Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis
Abstract
1. Introduction
2. Results
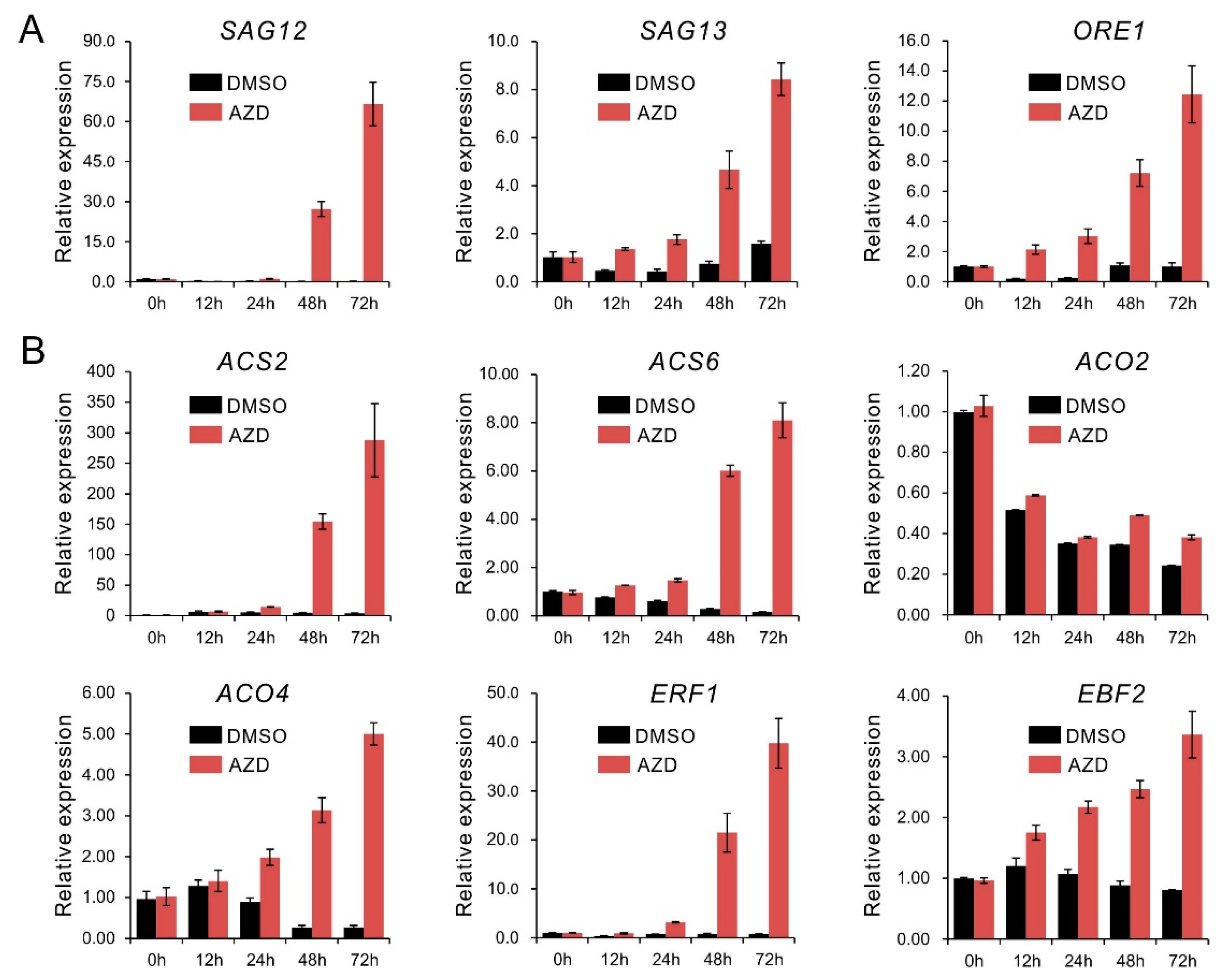
2.1. TOR Inhibition Upregulates Senescence- and Ethylene-Related Genes Expression
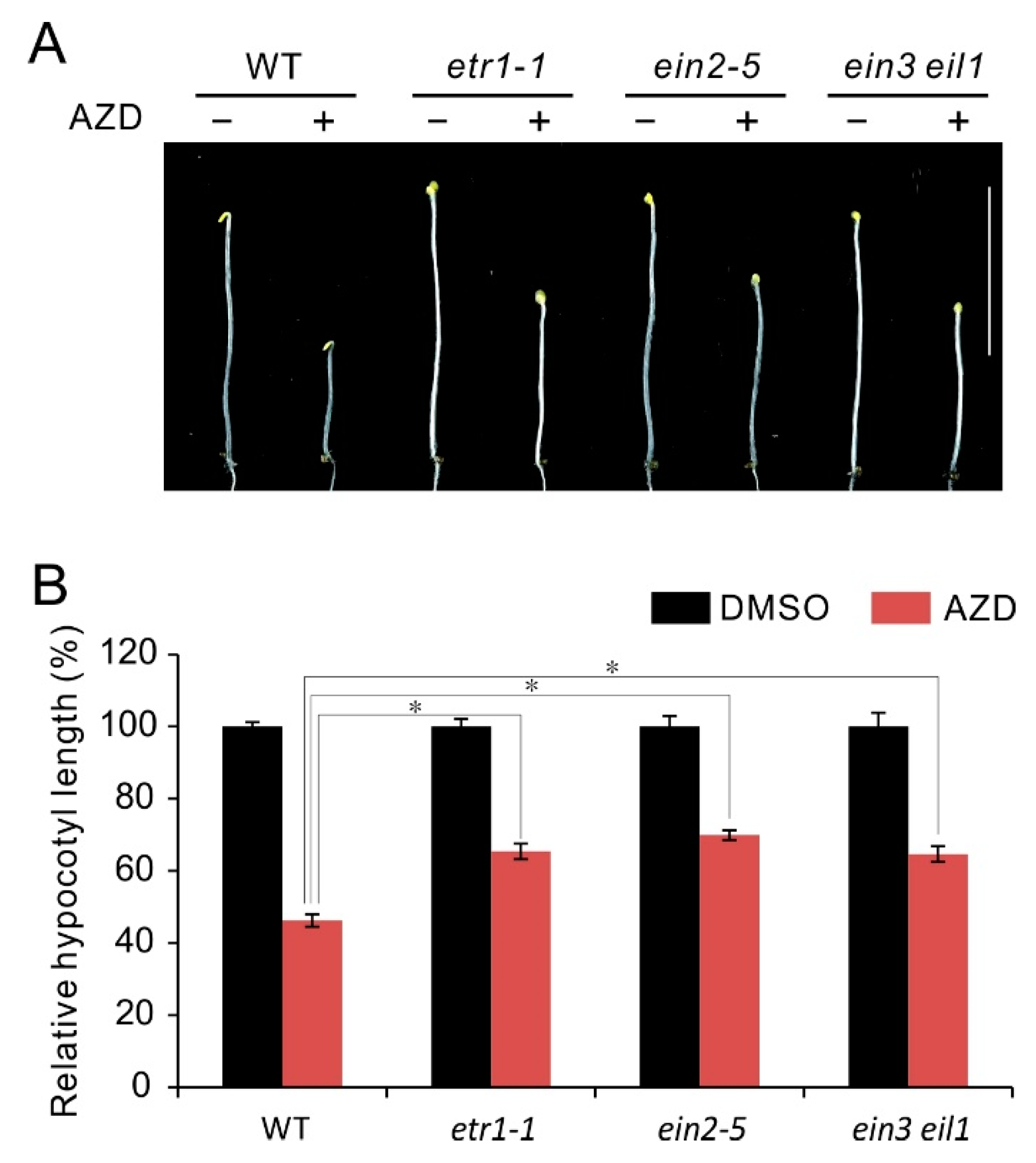
2.2. Ethylene Insensitive Mutants Display Hyposensitivity to AZD8055
2.3. Activated Ethylene Signal Is Required for Hypocotyl Growth Inhibition of TOR Inactivity
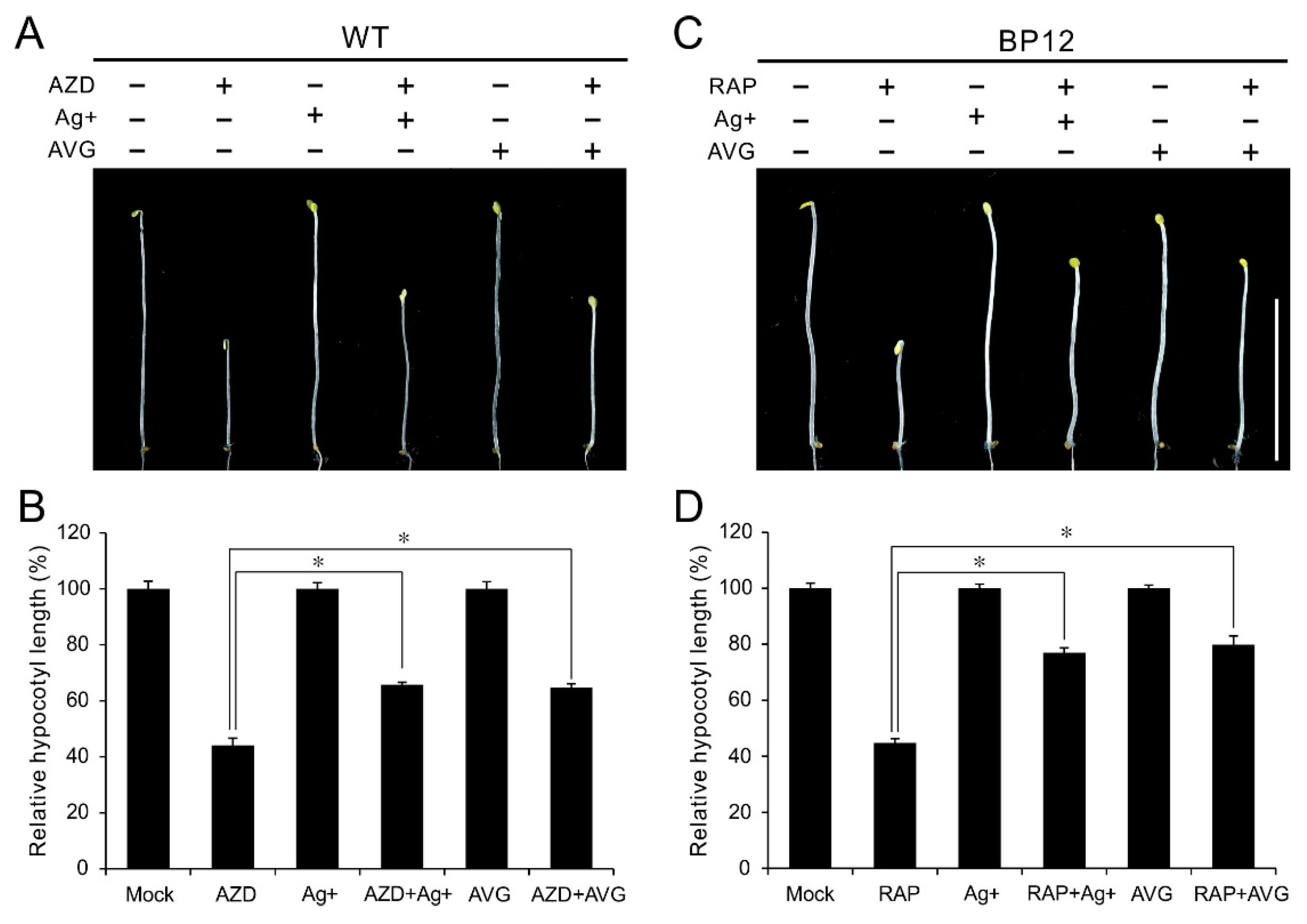
2.4. Ethylene Inhibitors Relieve Hypocotyl Growth Inhibition under TOR Inactivity
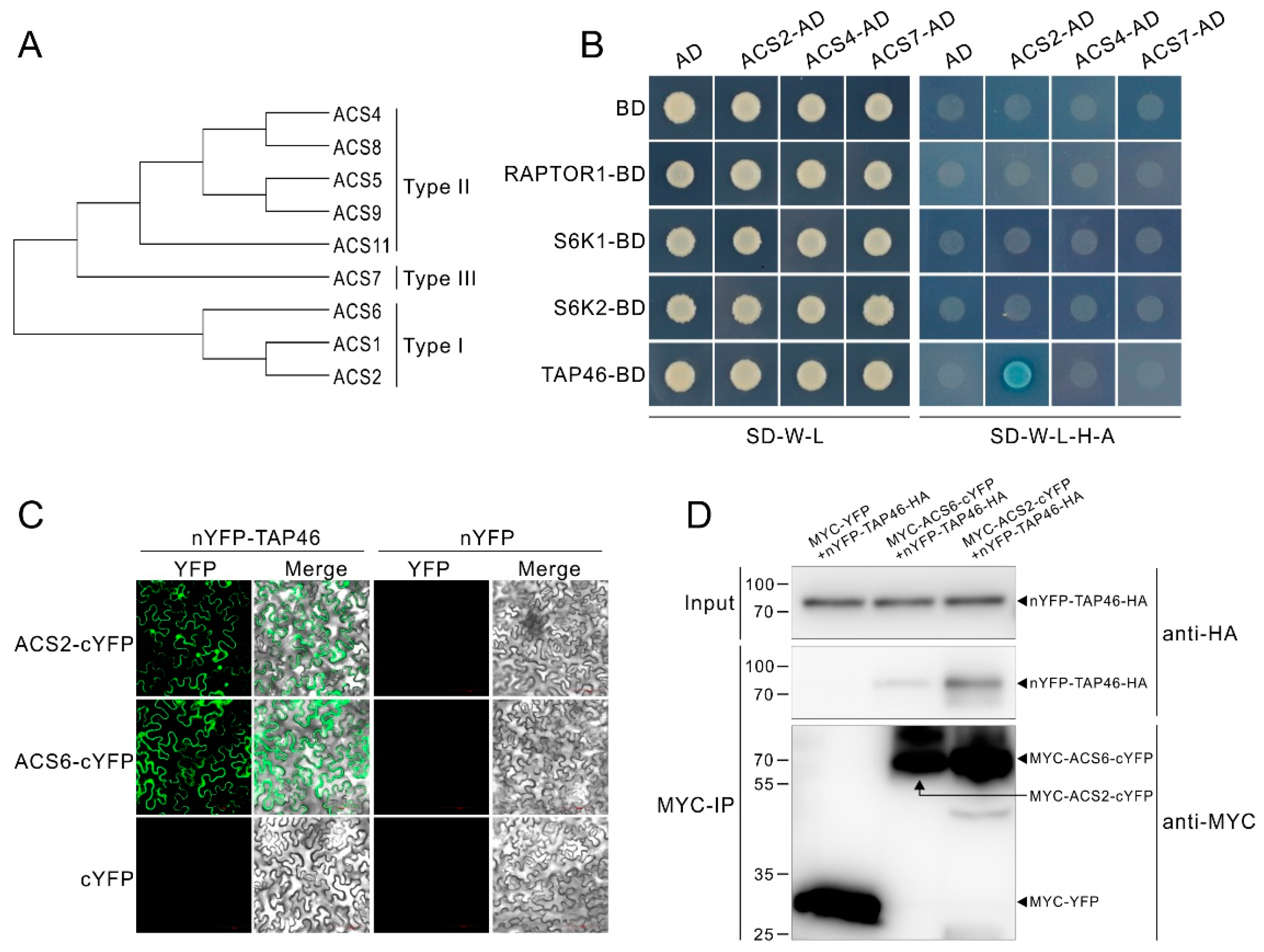
2.5. TAP46 Physically Interacts with ACS2/ACS6
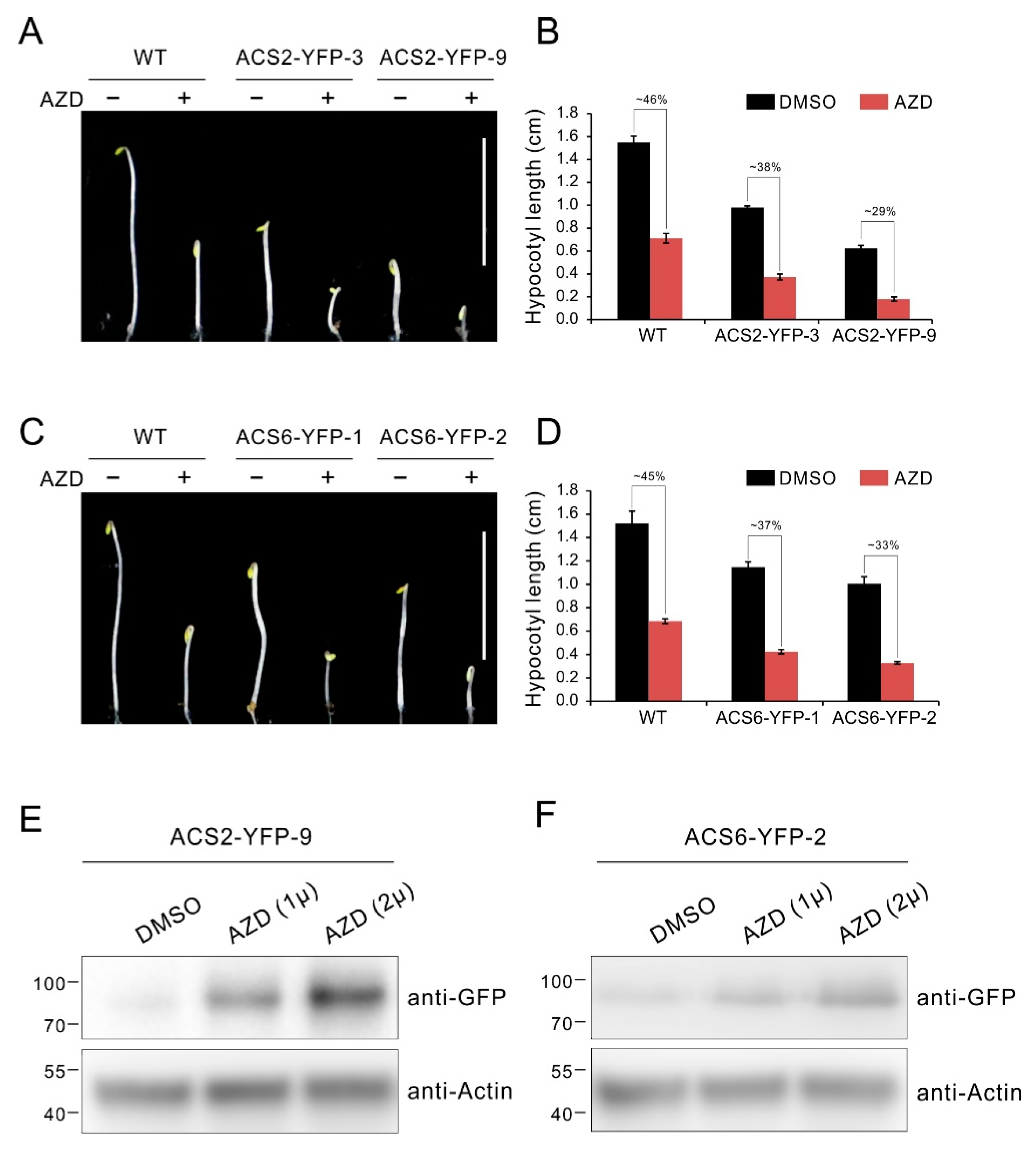
2.6. TOR Inhibition Causes Accumulation of ACS2/ACS6 Protein
3. Discussion
4. Materials and Methods
4.1. Arabidopsis Materials and Growth Conditions
4.2. Plasmids Construction and Arabidopsis Transformation
4.3. Inhibitor Treatments and Hypocotyl Growth inhibitionAssays
4.4. RNA Isolation and Gene Expression Analysis
4.5. Yeast Two-Hybrid Assay
4.6. BiFC Assays
4.7. Co-IP Assays
4.8. Western Blotting Analysis
4.9. Data Processing
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Baretic, D.; Williams, R.L. The structural basis for mTOR function. Semin. Cell Dev. Biol. 2014, 36, 91–101. [Google Scholar] [CrossRef]
- Rexin, D.; Meyer, C.; Robaglia, C.; Veit, B. TOR signalling in plants. J. Biochem. 2015, 470, 1–14. [Google Scholar] [CrossRef]
- Tatebe, H.; Shiozaki, K. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 2017, 7, 77. [Google Scholar] [CrossRef]
- Shi, L.; Wu, Y.; Sheen, J. TOR signaling in plants: Conservation and innovation. Development 2018, 145. [Google Scholar] [CrossRef]
- Cybulski, N.; Hall, M.N. TOR complex 2: A signaling pathway of its own. Trends Biochem. Sci. 2009, 34, 620–627. [Google Scholar] [CrossRef]
- Peterson, T.R.; Sengupta, S.S.; Harris, T.E.; Carmack, A.E.; Kang, S.A.; Balderas, E.; Guertin, D.A.; Madden, K.L.; Carpenter, A.E.; Finck, B.N.; et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011, 146, 408–420. [Google Scholar] [CrossRef]
- Oh, W.J.; Jacinto, E. mTOR complex 2 signaling and functions. Cell Cycle 2011, 10, 2305–2316. [Google Scholar] [CrossRef]
- Xiong, Y.; Sheen, J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Magyar, Z.; Monardes, A.; Khan, S.; Zalejski, C.; Orellana, J.; Szabados, L.; de la Torre, C.; Koncz, C.; Bogre, L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 2010, 29, 2979–2993. [Google Scholar] [CrossRef] [PubMed]
- Ahn, C.S.; Han, J.A.; Lee, H.S.; Lee, S.; Pai, H.S. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 2011, 23, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martinez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013, 32, 1087–1102. [Google Scholar] [CrossRef] [PubMed]
- Menand, B.; Desnos, T.; Nussaume, L.; Berger, F.; Bouchez, D.; Meyer, C.; Robaglia, C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 2002, 99, 6422–6427. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ichisaka, T.; Maeda, M.; Oshiro, N.; Hara, K.; Edenhofer, F.; Kiyama, H.; Yonezawa, K.; Yamanaka, S. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 2004, 24, 6710–6718. [Google Scholar] [CrossRef]
- Ren, M.; Qiu, S.; Venglat, P.; Xiang, D.; Feng, L.; Selvaraj, G.; Datla, R. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011, 155, 1367–1382. [Google Scholar] [CrossRef]
- Dowling, R.J.; Topisirovic, I.; Fonseca, B.D.; Sonenberg, N. Dissecting the role of mTOR: Lessons from mTOR inhibitors. BBA Proteins Proteom. 2010, 1804, 433–439. [Google Scholar] [CrossRef]
- Albert, S.; Serova, M.; Dreyer, C.; Sablin, M.P.; Faivre, S.; Raymond, E. New inhibitors of the mammalian target of rapamycin signaling pathway for cancer. Expert Opin. Investig. Drugs 2010, 19, 919–930. [Google Scholar] [CrossRef]
- Benjamin, D.; Colombi, M.; Moroni, C.; Hall, M.N. Rapamycin passes the torch: A new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 2011, 10, 868–880. [Google Scholar] [CrossRef]
- Montane, M.H.; Menand, B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J. Exp. Bot. 2013, 64, 4361–4374. [Google Scholar] [CrossRef] [PubMed]
- Chresta, C.M.; Davies, B.R.; Hickson, I.; Harding, T.; Cosulich, S.; Critchlow, S.E.; Vincent, J.P.; Ellston, R.; Jones, D.; Sini, P.; et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010, 70, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liang, S.; Kudla, J.; Luan, S. Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 1998, 15, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Sormani, R.; Yao, L.; Menand, B.; Ennar, N.; Lecampion, C.; Meyer, C.; Robaglia, C. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 2007, 7, 26. [Google Scholar] [CrossRef]
- Leiber, R.M.; John, F.; Verhertbruggen, Y.; Diet, A.; Knox, J.P.; Ringli, C. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 2010, 22, 1898–1908. [Google Scholar] [CrossRef]
- Xiong, Y.; Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 2012, 287, 2836–2842. [Google Scholar] [CrossRef]
- Xiong, F.; Dong, P.; Liu, M.; Xie, G.; Wang, K.; Zhuo, F.; Feng, L.; Yang, L.; Li, Z.; Ren, M. Tomato FK506 binding protein 12KD (FKBP12) mediates the interaction between Rapamycin and target of Rapamycin (TOR). Front. Plant Sci. 2016, 7, 1746. [Google Scholar] [CrossRef]
- Ren, M.; Venglat, P.; Qiu, S.; Feng, L.; Cao, Y.; Wang, E.; Xiang, D.; Wang, J.; Alexander, D.; Chalivendra, S.; et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 2012, 24, 4850–4874. [Google Scholar] [CrossRef]
- Xiong, F.; Zhang, R.; Meng, Z.; Deng, K.; Que, Y.; Zhuo, F.; Feng, L.; Guo, S.; Datla, R.; Ren, M. Brassinosteriod insensitive 2 (BIN2) acts as a downstream effector of the target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 2017, 213, 233–249. [Google Scholar] [CrossRef]
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 2013, 496, 181–186. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 2019, 70, 2227–2238. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Parola, R.; Andreola, S.; Pereyra, C.; Martinez-Noel, G. TOR and SnRK1 signaling pathways in plant response to abiotic stresses: Do they always act according to the “yin-yang” model? Plant Sci. 2019, 288, 110220. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, F.J.; Que, Y.M.; Wang, K.; Yu, L.H.; Li, Z.G.; Ren, M.Z. Expression profiling and functional analysis reveals that TOR is a key player in regulating photosynthesis and phytohormone signaling pathways in Arabidopsis. Front. Plant Sci. 2015, 6, 677. [Google Scholar] [CrossRef] [PubMed]
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTPase ROP2 binds and promotes activation of target of rapamycin, TOR, in response to auxin. EMBO J. 2017, 36, 886–903. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, J.Y.; Roh, J.; Marchive, C.; Kim, S.K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.Y. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in arabidopsis. Curr. Biol. 2016, 26, 1854–1860. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.C.; Liu, X.; Fu, L.; Hou, Y.J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 2018, 69, 100–112.e6. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, G.; Zhang, X.; Li, L.; Xiong, F.; Zhuo, F.; Zhang, C.; Yang, Z.; Datla, R.; Ren, M.; et al. The crosstalk between target of Rapamycin (TOR) and Jasmonic Acid (JA) signaling existing in Arabidopsis and cotton. Sci. Rep. 2017, 7, 45830. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.; Sheen, J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009, 14, 270–279. [Google Scholar] [CrossRef]
- Voesenek, L.; Pierik, R.; Sasidharan, R. Plant life without ethylene. Trends Plant Sci. 2015, 20, 783–786. [Google Scholar] [CrossRef]
- Dubois, M.; Van den, B.L.; Inze, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Garcia, M.J.; Romera, F.J.; Lucena, C.; Alcantara, E.; Perez-Vicente, R. Ethylene and the regulation of physiological and morphological responses to nutrient deficiencies. Plant Physiol. 2015, 169, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, W.; Schikora, A. Different pathways are involved in phosphate and iron stress-induced alterations of root epidermal cell development. Plant Physiol. 2001, 125, 2078–2084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romera, F.J.; Alcantara, E. Ethylene involvement in the regulation of Fe-deficiency stress responses by strategy I plants. Funct. Plant Biol. 2004, 31, 315–328. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Tena, G.; Xiong, Y.; Sheen, J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008, 451, 789–795. [Google Scholar] [CrossRef]
- Garcia, M.J.; Lucena, C.; Romera, F.J.; Alcantara, E.; Perez-Vicente, R. Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 2010, 61, 3885–3899. [Google Scholar] [CrossRef]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef]
- Chen, Y.F.; Etheridge, N.; Schaller, G.E. Ethylene signal transduction. Ann. Bot. 2005, 95, 901–915. [Google Scholar] [CrossRef]
- Chang, C.; Stadler, R. Ethylene hormone receptor action in Arabidopsis. BioEssays 2001, 23, 619–627. [Google Scholar] [CrossRef]
- Chen, Y.F.; Randlett, M.D.; Findell, J.L.; Schaller, G.E. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 2002, 277, 19861–19866. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef] [PubMed]
- Chao, Q.; Rothenberg, M.; Solano, R.; Roman, G.; Terzaghi, W.; Ecker, J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997, 89, 1133–1144. [Google Scholar] [CrossRef]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef] [PubMed]
- Bisson, M.M.; Bleckmann, A.; Allekotte, S.; Groth, G. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem. J. 2009, 424, 1–6. [Google Scholar] [CrossRef]
- Ju, C.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef]
- Jakubowicz, M. Structure, catalytic activity and evolutionary relationships of 1-aminocyclopropane-1-carboxylate synthase, the key enzyme of ethylene synthesis in higher plants. Acta Biochim. Pol. 2002, 49, 757–774. [Google Scholar] [CrossRef]
- Wang, K.L.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14 (Suppl. S14), S131–S151. [Google Scholar] [CrossRef]
- Guo, H.; Ecker, J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004, 7, 40–49. [Google Scholar] [CrossRef]
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Nicolai, M.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007, 8, 864–870. [Google Scholar] [CrossRef]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Kim, H.J.; Hong, S.H.; Kim, Y.W.; Lee, I.H.; Jun, J.H.; Phee, B.K.; Rupak, T.; Jeong, H.; Lee, Y.; Hong, B.S.; et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 4023–4036. [Google Scholar]
- Li, Z.; Peng, J.; Wen, X.; Guo, H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 2013, 25, 3311–3328. [Google Scholar] [CrossRef] [PubMed]
- Qiu, K.; Li, Z.; Yang, Z.; Chen, J.; Wu, S.; Zhu, X.; Gao, S.; Gao, J.; Ren, G.; Kuai, B.; et al. EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 2015, 11, e1005399. [Google Scholar] [CrossRef] [PubMed]
- Guzman, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523. [Google Scholar] [PubMed]
- Skottke, K.R.; Yoon, G.M.; Kieber, J.J.; DeLong, A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011, 7, e1001370. [Google Scholar] [CrossRef]
- Moreau, M.; Azzopardi, M.; Clement, G.; Dobrenel, T.; Marchive, C.; Renne, C.; Martin-Magniette, M.L.; Taconnat, L.; Renou, J.P.; Robaglia, C.; et al. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 2012, 24, 463–481. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Yoo, S.D.; Sheen, J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 2003, 425, 521–525. [Google Scholar] [CrossRef]
- Li, H.; Wong, W.S.; Zhu, L.; Guo, H.W.; Ecker, J.; Li, N. Phosphoproteomic analysis of ethylene-regulated protein phosphorylation in etiolated seedlings ofArabidopsis mutant ein2 using two-dimensional separations coupled with a hybrid quadrupoletime-of-flight mass spectrometer. Proteomics 2009, 9, 1646–1661. [Google Scholar] [CrossRef]
- Hu, R.; Zhu, Y.; Shen, G.; Zhang, H. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 2014, 164, 721–734. [Google Scholar] [CrossRef]
- Hall, A.E.; Chen, Q.G.; Findell, J.L.; Schaller, G.E.; Bleecker, A.B. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999, 121, 291–300. [Google Scholar] [CrossRef]
- Alonso, J.M.; Stepanova, A.N.; Solano, R.; Wisman, E.; Ferrari, S.; Ausubel, F.M.; Ecker, J.R. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 2992–2997. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Brumos, J.; Li, H.; Ji, Y.; Ke, M.; Gong, X.; Zeng, Q.; Li, W.; Zhang, X.; An, F.; et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 2011, 23, 3944–3960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, L.; Li, L.; Guo, M.; Chory, J.; Yin, Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2008, 105, 7618–7623. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, F.; Xiong, F.; Deng, K.; Li, Z.; Ren, M. Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 2680. https://doi.org/10.3390/ijms21082680
Zhuo F, Xiong F, Deng K, Li Z, Ren M. Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis. International Journal of Molecular Sciences. 2020; 21(8):2680. https://doi.org/10.3390/ijms21082680
Chicago/Turabian StyleZhuo, Fengping, Fangjie Xiong, Kexuan Deng, Zhengguo Li, and Maozhi Ren. 2020. "Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis" International Journal of Molecular Sciences 21, no. 8: 2680. https://doi.org/10.3390/ijms21082680
APA StyleZhuo, F., Xiong, F., Deng, K., Li, Z., & Ren, M. (2020). Target of Rapamycin (TOR) Negatively Regulates Ethylene Signals in Arabidopsis. International Journal of Molecular Sciences, 21(8), 2680. https://doi.org/10.3390/ijms21082680




