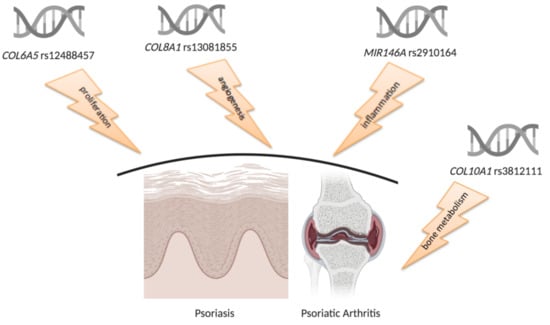
RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects
Abstract
1. Introduction
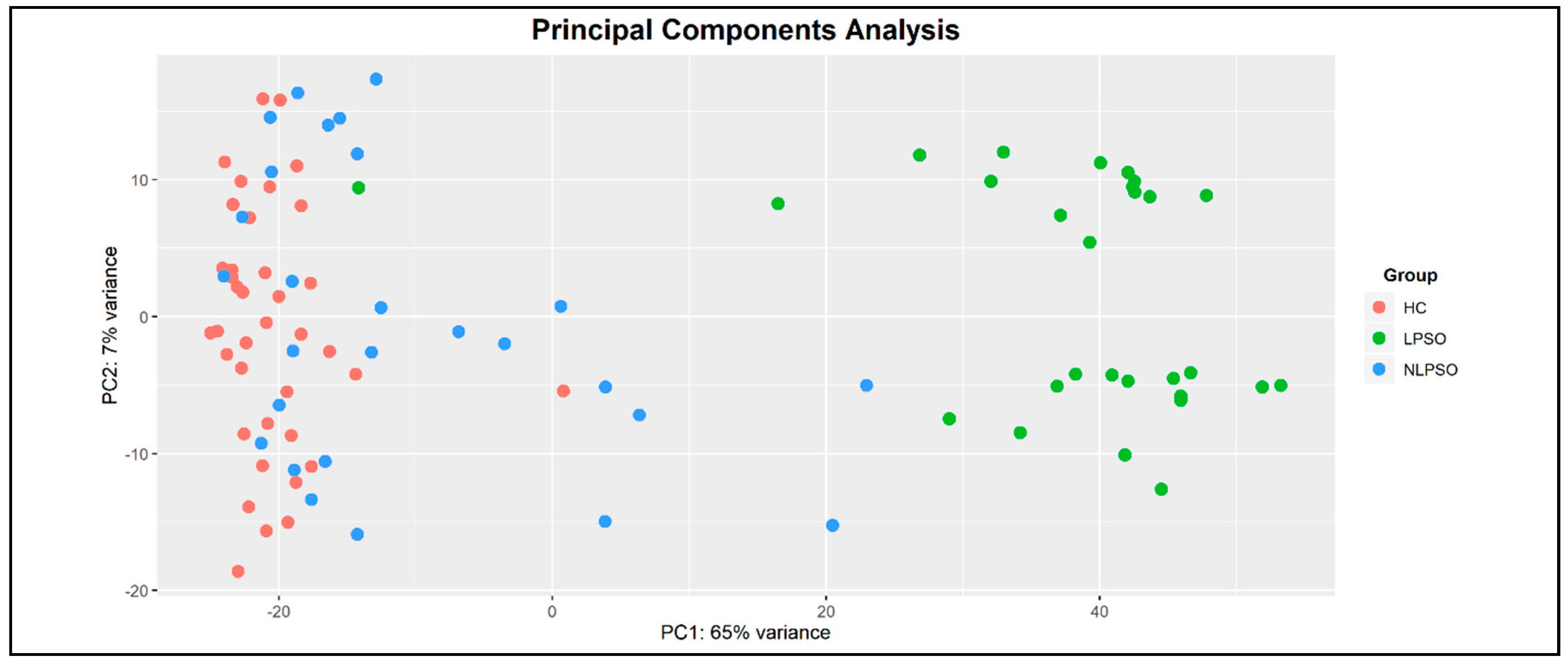
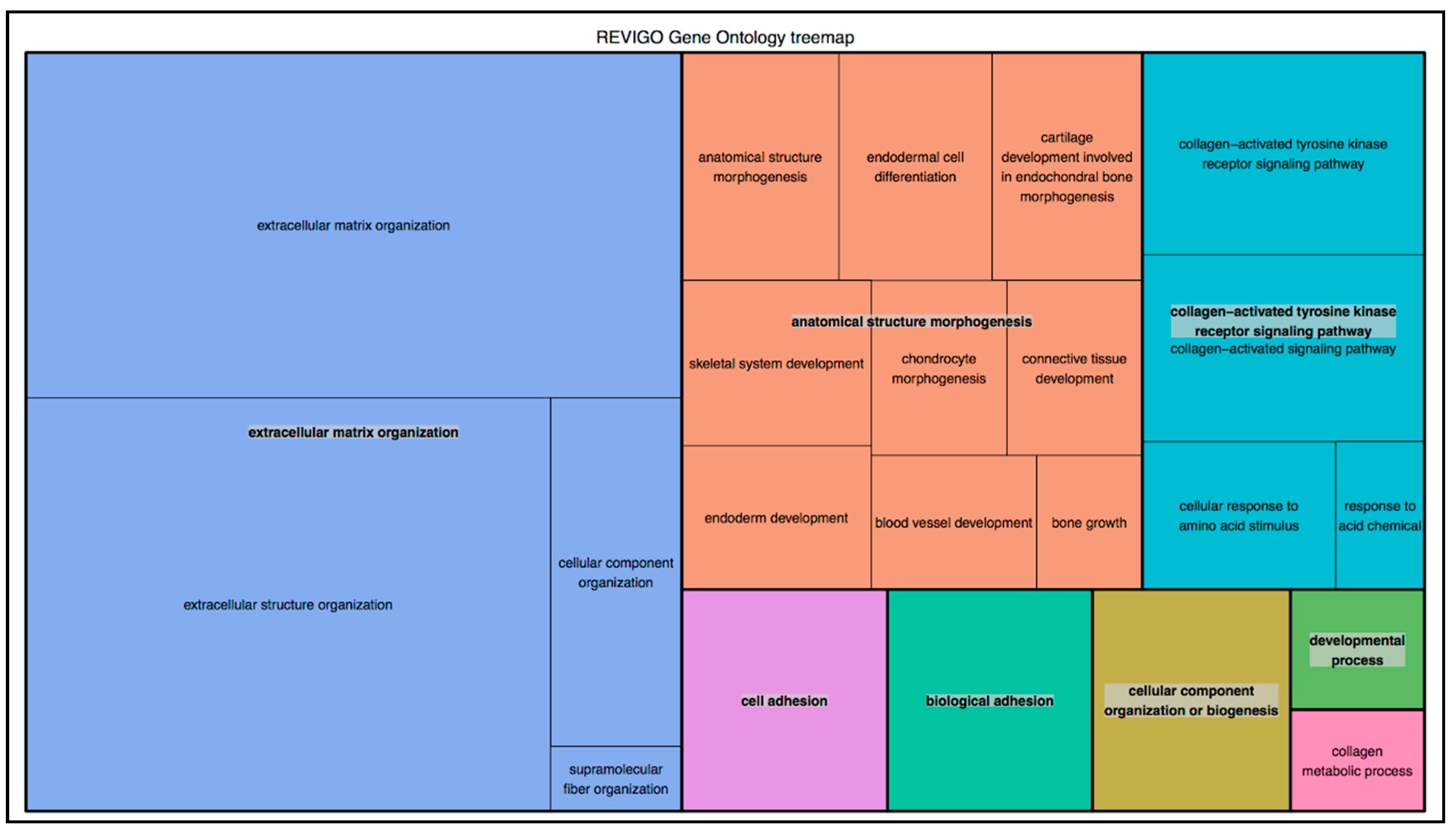
2. Results
3. Discussion
4. Materials and Methods
4.1. Prioritization of Collagen Genes
4.2. Study Cohorts
4.3. DNA Extraction and Genotyping Analysis
4.4. Biostatistical Analysis
4.5. Bioinformatic Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Giardina, E.; Sinibaldi, C.; Novelli, G. Mapping the future of common diseases: Lessons from psoriasis. Front. Biosci. 2007, 12, 1563–1573. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zeng, J.; Luo, S.; Huang, Y.; Lu, Q. Critical role of environmental factors in the pathogenesis of psoriasis. J. Dermatol. 2017, 44, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Cascella, R.; Strafella, C.; Longo, G.; Maccarone, M.; Borgiani, P.; Sangiuolo, F.; Novelli, G.; Giardina, E. Pharmacogenomics of multifactorial diseases: A focus on psoriatic arthritis. Pharmacogenomics 2016, 17, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Diani, M.; Altomare, G.; Reali, E. T cell responses in psoriasis and psoriatic arthritis. Autoimmun. Rev. 2015, 14, 286–292. [Google Scholar] [CrossRef]
- Coates, L.C.; Helliwell, P.S. Psoriatic arthritis: State of the art review. Clin. Med. (Lond.) 2017, 17, 65–70. [Google Scholar] [CrossRef] [PubMed]
- O’Rielly, D.D.; Rahman, P. Genetic, Epigenetic and Pharmacogenetic Aspects of Psoriasis and Psoriatic Arthritis. Rheum. Dis. Clin. N. Am. 2015, 41, 623–642. [Google Scholar] [CrossRef]
- Cascella, R.; Strafella, C.; Ragazzo, M.; Manzo, L.; Costanza, G.; Bowes, J.; Hüffmeier, U.; Potenza, S.; Sangiuolo, F.; Reis, A.; et al. KIF3A and IL-4 are disease-specific biomarkers for psoriatic arthritis susceptibility. Oncotarget 2017, 8, 95401–95411. [Google Scholar] [CrossRef]
- Pivarcsi, A.; Ståhle, M.; Sonkoly, E. Genetic polymorphisms altering microRNA activity in psoriasis—A key to solve the puzzle of missing heritability? Exp. Dermatol. 2014, 23, 620–624. [Google Scholar] [CrossRef]
- Ciancio, G.; Ferracin, M.; Saccenti, E.; Bagnari, V.; Farina, I.; Furini, F.; Galuppi, E.; Zagatti, B.; Trotta, F.; Negrini, M.; et al. Characterisation of peripheral blood mononuclear cell microRNA in early onset psoriatic arthritis. Clin. Exp. Rheumatol. 2017, 35, 113–121. [Google Scholar]
- Jobling, R.; D’Souza, R.; Baker, N.; Lara-Corrales, I.; Mendoza-Londono, R.; Dupuis, L.; Savarirayan, R.; Ala-Kokko, L.; Kannu, P. The collagenopathies: Review of clinical phenotypes and molecular correlations. Curr. Rheumatol. Rep. 2014, 16, 394. [Google Scholar] [CrossRef]
- Arseni, L.; Lombardi, A.; Orioli, D. From Structure to Phenotype: Impact of Collagen Alterations on Human Health. Int. J. Mol. Sci. 2018, 19, 1407. [Google Scholar] [CrossRef] [PubMed]
- Janeczko, Ł.; Janeczko, M.; Chrzanowski, R.; Zieliński, G. The role of polymorphisms of genes encoding collagen IX and XI in lumbar disc disease. Neurologia Neurochirurgia Polska 2014, 48, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Cascella, R.; Strafella, C.; Longo, G.; Ragazzo, M.; Manzo, L.; De Felici, C.; Errichiello, V.; Caputo, V.; Viola, F.; Eandi, C.M.; et al. Uncovering genetic and non-genetic biomarkers specific for exudative age-related macular degeneration: Significant association of twelve variants. Oncotarget 2018, 9, 7812–7821. [Google Scholar] [CrossRef] [PubMed]
- Strafella, C.; Caputo, V.; Minozzi, G.; Milano, F.; Arcangeli, M.; Sobhy, N.; Abdelmaksood, R.; Hashad, D.; Vakirlis, E.; Novelli, G.; et al. Atopic Eczema: Genetic Analysis of COL6A5, COL8A1, and COL10A1 in Mediterranean Populations. Biomed. Res. Int. 2019, 2019, 3457898. [Google Scholar] [CrossRef]
- Strafella, C.; Errichiello, V.; Caputo, V.; Aloe, G.; Ricci, F.; Cusumano, A.; Novelli, G.; Giardina, E.; Cascella, R. The Interplay between miRNA-Related Variants and Age-Related Macular Degeneration: EVIDENCE of Association of MIR146A and MIR27A. Int. J. Mol. Sci. 2019, 20, 1578. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Rodriguez, E.; Degenhardt, F.; Baurecht, H.; Wehkamp, U.; Volks, N.; Szymczak, S.; Swindell, W.R.; Sarkar, M.K.; Raja, K.; et al. Atopic Dermatitis Is an IL-13-Dominant Disease with Greater Molecular Heterogeneity Compared to Psoriasis. J. Investig. Dermatol. 2019, 139, 1480–1489. [Google Scholar] [CrossRef]
- Stallcup, W.B. NG2 Proteoglycan Enhances Brain Tumor Progression by Promoting Beta-1 Integrin Activation in both Cis and Trans Orientations. Cancers (Basel) 2017, 9, 31. [Google Scholar] [CrossRef]
- Cascella, R.; Strafella, C.; Caputo, V.; Errichiello, V.; Zampatti, S.; Milano, F.; Potenza, S.; Mauriello, S.; Novelli, G.; Ricci, F.; et al. Towards the application of precision medicine in Age-Related Macular Degeneration. Prog. Retin. Eye Res. 2018, 63, 132–146. [Google Scholar] [CrossRef]
- Gupta, S.; Kaur, M.; Gupta, R.; Singh, S.; Pant, L.; Singh, P.P. Dermal vasculature in psoriasis and psoriasiform dermatitis: A morphometric study. Indian J. Dermatol. 2011, 56, 647–649. [Google Scholar]
- Belasco, J.; Wei, N. Psoriatic Arthritis: What is Happening at the Joint? Rheumatol. Ther. 2019, 6, 305–315. [Google Scholar] [CrossRef]
- Gudmann, N.S.; Munk, H.L.; Christensen, A.F.; Ejstrup, L.; Sørensen, G.L.; Loft, A.G.; Karsdal, M.A.; Bay-Jensen, A.-C.; He, Y.; Siebuhr, A.S.; et al. Chondrocyte activity is increased in psoriatic arthritis and axial spondyloarthritis. Arthritis Res. Ther. 2016, 18, 141. [Google Scholar] [CrossRef]
- Gong, H.-B.; Zhang, S.-L.; Wu, X.-J.; Pu, X.-M.; Kang, X.-J. Association of rs2910164 polymorphism in MiR-146a gene with psoriasis susceptibility: A meta-analysis. Medicine (Baltimore) 2019, 98, e14401. [Google Scholar] [CrossRef]
- Srivastava, A.; Nikamo, P.; Lohcharoenkal, W.; Li, D.; Meisgen, F.; Xu Landén, N.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. MicroRNA-146a suppresses IL-17-mediated skin inflammation and is genetically associated with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Eder, L.; Chandran, V.; Pellett, F.; Pollock, R.; Shanmugarajah, S.; Rosen, C.F.; Rahman, P.; Gladman, D.D. IL13 gene polymorphism is a marker for psoriatic arthritis among psoriasis patients. Ann. Rheum. Dis. 2011, 70, 1594–1598. [Google Scholar] [CrossRef] [PubMed]
- Luan, L.; Shi, J.; Yu, Z.; Andl, T. The major miR-31 target genes STK40 and LATS2 and their implications in the regulation of keratinocyte growth and hair differentiation. Exp. Dermatol. 2017, 26, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Abreu-Velez, A.M.; Howard, M.S. Collagen IV in Normal Skin and in Pathological Processes. N. Am. J. Med. Sci. 2012, 4, 1–8. [Google Scholar] [CrossRef]
- Zuo, X.; Sun, L.; Yin, X.; Gao, J.; Sheng, Y.; Xu, J.; Zhang, J.; He, C.; Qiu, Y.; Wen, G.; et al. Whole-exome SNP array identifies 15 new susceptibility loci for psoriasis. Nat. Commun. 2015, 6, 6793. [Google Scholar] [CrossRef]
- Sonkoly, E.; Ståhle, M.; Pivarcsi, A. MicroRNAs: Novel regulators in skin inflammation. Clin. Exp. Dermatol. 2008, 33, 312–315. [Google Scholar] [CrossRef]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, F.; Wei, M.; Qiu, Y.; Ma, C.; Shen, L.; Huang, Y. Chronic constriction injury-induced microRNA-146a-5p alleviates neuropathic pain through suppression of IRAK1/TRAF6 signaling pathway. J. Neuroinflamm. 2018, 15, 179. [Google Scholar] [CrossRef]
- Strafella, C.; Caputo, V.; Galota, M.R.; Zampatti, S.; Marella, G.; Mauriello, S.; Cascella, R.; Giardina, E. Application of Precision Medicine in Neurodegenerative Diseases. Front. Neurol. 2018, 9, 701. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Taylor, W.; Gladman, D.; Helliwell, P.; Marchesoni, A.; Mease, P.; Mielants, H.; CASPAR Study Group. Classification criteria for psoriatic arthritis: Development of new criteria from a large international study. Arthritis Rheum. 2006, 54, 2665–2673. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 7, 1–41. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2012, 41, D344–D347. [Google Scholar] [CrossRef]
- Desmet, F.-O.; Hamroun, D.; Lalande, M.; Collod-Béroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42, D86–D91. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]

| Gene | SNP | Allele Frequencies (Ps) | Allele Frequencies (PsA) | Allele Frequencies (Italian Controls) | AMR Allele Frequencies | EAS Allele Frequencies | EUR Allele Frequencies | AFR Allele Frequencies |
|---|---|---|---|---|---|---|---|---|
| COL6A5 | rs12488457 A/C | A: 0.31 | A: 0.35 | A: 0.43 | A: 0.57 | A: 0.83 | A: 0.28 | A: 0.95 |
| C: 0.69 | C: 0.65 | C: 0.57 | C: 0.43 | C: 0.17 | C: 0.72 | C: 0.05 | ||
| COL8A1 | rs13081855 G/T | G: 0.90 | G: 0.89 | G: 0.95 | G: 0.95 | G: 0.95 | G: 0.90 | G: 0.97 |
| T: 0.10 | T: 0.11 | T: 0.05 | T: 0.05 | T: 0.05 | T: 0.10 | T: 0.03 | ||
| COL10A1 | rs3812111 A/T | A: 0.37 | A: 0.34 | A: 0.40 | A: 0.33 | A: 0.26 | A: 0.40 | A: 0.74 |
| T: 0.63 | T: 0.66 | T: 0.60 | T: 0.67 | T: 0.74 | T: 0.60 | T: 0.26 | ||
| MIR146A | rs2910164 C/G | C: 0.26 | C: 0.25 | C: 0.30 | C: 0.31 | C: 0.63 | * C: 0.23 | C: 0.40 |
| G: 0.74 | G: 0.75 | G: 0.70 | G: 0.69 | G: 0.37 | * G: 0.77 | G: 0.60 |
| Gene | SNP | Disease | p | OR (95% C.I.) |
|---|---|---|---|---|
| COL6A5 | rs12488457 A/C | Ps | 1.39 × 10−8 | C: 1.74 (1.43–2.10) |
| PsA | 5.12 × 10−5 | C: 1.46 (1.21–1.75) | ||
| COL8A1 | rs13081855 G/T | Ps | 4.52 × 10−4 | T: 1.85 (1.31–2.64) |
| PsA | 1.19 × 10−6 | T: 2.24 (1.61–3.12) | ||
| COL10A1 | rs3812111 A/T | Ps | ns | - |
| PsA | 0.005 | T: 1.30 (1.08–1.56) | ||
| MIR146A | rs2910164 C/G | Ps | 0.04 | G: 1.23 (1.00–1.51) |
| PsA | 0.01 | G: 1.29 (1.04–1.61) |
| Gene | SNP | Effect Allele | Predicted Functional Effect | Target Gene | Gene Molecular Function |
|---|---|---|---|---|---|
| MIR146A | rs2910164 C/G | G | Target sites disrupted | IL13 | Cytokine signaling |
| TNFAIP3 | Cytokine signaling | ||||
| STK40 | Keratinocyte proliferation | ||||
| KIF3A | Bone metabolism | ||||
| Target sites created | COL4A3 | ECM remodeling | |||
| LCE3D | Epidermal barrier |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caputo, V.; Strafella, C.; Termine, A.; Campione, E.; Bianchi, L.; Novelli, G.; Giardina, E.; Cascella, R. RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects. Int. J. Mol. Sci. 2020, 21, 2740. https://doi.org/10.3390/ijms21082740
Caputo V, Strafella C, Termine A, Campione E, Bianchi L, Novelli G, Giardina E, Cascella R. RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects. International Journal of Molecular Sciences. 2020; 21(8):2740. https://doi.org/10.3390/ijms21082740
Chicago/Turabian StyleCaputo, Valerio, Claudia Strafella, Andrea Termine, Elena Campione, Luca Bianchi, Giuseppe Novelli, Emiliano Giardina, and Raffaella Cascella. 2020. "RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects" International Journal of Molecular Sciences 21, no. 8: 2740. https://doi.org/10.3390/ijms21082740
APA StyleCaputo, V., Strafella, C., Termine, A., Campione, E., Bianchi, L., Novelli, G., Giardina, E., & Cascella, R. (2020). RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects. International Journal of Molecular Sciences, 21(8), 2740. https://doi.org/10.3390/ijms21082740