Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis
Abstract
1. Introduction
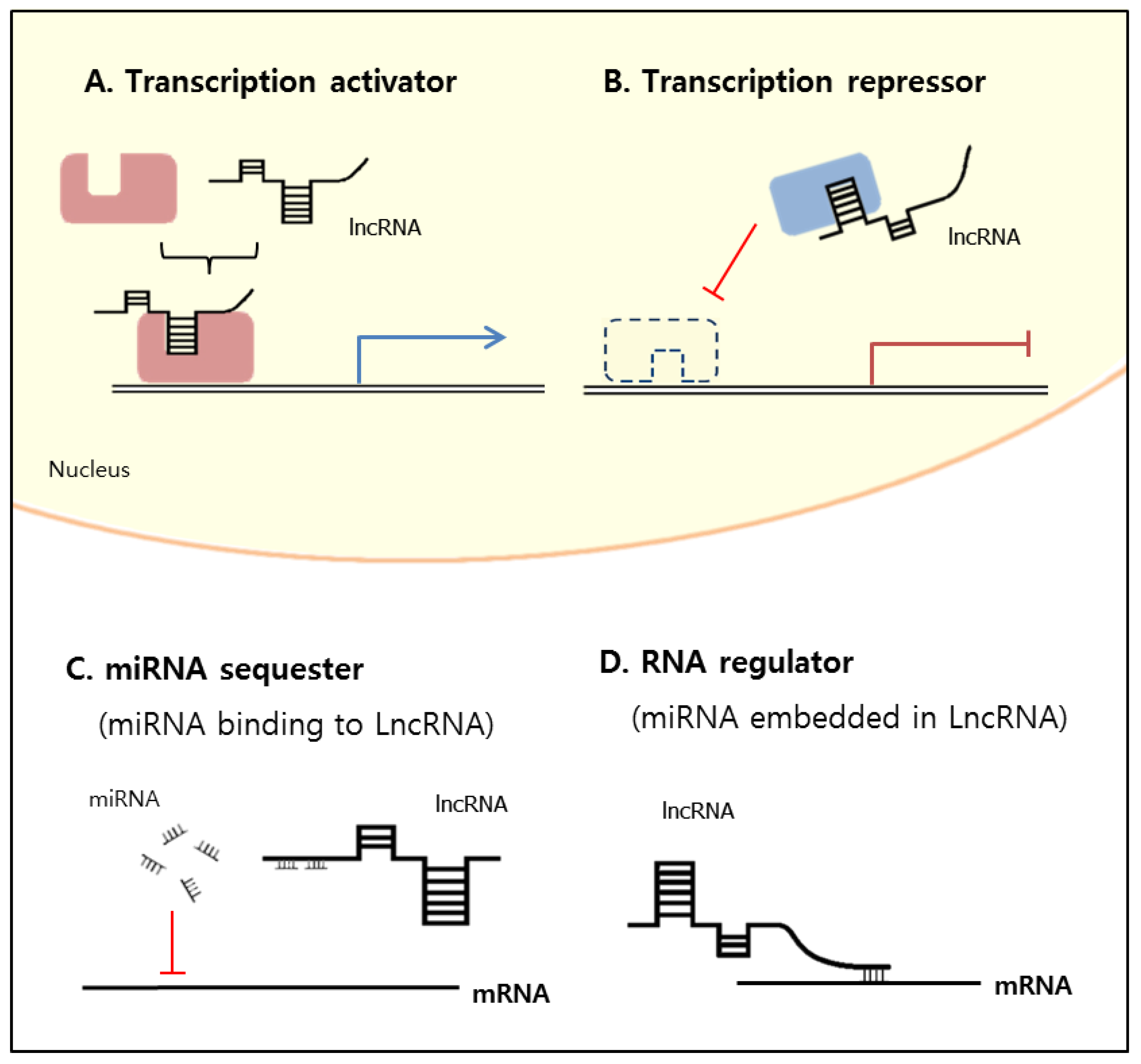
2. Classification and Mechanisms of lncRNA Functions
3. LncRNAs Related to Renal Fibrosis
3.1. Arid2-IR
3.2. CHCHD4P4 (+EMT)
3.3. CYP4B1-PS1-001
3.4. ENSMUST00000147869
3.5. Erbb4-IR
3.6. H19
3.7. HOTAIR
3.8. LINC00936
3.9. LncRNA-ATB
3.10. MALAT1
3.11. MIAT
3.12. NEAT1
3.13. RANTES
3.14. PVT1
3.15. TapSAKI
3.16. Tug1
3.17. Xist
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kapranov, P.; St Laurent, G.; Raz, T.; Ozsolak, F.; Reynolds, C.P.; Sorensen, P.H.; Reaman, G.; Milos, P.; Arceci, R.J.; Thompson, J.F.; et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ’dark matter’ un-annotated RNA. BMC Biol. 2010, 8, 149. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Guttman, M.; Russell, P.; Ingolia, N.T.; Weissman, J.S.; Lander, E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013, 154, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Liu, Y.; Jiang, L.; Zeng, Y.; Tang, W. High expression of long non-coding RNA lncRNA-ATB is correlated with metastases and promotes cell migration and invasion in renal cell carcinoma. Jpn. J. Clin. Oncol. 2016, 46, 378–384. [Google Scholar] [CrossRef]
- Nguyen, Q.; Carninci, P. Expression Specificity of Disease-Associated lncRNAs: Toward Personalized Medicine. Curr. Top. Microbiol. Immunol. 2016, 394, 237–258. [Google Scholar]
- Maass, P.G.; Luft, F.C.; Bahring, S. Long non-coding RNA in health and disease. J. Mol. Med. 2014, 92, 337–346. [Google Scholar] [CrossRef]
- Xie, H.; Xue, J.D.; Chao, F.; Jin, Y.F.; Fu, Q. Long non-coding RNA-H19 antagonism protects against renal fibrosis. Oncotarget 2016, 7, 51473–51481. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Tang, P.M.; Tang, P.C.; Chung, J.Y.; Lan, H.Y. TGF-beta1 signaling in kidney disease: From Smads to long non-coding RNAs. Non-Coding RNA Res. 2017, 2, 68–73. [Google Scholar] [CrossRef]
- Kumar, M.M.; Goyal, R. LncRNA as a Therapeutic Target for Angiogenesis. Curr. Top. Med. Chem. 2017, 17, 1750–1757. [Google Scholar] [CrossRef]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120 Pt 15, 2498–2506. [Google Scholar] [CrossRef]
- Hajjari, M.; Khoshnevisan, A.; Shin, Y.K. Molecular function and regulation of long non-coding RNAs: Paradigms with potential roles in cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 10645–10663. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Pan, H.F.; Leng, R.X.; Wang, D.G.; Li, X.P.; Li, X.M.; Ye, D.Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015, 14, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Ounzain, S.; Burdet, F.; Ibberson, M.; Pedrazzini, T. Discovery and functional characterization of cardiovascular long noncoding RNAs. J. Mol. Cell. Cardiol. 2015, 89 Pt A, 17–26. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Thum, T. Long noncoding RNAs in kidney and cardiovascular diseases. Nat. Rev. Nephrol. 2016, 12, 360–373. [Google Scholar] [CrossRef]
- Ignarski, M.; Islam, R.; Müller, R.-U. Long non-coding RNAs in kidney disease. Int. J. Mol. Sci. 2019, 20, 3276. [Google Scholar] [CrossRef]
- Van der Hauwaert, C.; Glowacki, F.; Pottier, N.; Cauffiez, C. Non-Coding RNAs as New Therapeutic Targets in the Context of Renal Fibrosis. Int. J. Mol. Sci. 2019, 20, 1977. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Y.; Shu, S.; Cai, J.; Tang, C.; Dong, Z. Non-coding RNAs in kidney injury and repair. Am. J. Physiol. Cell Physiol. 2019, 317, C177–C188. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Hsieh, H.Y.; Shih, H.M.; Sytwu, H.K.; Wu, C.C. Urinary Xist is a potential biomarker for membranous nephropathy. Biochem. Biophys. Res. Commun. 2014, 452, 415–421. [Google Scholar] [CrossRef]
- Niu, Z.S.; Niu, X.J.; Wang, W.H. Long non-coding RNAs in hepatocellular carcinoma: Potential roles and clinical implications. World J. Gastroenterol. 2017, 23, 5860–5874. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Huang, X.R.; Yu, J.; Yu, X.; Lan, H.Y. Long Noncoding RNA Arid2-IR Is a Novel Therapeutic Target for Renal Inflammation. Mol. Ther. J. Am. Soc. Gene Ther. 2015, 23, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chung, A.C.; Huang, X.R.; Dong, Y.; Yu, X.; Lan, H.Y. Identification of novel long noncoding RNAs associated with TGF-beta/Smad3-mediated renal inflammation and fibrosis by RNA sequencing. Am. J. Pathol. 2014, 184, 409–417. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15 (Suppl. 1), R17–R29. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Pan, Y.; Han, S.; Feng, B.; Gao, Y.; Chen, J.; Zhang, K.; Wang, R.; Chen, L. The Emerging Role and Promise of Long Noncoding RNAs in Lung Cancer Treatment. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2016, 38, 2194–2206. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 2014, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Saayman, S.; Ackley, A.; Turner, A.W.; Famiglietti, M.; Bosque, A.; Clemson, M.; Planelles, V.; Morris, K.V. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 1164–1175. [Google Scholar] [CrossRef]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, L.Q.; Lu, W.W.; Zhang, J.; Zhu, J.S. LncRNAs and cancer. Oncol. Lett. 2016, 12, 1233–1239. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, S. Long noncoding RNAs in cell differentiation and pluripotency. Cell Tissue Res. 2016, 366, 509–521. [Google Scholar] [CrossRef]
- Quan, M.; Chen, J.; Zhang, D. Exploring the secrets of long noncoding RNAs. Int. J. Mol. Sci. 2015, 16, 5467–5496. [Google Scholar] [CrossRef]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Arai, S.; Song, X.; Reichart, D.; Du, K.; Pascual, G.; Tempst, P.; Rosenfeld, M.G.; Glass, C.K.; Kurokawa, R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 2008, 454, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, J.H.; Spector, D.L. Long non-coding RNAs: Modulators of nuclear structure and function. Curr. Opin. Cell Biol. 2014, 26, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Tian, J.; Tang, X.; Ma, J.; Wang, S. Long non-coding RNAs in the regulation of myeloid cells. J. Hematol. Oncol. 2016, 9, 99. [Google Scholar] [CrossRef]
- van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef]
- Moghaddas Sani, H.; Hejazian, M.; Hosseinian Khatibi, S.M.; Ardalan, M.; Zununi Vahed, S. Long non-coding RNAs: An essential emerging field in kidney pathogenesis. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 99, 755–765. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Schauerte, C.; Kolling, M.; Hubner, A.; Knapp, M.; Haller, H.; Thum, T. Long Noncoding RNAs in Urine Are Detectable and May Enable Early Detection of Acute T Cell-Mediated Rejection of Renal Allografts. Clin. Chem. 2015, 61, 1505–1514. [Google Scholar] [CrossRef]
- Chen, W.; Peng, W.; Huang, J.; Yu, X.; Tan, K.; Chen, Y.; Lin, X.; Chen, D.; Dai, Y. Microarray analysis of long non-coding RNA expression in human acute rejection biopsy samples following renal transplantation. Mol. Med. Rep. 2014, 10, 2210–2216. [Google Scholar] [CrossRef]
- Sui, W.; Lin, H.; Peng, W.; Huang, Y.; Chen, J.; Zhang, Y.; Dai, Y. Molecular dysfunctions in acute rejection after renal transplantation revealed by integrated analysis of transcription factor, microRNA and long noncoding RNA. Genomics 2013, 102, 310–322. [Google Scholar] [CrossRef]
- Donate-Correa, J.; Martin-Nunez, E.; Muros-de-Fuentes, M.; Mora-Fernandez, C.; Navarro-Gonzalez, J.F. Inflammatory cytokines in diabetic nephropathy. J. Diabetes Res. 2015, 2015, 948417. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.F.; Tang, P.M.K.; Feng, M.; Xiao, J.; Huang, X.R.; Li, P.; Ma, R.C.W.; Lan, H.Y. Novel lncRNA Erbb4-IR Promotes Diabetic Kidney Injury in db/db Mice by Targeting miR-29b. Diabetes 2018, 67, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yao, D.; Wang, S.; Yan, Q.; Lu, W. Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine 2016, 54, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Inazaki, K.; Kanamaru, Y.; Kojima, Y.; Sueyoshi, N.; Okumura, K.; Kaneko, K.; Yamashiro, Y.; Ogawa, H.; Nakao, A. Smad3 deficiency attenuates renal fibrosis, inflammation, and apoptosis after unilateral ureteral obstruction. Kidney Int. 2004, 66, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Huang, X.R.; Lan, H.Y. Smad3 mediates ANG II-induced hypertensive kidney disease in mice. Am. J. Physiol. Ren. Physiol. 2012, 302, F986–F997. [Google Scholar] [CrossRef]
- Yang, X.; Letterio, J.J.; Lechleider, R.J.; Chen, L.; Hayman, R.; Gu, H.; Roberts, A.B.; Deng, C. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 1999, 18, 1280–1291. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, J.; Hu, H.; Chen, W.; Liu, M.; Zhang, J.; Sun, S.; Guo, Z. Long non-coding RNA CHCHD4P4 promotes epithelial-mesenchymal transition and inhibits cell proliferation in calcium oxalate-induced kidney damage. Braz. J. Med. Biol. Res. 2017, 51, e6536. [Google Scholar] [CrossRef]
- Wang, M.; Wang, S.; Yao, D.; Yan, Q.; Lu, W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol. Cell. Endocrinol. 2016, 426, 136–145. [Google Scholar] [CrossRef]
- Feng, M.; Tang, P.M.; Huang, X.R.; Sun, S.F.; You, Y.K.; Xiao, J.; Lv, L.L.; Xu, A.P.; Lan, H.Y. TGF-beta Mediates Renal Fibrosis via the Smad3-Erbb4-IR Long Noncoding RNA Axis. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 148–161. [Google Scholar] [CrossRef]
- Kanwar, Y.S.; Pan, X.; Lin, S.; Kumar, A.; Wada, J.; Haas, C.S.; Liau, G.; Lomasney, J.W. Imprinted mesodermal specific transcript (MEST) and H19 genes in renal development and diabetes. Kidney Int. 2003, 63, 1658–1670. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okamoto, K.; Morison, I.M.; Taniguchi, T.; Reeve, A.E. Epigenetic changes at the insulin-like growth factor II/H19 locus in developing kidney is an early event in Wilms tumorigenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 5367–5371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, G.; Zhang, M.; Zhou, J.; Gao, W.; Xuan, X.; Yang, X.; Yang, D.; Tian, Z.; Ni, B.; et al. Critical effects of long non-coding RNA on fibrosis diseases. Exp. Mol. Med. 2018, 50, e428. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhang, M.; Fan, Z.; Sun, W.; Tang, Y. Influence of lncRNA HOTAIR on acute kidney injury in sepsis rats through regulating miR-34a/Bcl-2 pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3512–3519. [Google Scholar]
- Shen, J.; Zhang, J.; Jiang, X.; Wang, H.; Pan, G. LncRNA HOX transcript antisense RNA accelerated kidney injury induced by urine-derived sepsis through the miR-22/high mobility group box 1 pathway. Life Sci. 2018, 210, 185–191. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, L.; Yu, Z.H.; Hong, S.J.; Zhang, Z.W.; Qiu, Z.Z. LncRNA HOTAIR promotes renal interstitial fibrosis by regulating Notch1 pathway via the modulation of miR-124. Nephrology 2019, 24, 472–480. [Google Scholar] [CrossRef]
- Majumder, S.; Hadden, M.J.; Thieme, K.; Batchu, S.N.; Niveditha, D.; Chowdhury, S.; Yerra, V.G.; Advani, S.L.; Bowskill, B.B.; Liu, Y. Dysregulated expression but redundant function of the long non-coding RNA HOTAIR in diabetic kidney disease. Diabetologia 2019, 62, 2129–2142. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, L.; Zhou, Z.Q.; Ren, Y.Q.; Sun, L.N.; Man, Y.L.; Ma, Z.W.; Wang, Z.K. Effects of Long Non-Coding RNA LINC00963 on Renal Interstitial Fibrosis and Oxidative Stress of Rats with Chronic Renal Failure via the Foxo Signaling Pathway. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 815–828. [Google Scholar] [CrossRef]
- Qiu, J.; Chen, Y.; Huang, G.; Zhang, Z.; Chen, L.; Na, N. Transforming growth factor-beta activated long non-coding RNA ATB plays an important role in acute rejection of renal allografts and may impacts the postoperative pharmaceutical immunosuppression therapy. Nephrology 2017, 22, 796–803. [Google Scholar] [CrossRef]
- Liu, B.; Qiang, L.; Wang, G.D.; Duan, Q.; Liu, J. LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3478–3486. [Google Scholar] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Yao, J.; Li, X.M.; Song, Y.C.; Wang, X.Q.; Li, Y.J.; Yan, B.; Jiang, Q. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis. 2014, 5, e1506. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J. Cell. Mol. Med. 2017, 21, 2732–2747. [Google Scholar] [CrossRef]
- Yan, B.; Yao, J.; Liu, J.Y.; Li, X.M.; Wang, X.Q.; Li, Y.J.; Tao, Z.F.; Song, Y.C.; Chen, Q.; Jiang, Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 2015, 116, 1143–1156. [Google Scholar] [CrossRef]
- Bijkerk, R.; Au, Y.W.; Stam, W.; Duijs, J.; Koudijs, A.; Lievers, E.; Rabelink, T.J.; van Zonneveld, A.J. Long Non-coding RNAs Rian and Miat Mediate Myofibroblast Formation in Kidney Fibrosis. Front. Pharmacol. 2019, 10, 215. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, D.Y.; Sha, W.G.; Shen, L.; Lu, G.Y.; Yin, X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem. Biophys. Res. Commun. 2015, 468, 726–732. [Google Scholar] [CrossRef]
- Chen, Y.; Qiu, J.; Chen, B.; Lin, Y.; Chen, Y.; Xie, G.; Qiu, J.; Tong, H.; Jiang, D. Long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int. Immunopharmacol. 2018, 59, 252–260. [Google Scholar] [CrossRef]
- Jiang, X.; Li, D.; Shen, W.; Shen, X.; Liu, Y. LncRNA NEAT1 promotes hypoxia-induced renal tubular epithelial apoptosis through downregulating miR-27a-3p. J. Cell. Biochem. 2019, 120, 16273–16282. [Google Scholar] [CrossRef]
- Li, N.; Jia, T.; Li, Y. LncRNA NEAT1 accelerates the occurrence and development of diabetic nephropathy by sponging miR-23c. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1325–1337. [Google Scholar]
- Yu, T.M.; Palanisamy, K.; Sun, K.T.; Day, Y.J.; Shu, K.H.; Wang, I.K.; Shyu, W.C.; Chen, P.; Chen, Y.L.; Li, C.Y. RANTES mediates kidney ischemia reperfusion injury through a possible role of HIF-1alpha and LncRNA PRINS. Sci. Rep. 2016, 6, 18424. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.L.; Craig, D.W.; Millis, M.P.; Yeatts, K.A.; Kobes, S.; Pearson, J.V.; Lee, A.M.; Knowler, W.C.; Nelson, R.G.; Wolford, J.K. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 2007, 56, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Millis, M.P.; Bowen, D.; Kingsley, C.; Watanabe, R.M.; Wolford, J.K. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007, 56, 3027–3032. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.L.; DiStefano, J.K. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 2011, 6, e18671. [Google Scholar] [CrossRef]
- Chun-Mei, H.; Qin-Min, G.; Shu-Ming, P.; Xiang-Yang, Z. Expression profiling and ontology analysis of circulating long non-coding RNAs in septic acute kidney injury patients. Clin. Chem. Lab. Med. 2016, 54, e395–e399. [Google Scholar] [CrossRef]
- Zhou, P.; Chen, Z.; Zou, Y.; Wan, X. Roles of Non-Coding RNAs in Acute Kidney Injury. Kidney Blood Press. Res. 2016, 41, 757–769. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Schauerte, C.; Kielstein, J.T.; Hubner, A.; Martino, F.; Fiedler, J.; Gupta, S.K.; Faulhaber-Walter, R.; Kumarswamy, R.; Hafer, C.; et al. Circulating long noncoding RNATapSaki is a predictor of mortality in critically ill patients with acute kidney injury. Clin. Chem. 2015, 61, 191–201. [Google Scholar] [CrossRef]
- Kato, M.; Wang, M.; Chen, Z.; Bhatt, K.; Oh, H.J.; Lanting, L.; Deshpande, S.; Jia, Y.; Lai, J.Y.; O’Connor, C.L.; et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat. Commun. 2016, 7, 12864. [Google Scholar] [CrossRef]
- Li, S.Y.; Susztak, K. The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes. J. Clin. Investig. 2016, 126, 4072–4075. [Google Scholar] [CrossRef]
- Long, J.; Badal, S.S.; Ye, Z.; Wang, Y.; Ayanga, B.A.; Galvan, D.L.; Green, N.H.; Chang, B.H.; Overbeek, P.A.; Danesh, F.R. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Investig. 2016, 126, 4205–4218. [Google Scholar] [CrossRef]
- Zang, X.J.; Li, L.; Du, X.; Yang, B.; Mei, C.L. LncRNA TUG1 inhibits the proliferation and fibrosis of mesangial cells in diabetic nephropathy via inhibiting the PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7519–7525. [Google Scholar] [PubMed]
| LncRNA | Dysregulation | Related Disease/Experiment | Biological Role | Signal/Target Cell |
|---|---|---|---|---|
| Arid2-IR | Upregulation | UUO kidney | TGF-β/Smad3-associated renal fibrosis | NF-κB-dependent inflammation |
| CHCHD4P4 | Upregulation | Renal damage (fibrosis) | Epithelial-to mesenchymal transition | Tubular epithelial cell |
| CYP4B1-PS1-001 | Upregulation | Diabetic nephropathy | Regulation of fibrosis | Mesangial cell |
| ENSMUST00000147869 | Upregulation | Diabetic nephropathy | Decreased PCNA and cyclin D1 | Mesangial cell |
| Erbb4-IR | Upregulation | UUO kidney, diabetic nephropathy | Downregulation of Smad7 | TGF-β1/Smad3 signaling |
| H19 | Upregulation | UUO kidney | Downregulation of miR-17 expression | Tubular epithelial cell |
| HOTAIR | Upregulation | Acute kidney injury | Promotion of apoptosis | Notch1 pathway |
| LINC00936 | Downregulation | Chronic renal failure | Regulation of fibrosis | FoxO signaling |
| LncRNA-ATB | Upregulation | Acute rejection | Loss of kidney function | Activated by TGF-β1 |
| MALAT1 | Upregulation | Diabetic nephropathy | Translocation of β-catenin; SRSF1 overexpression | TGF-β1-mediated fibrosis |
| MIAT lncRNA | Upregulation | Diabetic nephropathy | Nrf2 regulation | Tubular epithelial cell |
| NEAT1 | Upregulation | Acute kidney disease, diabetic nephropathy | Proliferation of Mesangial cell | NF-κB pathway |
| PVT1 | Upregulation | Diabetic nephropathy | ECM accumulation | TGF-β1-mediated fibrosis |
| Rantes lncRNA | Upregulation | Acute kidney injury, ischemic reperfusion | Produced by HIF-1α | Tubular epithelial cell |
| TapSAKI | Upregulation | Acute kidney injury, hypoxia | Predictor of prognosis | Tubular epithelial cell |
| Tug1 | Upregulation | Diabetic nephropathy | Mitochondria-dependent mechanism | Podocyte |
| Xist | Upregulation | Glomerular disease | Urinary biomarker | Tubular epithelial and glomerular cell |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, H.J.; Kim, H.-J.; Park, K.-K. Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis. Int. J. Mol. Sci. 2020, 21, 2698. https://doi.org/10.3390/ijms21082698
Jung HJ, Kim H-J, Park K-K. Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis. International Journal of Molecular Sciences. 2020; 21(8):2698. https://doi.org/10.3390/ijms21082698
Chicago/Turabian StyleJung, Hyun Jin, Hyun-Ju Kim, and Kwan-Kyu Park. 2020. "Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis" International Journal of Molecular Sciences 21, no. 8: 2698. https://doi.org/10.3390/ijms21082698
APA StyleJung, H. J., Kim, H.-J., & Park, K.-K. (2020). Potential Roles of Long Noncoding RNAs as Therapeutic Targets in Renal Fibrosis. International Journal of Molecular Sciences, 21(8), 2698. https://doi.org/10.3390/ijms21082698




