [CrossRef]
- Haneklaus, S.; Bloem, E.; Schnug, E. History of Sulfur Deficiency in Crops. In Sulfur: A Missing Link between Soils, Crops, and Nutrition; Jez, J., Ed.; American Society of Agronomy, Crop Science Society of America and Soil Science Society of America: Madison, WI, USA, 2008; pp. 45–58. [Google Scholar]
- Lucheta, A.R.; Lambais, M.R. Sulfur in agriculture. Rev. Bras. Ciência Solo 2012, 36, 1369–1379. [Google Scholar] [CrossRef]
- Zhao, F.; Tausz, M.; De Kok, L.J. Role of sulfur for plant production in agricultural and natural ecosystems. In Sulfur Metabolism in Phototrophic Organisms; Hell, R., Dahl, C., Knaff, D., Leustek, T., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 417–435. ISBN 978-1-4020-6863-8. [Google Scholar]
- Yoshimoto, N.; Inoue, E.; Watanabe-Takahashi, A.; Saito, K.; Takahashi, H. Posttranscriptional regulation of high-affinity sulfate transporters in arabidopsis by sulfur nutrition. Plant Physiol. 2007, 145, 378–388. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.H.; Shang, Y.T.; Wang, W.H.; Hu, W.J.; Simon, M.; Liu, X.; Shangguan, Z.P.; Zheng, H.L. Hydrogen sulphide improves adaptation of Zea mays seedlings to iron deficiency. J. Exp. Bot. 2015, 66, 6605–6622. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Zhang, L.; Zhao, H.; Li, H. Hydrogen sulphide improves iron homeostasis in wheat under iron-deficiency. J. Plant Sci. 2017, 5, 170. [Google Scholar]
- Lisjak, M.; Teklic, T.; Wilson, I.D.; Whiteman, M.; Hancock, J.T. Hydrogen sulfide: Environmental factor or signalling molecule? Plant. Cell Environ. 2013, 36, 1607–1616. [Google Scholar] [CrossRef]
- Chen, J.; Shangguan, Z.P.; Zheng, H.L. The function of hydrogen sulphide in iron availability: Sulfur nutrient or signaling molecule? Plant Signal. Behav. 2016, 11, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Itai, R.N.; Kobayashi, T.; Aung, M.S.; Nakanishi, H.; Nishizawa, N.K. OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 2011, 75, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Vámos, R. H2S, the cause of the bruzone (Akiochi) disease of rice. Soil Sci. Plant Nutr. 1958, 4, 37–40. [Google Scholar] [CrossRef]
- Dobermann, A.; Fairhurst, T. Rice: Nutrient Disorders & Nutrient Management, 1st ed.; Potash & Phosphate Institute and International Rice Research Institute: Manila, Philippines, 2000. [Google Scholar]
- Armstrong, J.; Armstrong, W. Rice: Sulfide-induced barriers to root radial oxygen loss, Fe2+ and water uptake, and lateral root emergence. Ann. Bot. 2005, 96, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Groth, D.; Lee, F. Rice diseases. In Rice: Origin, History, Technology, and Production; Smith, C.W., Dilday, R.H., Eds.; John and Wiley and Sons: Hoboken, NJ, USA, 2002; pp. 413–436. ISBN 978-0-471-34516-9. [Google Scholar]
- Joshi, M.M. Hydrogen sulfide: Effects on the physiology of rice plants and relation to straighthead disease. Phytopathology 1975, 65, 1165. [Google Scholar] [CrossRef]
- Yang, J.X.; Liu, Y.; Ye, Z.H. Root-induced changes of pH, Eh, Fe(II) and fractions of Pb and Zn in rhizosphere soils of four wetland plants with different radial oxygen losses. Pedosphere 2012, 22, 518–527. [Google Scholar] [CrossRef]
- LaFond-Hudson, S.; Johnson, N.W.; Pastor, J.; Dewey, B. Iron sulfide formation on root surfaces controlled by the life cycle of wild rice (Zizania palustris). Biogeochemistry 2018, 141, 95–106. [Google Scholar] [CrossRef]
- Schmidt, H.; Eickhorst, T.; Tippkötter, R. Monitoring of root growth and redox conditions in paddy soil rhizotrons by redox electrodes and image analysis. Plant Soil 2011, 341, 221–232. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Z.; Wan, X.; Zheng, G.; Yang, J.; Zhang, H.; Guo, L.; Wang, X.; Zhou, X.; Guo, Q.; et al. Interaction between sulfur and lead in toxicity, iron plaque formation and lead accumulation in rice plant. Ecotoxicol. Environ. Saf. 2016, 128, 206–212. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, F.; Mao, D. Effect of iron plaque outside roots on nutrient uptake by rice (Oryza sativa L.): Phosphorus uptake. Plant Soil 1999, 209, 187–192. [Google Scholar] [CrossRef]
- Zhou, X.-B.; Shi, W.-M. Effect of root surface iron plaque on Se translocation and uptake by Fe-deficient rice. Pedosphere 2007, 17, 580–587. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, F.; Mao, D. Effect of iron plaque outside roots on nutrient uptake by rice (Oryza sativa L.). Zinc uptake by Fe-deficient rice. Plant Soil 1998, 202, 33–39. [Google Scholar] [CrossRef]
- Nozoye, T.; Itai, R.N.; Nagasaka, S.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Diurnal changes in the expression of genes that participate im phytosiderophore synthesis in rice. Soil Sci. Plant Nutr. 2004, 50, 1125–1131. [Google Scholar] [CrossRef]
- Murakami, T.; Ise, K.; Hayakawa, M.; Kamei, S.; Takagi, S. Stabilities of Metal Complexes of Mugineic Acids and Their Specific Affinities for Iron(III). Chem. Lett. 1989, 18, 2137–2140. [Google Scholar] [CrossRef]
- Qin, Y.; Song, F.; Ai, Z.; Zhang, P.; Zhang, L. Protocatechuic acid promoted alachlor degradation in Fe(III)/H2O2 Fenton system. Environ. Sci. Technol. 2015, 49, 7948–7956. [Google Scholar] [CrossRef]
- Freney, J.R.; Jacq, V.A.; Baldensperger, J.F. The significance of the biological sulfur cycle in rice production. In Microbiology of Tropical Soils and Plant Productivity; Springer: Dordrecht, The Netherlands, 1982; pp. 271–317. [Google Scholar]
- Attanandana, T.; Vacharotayan, S. Acid sulfate soils: Their characteristics, genesis, amelioration and utilization. Southeast Asian Stud. 1986, 24, 154–180. [Google Scholar]
- Lee, S.; Persson, D.P.; Hansen, T.H.; Husted, S.; Schjoerring, J.K.; Kim, Y.S.; Jeon, U.S.; Kim, Y.K.; Kakei, Y.; Masuda, H.; et al. Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 2011, 9, 865–873. [Google Scholar] [CrossRef]
- Broadley, M.; Brown, P.; Cakmak, I.; Rengel, Z.; Zhao, F. Function of Nutrients: Micronutrients. In Marschner’s Mineral Nutrition of Higher Plants; Marschner, P., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 191–248. ISBN 9780123849052. [Google Scholar]
- Lin, Y.F.; Aarts, M.G.M. The molecular mechanism of zinc and cadmium stress response in plants. Cell. Mol. Life Sci. 2012, 69, 3187–3206. [Google Scholar] [CrossRef]
- Darbani, B.; Noeparvar, S.; Borg, S. Deciphering mineral homeostasis in barley seed transfer cells at transcriptional level. PLoS ONE 2015, 10, e0141398. [Google Scholar] [CrossRef]
- Uraguchi, S.; Fujiwara, T. Cadmium transport and tolerance in rice: Perspectives for reducing grain cadmium accumulation. Rice 2012, 5, 1–8. [Google Scholar] [CrossRef]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Tauris, B.; Borg, S.; Gregersen, P.L.; Holm, P.B. A roadmap for zinc trafficking in the developing barley grain based on laser capture microdissection and gene expression profiling. J. Exp. Bot. 2009, 60, 1333–1347. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawakami, Y.; Bhullar, N.K. Potential Implications of Interactions between Fe and S on Cereal Fe Biofortification. Int. J. Mol. Sci. 2020, 21, 2827. https://doi.org/10.3390/ijms21082827
Kawakami Y, Bhullar NK. Potential Implications of Interactions between Fe and S on Cereal Fe Biofortification. International Journal of Molecular Sciences. 2020; 21(8):2827. https://doi.org/10.3390/ijms21082827
Chicago/Turabian StyleKawakami, Yuta, and Navreet K. Bhullar. 2020. "Potential Implications of Interactions between Fe and S on Cereal Fe Biofortification" International Journal of Molecular Sciences 21, no. 8: 2827. https://doi.org/10.3390/ijms21082827
APA StyleKawakami, Y., & Bhullar, N. K. (2020). Potential Implications of Interactions between Fe and S on Cereal Fe Biofortification. International Journal of Molecular Sciences, 21(8), 2827. https://doi.org/10.3390/ijms21082827



