Abstract
N6-methyladenosine (m6A) is the most prevalent internal modification present in the mRNAs of all higher eukaryotes, where it is present within both coding and noncoding regions. In mammals, methylation requires the catalysis of a multicomponent m6A methyltransferase complex. Proposed biological functions for m6A modification include pre-mRNA splicing, RNA stability, cell fate regulation, and embryonic development. However, few studies have been conducted on m6A modification in trees. In particular, the regulation mechanism of RNA m6A in Populus development remains to be further elucidated. Here, we show that PtrMTA (Populus trichocarpa methyltransferase) was colocalized with PtrFIP37 in the nucleus. Importantly, the PtrMTA-overexpressing plants significantly increased the density of trichomes and exhibited a more developed root system than that of wild-type controls. Moreover, we found that PtrMTA-overexpressing plants had better tolerance to drought stress. We also found PtrMTA was a component of the m6A methyltransferase complex, which participated in the formation of m6A methylation in poplar. Taken together, these results demonstrate that PtrMTA is involved in drought resistance by affecting the development of trichomes and roots, which will provide new clues for the study of RNA m6A modification and expand our understanding of the epigenetic molecular mechanism in woody plants.
1. Introduction
More than 150 nucleotide modifications of RNA have been described, and more than 10 of them have been reported in mRNA [1]. RNA methylation, especially N6-methyladenosine (m6A), is an abundant internal modification of mRNAs in eukaryotes [2,3]. N6-methyladenosine was first discovered in bacteria DNA in 1955 [3]. Since the discovery of m6A methylation, m6A has been found in viral RNAs and various other eukaryotes, including mammals [4,5,6,7,8], yeast [9], fruit flies [10], and plants, such as oats [11], maize [12], Arabidopsis [13], and wheat [14]. m6A modifications are mainly distributed in mRNA and long non-coding RNA. In addition, m6A also exists in other types of eukaryotic RNA, such as tRNA (transport RNA) [15], rRNA (ribosomal RNA) [16], and snRNA (small nuclear RNA) [17].
In mammals, m6A mRNA methylation is catalyzed by the methyltransferase complex containing METTL3 (methyltransferase 3) [17], METTL14 (methyltransferase 14) [18] and WTAP (Wilms’ tumor 1-related protein) [19,20]. Methyltransferases may be conserved in eukaryotes [21,22]. Recently, it has been found that KIAA1429 may be a new element of the m6A methyltransferase complex, which may be related to the formation of m6A [21]. The m6A modification can be demethylated by m6A demethylases, such as fat and obesity-related protein (FTO) [23] and its homolog alkylated DNA repair protein ALkB homolog 5 (ALKBH5) [24]. Therefore, m6A modification is a reversible biological process.
The m6A-modified transcripts can be recognized by “reader” proteins, such as the YTH (YT512-B homology) domain protein family in animals [25,26,27,28]. To date, 13 proteins with YTH domains have been reported in Arabidopsis, including ECT1-11 (Evolutionarily Conserved C-Terminal Region1-11) [25,26,27,28]. Recently, studies have shown that ECT2 (Evolutionarily Conserved C-Terminal Region2) can regulate trichome morphology by affecting the stability of mRNA and plays an important role in trichome branching in Arabidopsis [29,30]. In addition, ECT2 can control the developmental timing of leaf formation in Arabidopsis [31]. More importantly, the m6A-modified transcripts appear to be involved in a wide range of biological processes, including mRNA translatability and stability [22,26,27,32,33], mRNA export from the nucleus [34], alternative polyadenylate site selection [35], splicing regulation [36,37,38], and other mechanisms that accompany RNA maturation [2,3].
At present, the functional studies of methyltransferase plant homologs are mainly concentrated on the herbaceous model plant—Arabidopsis thaliana. Luo et al. found that m6A methylation was highly conserved and abundant in mRNA modification by comparing the m6A methylomes of two different Arabidopsis lines [39]. Arabidopsis MTA (N6-adenosine-methyltransferase MT-A70-like, a homolog of human METTL3) is expressed at relatively low levels in most tissues but is highly expressed in flowers, meristems, seeds, and microspores [13]. In addition, AtMTA also affects the development of embryos and trichomes and is required for m6A mRNA methylation [13]. Embryos cannot develop from the globular to the heart stage when AtMTA is knocked out in Arabidopsis [13]. FIP37 (FKBP12 interacting protein 37 KD) has been found to be a homolog of mammalian WTAP in Arabidopsis, and embryo development in the fip37 mutant stopped at the globular stage [40]. Recently, Shen et al. found that AtFIP37 can regulate the shoot stem cell fate in Arabidopsis [41]. AtFIP37-overexpressing plants showed increased trichome branches, suggesting that AtFIP37 affects the development of trichomes [40]. Plant trichomes play an important role in buffering environmental and plant interactions [42]. In addition, plant trichomes have biological functions, such as reducing transpiration and drought resistance [42,43,44].
Although functional studies of MTA have been reported in herbaceous plants, its potential functions in ligneous plants, especially in trees, remain unknown. Poplar is the dominant species of forest ecosystems, which possesses great economic and ecological value [45]. Poplar has become a model ligneous plant to explore gene function because of its fast growth and other advantages. In this study, we demonstrated that Populus trichocarpa methyltransferase (PtrMTA) and PtrFIP37 (FKBP12 interacting protein 37 KD form Populus trichocarpa) were co-localized in the nucleus. Moreover, we revealed PtrMTA-overexpressing plants had a higher density of trichomes and a more developed root system. In addition, PtrMTA enhanced drought resistance in poplar. We also revealed that PtrMTA participated in the formation of m6A methylation in 84K poplar. In conclusion, we found that PtrMTA plays an indispensable role in the growth and resistance of poplar, which provides a basis for systematic research on m6A in the future.
2. Results
2.1. Cloning and Obtaining Transgenic Plants of PtrMTA
To elucidate the potential function of PtrMTA in ligneous plants, we constructed a phylogenetic tree of MTA using DNAMAN software. We found that PtrMTA (Potri.001G085200.1) had 99% homology with PagMTA and PeMTA (Figure S1). We further found that MTA was highly conserved among a variety of plant species through multiple sequence alignments, such as Populus trichocarpa, 84K poplar, Populus euphratica, Oryza sativa, Prunus persica, Glycine max, Solanum lycopersicum, Fragaria vesca, and Arabidopsis thaliana. (Figure S2).
To further study the potential biological functions of PtrMTA, we cloned the MTA gene from Populus trichocarpa and constructed a 35S overexpression vector. Next, we performed genetic transformation through the leaf disc method [46] (Figure 1A). The leaves can differentiate into adventitious buds under light culture conditions (Figure 1B). Through screening for resistance, we initially obtained PtrMTA-overexpressing transgenic plants (Figure 1C). Every transgenic line was verified by PCR (polymerase chain reaction) and qRT-PCR (quantitative real-time PCR) (Figure 1D, E). In the literature, these two methods are usually used to verify transgenic plants. Through the verification of the two methods, we obtained transgenic poplar 84K (P. alba X P. glandulosa), i.e., 14 lines that overexpressed PtrMTA.
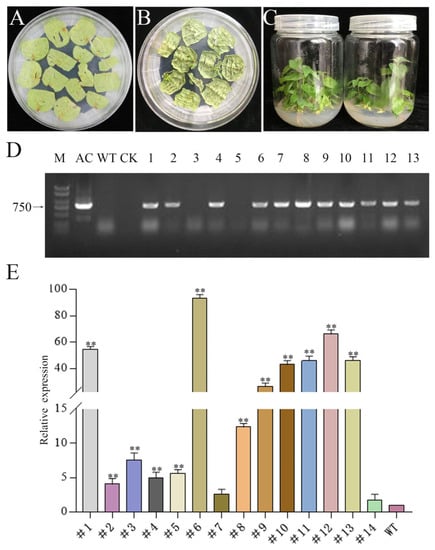
Figure 1.
Obtaining and identification of Populus trichocarpa methyltransferase (PtrMTA) transgenic plants. (A–C) Genetic transformation of 84K poplar and obtained transgenic seedlings. (D) PCR detection of transgenic plants (M: DNA marker 2000+; AC: active control; WT: 84K wild type; CK: control check). (E) Expression level analysis of PtrMTA transgenic plants by qRT-PCR. Error bars are means ± SE (n = 3). All asterisks denote significant differences: ** p < 0.01.
2.2. Subcellular Localization of PtrMTA
Previous studies indicated that Arabidopsis MTA, an m6A methyltransferase, was localized in the nucleus [13]. Therefore, PtrMTA, a homolog of MELLT3, might localize to the same cellular structures. To determine the subcellular localization of PtrMTA, we constructed a 35S::PtrMTA-GFP fusion protein (green fluorescent protein) expression vector and transiently transformed it into tobacco leaves. Using laser scanning confocal microscopy (LSCM), we found that PtrMTA-GFP was localized in the nucleus (Figure 2A). We further transiently transformed tobacco leaves with the 35S::PtrMTA-GFP fusion protein and 35S::PtrFIP37-mCherry fusion protein. LSCM observation showed that 35S::PtrMTA-GFP and 35S:: PtrFIP37-mCherry were co-localized in the nucleus (Figure 2B).
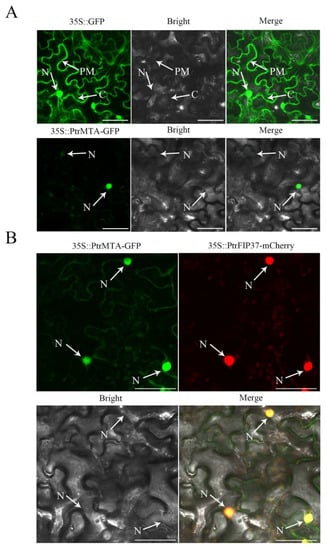
Figure 2.
PtrMTA was targeted to the nucleus. (A) Subcellular localization of 35S::GFP and 35S::PtrMTA-GFP in transiently expressed tobacco leaves. Bar = 15 μm. (B) Subcellular co-localization of 35S::PtrMTA-GFP and 35S::PtrFIP37-mCherry in transiently expressed tobacco leaves. Bright: bright-field image; PM: plasma membrane; C: cytoplasm; N: nucleus. Bars = 18 μm.
2.3. Overexpression of PtrMTA Affected Trichome Development
To determine whether PtrMTA affected the development of trichomes in poplar, we selected PtrMTA-overexpressing plants (OE-PtrMTA-14, OE-PtrMTA-10, OE-PtrMTA-6) and wild type (WT) plants for observation of leaf trichomes. Using scanning electron microscopy (SEM), we found that the trichomes of poplar were single-celled, non-glandular trichomes that did not have branches (Figure 3A). Furthermore, we counted the number of trichomes in the 20 areas (1500 μm * 1000 μm) of leaves in each line. In the transgenic line OE-PtrMTA-14, the number of trichomes was about 13, which was slightly more than in the WT (Figure 3B). The number of trichomes of transgenic line OE-PtrMTA-10 was about 18 (Figure 3C). Compared with the WT, the number of trichomes in the transgenic line OE-PtrMTA-6 was significantly increased and was about two times that of the WT (Figure 3D). More significantly, through statistical analysis, we found the number of trichomes in the PtrMTA-overexpressing plants was significantly greater than that in the WT (Figure 3E and Table S1). Our results illustrated that overexpression of PtrMTA can increase the density of trichomes in poplar.
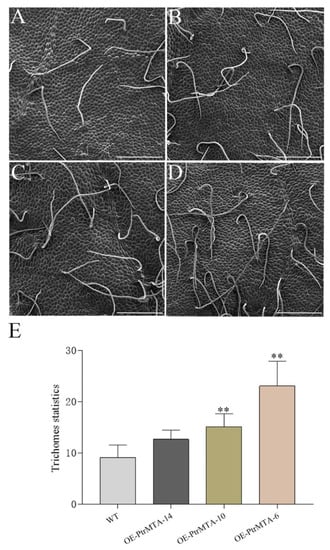
Figure 3.
PtrMTA increases trichome density in transgenic 84K poplar plants. (A) Illustration of trichome density in wild type (WT) plants. (B–D) Illustration of the increase in trichome density in 35S::PtrMTA lines OE-PtrMTA-14, OE-PtrMTA-10, and OE-PtrMTA-6, respectively. (E) Statistics on the number of trichomes in the 20 areas (1500 μm * 1000 μm) of WT and transgenic 84K poplar leaves. Bars = 200 μm. Error bars are means ± SE (n = 20). All asterisks denote significant differences: **p < 0.01.
2.4. PtrMTA-Overexpressing Poplar had a More Developed Root System
To investigate whether PtrMTA affected root development in poplar, we selected WT, OE-PtrMTA-14, OE-PtrMTA-10, and OE-PtrMTA-6 seedlings aged 20 days and potted the seedlings for 50 days. At the seedling stage, the PtrMTA-overexpressing plants showed a more developed root system than that of the WT (Figure 4A). The root length of PtrMTA-overexpressing plants was significantly longer than that of the WT (Figure 4B). In addition, the number of root tips in the PtrMTA-overexpressing plants was about five, which was also significantly higher than that of the WT (Figure 4C). Moreover, we examined the PtrMTA-overexpressing poplars that had been grown for 50 days and found that their root systems were significantly more developed than those of the WT (Figure 4D). Compared with the WT roots, the root length of PtrMTA-overexpressing plants (about 10 cm) was longer (Figure 4E). The root fresh and dry weights of different lines were measured. We found that the root weight of PtrMTA-overexpressing plants was significantly higher than that of the WT (Figure 4F). In summary, PtrMTA-overexpressing plants had a more developed root system compared with the WT.
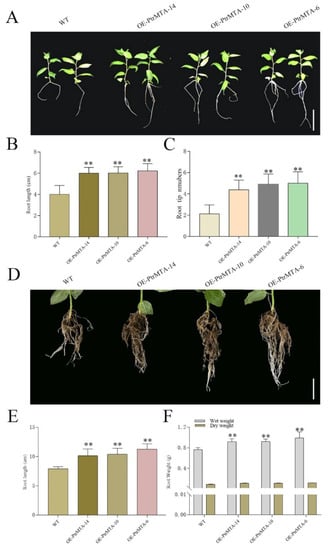
Figure 4.
Root length, root tip number, and root weight of the WT and PtrMTA-overexpressing plants were compared. (A) Morphological differences between WT and transgenic plants grown for 20 days. (B) Comparison of root lengths between wild-type and PtrMTA-overexpressing plants ogrown for 20 days. (C) Comparison of root tip numbers between wild-type and transgenic plants grown for 20 days. (D) Morphological differences between wild-type and transgenic plants grown for 50 days in pots. (E) Comparison of root lengths between wild type and PtrMTA-overexpressing plants grown for 50 days. (F) Comparison of root fresh weight and dry weight in WT and transgenic plants. Bars = 3 cm, A; 5 cm, D. Error bars are means ± SE (n = 10). All asterisks denote significant differences: **p < 0.01.
2.5. OE-PtrMTA Poplar Possessed Drought Tolerance
We further analyzed the approximately 3000 bp promoter sequence upstream of the PtrMTA ATG using the https://www.ncbi.nlm.nih.gov/pubmed/11752327 website. Interestingly, we found that the PtrMTA promoter contained MYB binding sites involved in drought induction and cis-acting elements involved in stress responses (Table S2). Furthermore, to investigate whether PtrMTA increased the drought resistance of poplar, PtrMTA-overexpressing poplar and WT poplar plants were cultured under the same conditions and were then subjected to drought treatment to reduce the soil RWC (relative water content) from 70%. On day 2, some leaves of the WT poplars were slightly withered, while the PtrMTA-overexpressing (OE-PtrMTA) poplars appeared normal (Figure 5A). On day 5, the leaves of the WT poplars were more seriously withered than those of the PtrMTA-overexpressing poplars (Figure 5A).
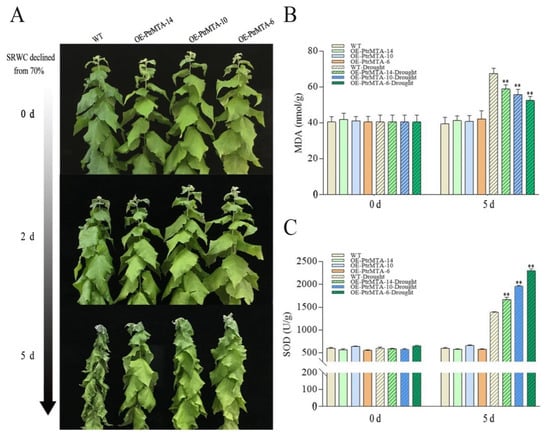
Figure 5.
OE-PtrMTA plants had increased tolerance under drought treatment. (A) Phenotypic differences in drought conditions. (B) Analysis of the malondialdehyde (MDA) content. (C) Superoxide dismutase (SOD) activity determination. Error bars are means ± SE (n = 4). All asterisks denote significant differences: **p < 0.01.
As the malondialdehyde (MDA) content is an effective indicator of the oxidative damage of the cytomembrane, we measured the changes in the MDA content in leaves of WT and PtrMTA-overexpressing poplars under drought stress. The results showed that the MDA content of OE-PtrMTA poplars was lower than that of WT poplar under drought stress (Figure 5B). Superoxide dismutase (SOD) is an important antioxidant that removes reactive oxygen species. Therefore, we tested the activities of SOD in OE-PtrMTA and WT poplars before and after drought treatment. The results showed that the activity of SOD was significantly higher in OE-PtrMTA poplars than in the WT under drought stress (Figure 5C). Except that the soil RWC remained at 70%, the control plants were maintained under the same culture conditions, and no significant differences in stem diameter and plant height were found between OE-PtrMTA poplars and the WT (Figure S3). Therefore, we can conclude that OE-PtrMTA transgenic poplars had better tolerance to drought stress.
2.6. PtrMTA Affected the Level of m6A
To study whether PtrMTA affected the level of m6A, we selected WT, OE-PtrMTA-14, OE-PtrMTA-10, and OE-PtrMTA-6 seedlings aged 30 days and extracted RNA to measure the level of m6A. Compared to the WT, the level of m6A was significantly higher in PtrMTA-overexpressing plants (Figure 6A). Furthermore, we also detected the m6A level in the roots, stems, and leaves of the WT and overexpressing transgenic seedlings. Importantly, we found that the level of m6A in the roots of PtrMTA-overexpressing plants was higher than that of the WT, and the m6A content in the roots of OE-PtrMTA-6 was about twice as high as that of the WT (Figure 6B). It is worth mentioning that the level of m6A in stems of PtrMTA-overexpressing plants was much higher than that of the WT (Figure 6C). In addition, the m6A content in leaves was lower than that in the roots and stems, but the m6A content in PtrMTA-overexpressing plants was still significantly higher than that in the WT (Figure 6D). These results indicate that PtrMTA is a component of m6A methyltransferase and affects the level of m6A.
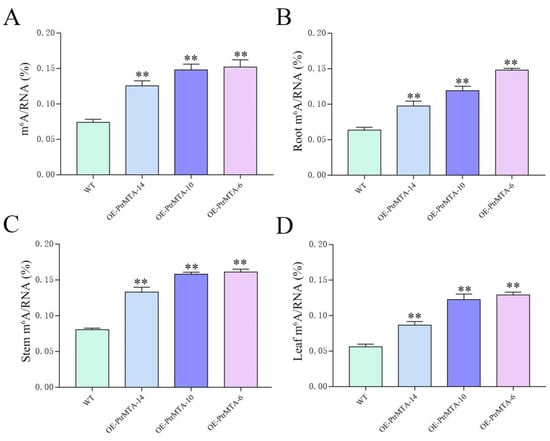
Figure 6.
N6-methyladenosine (m6A) content in WT and PtrMTA-overexpressing plants under normal conditions. (A) m6A content in WT and PtrMTA-overexpressing plants at 30 days. (B) Analysis of the m6A content in the roots of WT and PtrMTA-overexpressing plants. (C) m6A content statistics in the stems of WT and PtrMTA-overexpressing plants. (D) Analysis of the m6A content in the leaves of WT and PtrMTA-overexpressing plants. Error bars are means ± SE (n = 6). All asterisks denote significant differences: ** p < 0.01.
3. Discussion
RNA m6A is a conserved and abundant mRNA modification that affects many aspects of RNA metabolism [34]. In 2015, Chen et al. found that when the level of m6A modification was lowered. It was found to impede cell reprogramming seriously [47]. In mammals, loss of function in the core component of the m6A methyltransferase complex, including METTL3 and WTAP, leads to embryonic death and reduces the levels of m6A [48,49]. In Arabidopsis, AtMTA (a homolog of METTL3) and AtFIP37 (a homolog of WTAP) are localized in the nucleus [13]. Moreover, when the fluorescent fusion proteins MTA-YFP and FIP37-CFP were transiently transformed into Allium cepa epidermal cells, it was found that the two proteins were localized to the same nuclear position [13]. Therefore, AtMTA and AtFIP37 were co-localized in the nucleus. Importantly, AtMTA and AtFIP37 are critical for early embryonic development [13,40,41]. T-DNA knockout of MTA or FIP37 led to the stagnation of embryonic development during the global stage and also reduced levels of m6A in different tissues [13,40]. In Arabidopsis, the demethylases include an AlkB family of 13 homologous proteins [29,50,51]. ALKBH9B participates in defense against viral infections [50]. ALKBH10B is an mRNA m6A demethylase that affects vegetative growth and floral transition [51]. In our study, we transiently transformed the 35S::PtrMTA-GFP fusion protein into tobacco leaves and found that PtrMTA targeted the nuclear spots. In addition, we simultaneously transferred 35S::PtrMTA-GFP and 35S::PtrFIP37-mCherry into tobacco leaves and found that the two proteins were co-localized in the nucleus. In addition, we found that the level of m6A in PtrMTA-overexpressing plants increased significantly compared to the WT. This indicated that PtrMTA is a component of m6A methyltransferase and can affect the level of m6A.
Trichomes and roots are often associated with plant resistance [52]. A trichome is a hairy structural appendage extending from the epidermal tissues of aerial parts; trichomes include glandular and non-glandular hairs, linear trichomes, and branched trichomes [53]. Trichomes have a variety of biological functions, such as reducing transpiration. Moreover, trichomes are involved in defense against adverse conditions, such as cold, drought, high temperature, strong radiation, as well as disease infestation [53]. Glandular hairs can secrete chemicals, such as alkaloids, aromatic oils, and resins, to defend against biotic and abiotic stresses [53]. Non-glandular hairs play an important role in resisting drought and mechanical damage [54]. The root system is the organ through which a plant directly senses water signals from the soil and absorbs soil water [55]. The root length and depth directly affect the absorption and utilization of soil water and nutrients [56]. In addition, the root tip number affects the range of the root system depth and breadth in space and also affects the ability to absorb water [57]. Root length, root tip number, and root weight are all closely related to plant drought resistance [58]. In this study, the results of SEM showed that overexpression of PtrMTA significantly increased the density of trichomes in poplar. In addition, we found that the root length, root tip number, and root weight of PtrMTA-overexpressing plants were significantly increased compared to WT plants. These results indicate that PtrMTA is closely related to the development of trichomes and plant roots. More importantly, we found that PtrMTA-overexpressing plants had stronger drought resistance than the WT under drought treatment. We speculate that PtrMTA may be involved in the regulation of drought resistance.
The balance between the generation and elimination of reactive oxygen species in a cell would be broken under stress conditions [59]. Excess reactive oxygen species first attack the cell membrane system, thus destroying the cell membrane [60,61]. MDA is the final product of cytoplasmic membrane peroxidation. MDA can combine with proteins on the cell membrane to inactivate it, thereby destroying the structure and function of the biological membranes [60]. Plants have a membrane protection system that can remove excess reactive oxygen species from cells [60,62]. This membrane protective system is an antioxidant system composed of many enzymes, such as SOD [62]. SOD can protect the biological membrane system by removing harmful reactive oxygen species [63]. Therefore, the MDA content and SOD activity can reflect the resistance of plants to a certain extent [59]. In this study, we found that PtrMTA-overexpressing plants were more resistant to drought stress compared to WT plants. Therefore, we measured the MDA content and SOD activities of PtrMTA-overexpressing plants before and after drought treatment. After drought treatment, the MDA content was significantly lower than that of the WT, which may be related to the strong drought tolerance of PtrMTA-overexpressing plants. In addition, the SOD activity of PtrMTA-overexpressing plants was obviously higher than that of the WT, which may be related to resistance to stress.
4. Materials and Methods
4.1. Plant Materials and Plant Growth Conditions
In this study, the experimental materials—Tobacco, Populus trichocarpa, and 84K poplar—were planted in a greenhouse of the Beijing Forestry University at 25°C under a 16 h light (240 μmol m –2 s –1) and 8 h dark. Potted Populus trichocarpa were watered based on evaporation demand and were regularly irrigated with water.
4.2. cDNA Cloning and Plasmid Construction
Whole seedlings of poplar were collected and stored in liquid nitrogen. The total RNA was isolated using an RNA Isolation Kit (TIANGEN, Beijing, China). In the last step, potentially contaminating DNA was removed by treatment with DNase I. Four micrograms of total RNA was used for the reverse transcription reaction with a First-Strand cDNA Synthesis Kit (TIANGEN, Beijing, China) according to the protocol. The cDNA of PtrMTA and PtrFIP37 was cloned by PCR. The DNA polymerase we used was ex-Taq (Takara, Japan). The initial denaturing time was 95 °C for 5 min, followed by 35 cycles at 95 °C for 30 s, 56 °C for 30 s and 72 °C for 2.5 min, with a final extension at 72 °C for 5 min. Furthermore, the PtrMTA sequence was inserted between the KpnI (Takara, Japan) and BamHI (Takara, Japan) sites of the pCAMBIA-2301 and pCAMBIA-2301-GFP vectors. We transformed the constructed pCAMBIA-2301-MTA and pCAMBIA-2301-MTA-GFP vectors into Agrobacterium tumefaciens GV3101. The transformed Agrobacterium was then applied to Luria-Bertani (LB) solid plates with 50 mg/L kanamycin and 50 mg/L rifampicin antibiotics and incubated at 28 °C for 2–3 days. A single clone was picked and subsequently verified by PCR. The PtrFIP37 sequence was inserted between the KpnI (Takara, Japan) and XbaI (Takara, Japan) sites of the pCAMBIA-1300-mCherry vector. pCAMBIA-1300-FIP37-mCherry Agrobacterium transformation and PCR were performed using the same method as above. The primers are in Table S3.
4.3. qRT-PCR (Quantitative Real-Time Polymerase Chain Reaction) Analysis
RNA from transgenic plants and WT plants was extracted using an Adelaide RNA Kit (Adelaide, China). The RNA was reverse transcribed into cDNA according to the method provided by the One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, China). qRT-PCR was performed using SYBR Green Mix (Takara, Japan) in an optical 96-well plate. As an internal control, 18S rRNA was used in qRT-PCR. The qRT-PCR instrument we use was Bio-Rad-CFX96 Touch, America. The 2-DDCt analysis method was used to analyze the relative expression of PtrMTA. The initial denaturing time was 95 °C for 2 min, followed by 40 cycles at 95 °C for 5 s, 60 °C for 5 s, and 95 °C for 5 s. The primers are in Table S3.
4.4. Phylogenetic and Domain Analyses of PtrMTA
The homologous amino acid sequences of MTA in different plant species were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/) and Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). In NCBI, PtrMTA homologous sequences were obtained by the BLASTn method (Table S4). In addition, the phylogenetic tree of PtrMTA was simply created using DNAMAN. DNAMAN was used for comparison of PtrMTA amino acid sequences and confirmed conserved structures.
4.5. Genetic Transformation of Poplar and Identification of Transgenic Seedlings
A single colony of Agrobacterium was shaken in LB medium at 28 °C overnight until the culture density reached an OD600 of 0.6–0.8. Agrobacterium cells were harvested by centrifugation at 5000 rpm for 5–8 min. The harvested cells were suspended in a ½-strength MS solution (pH 5.8) containing 5% (w/v) sucrose and acetosyringone (100 μM) as a conversion solution (Song et al., 2019). The pretreated leaves were placed in the transformation solution and soaked for about 15 min, during which time shaking was required every 3 min. The infested leaves were placed on sterile filter paper, and extra Agrobacterium was aspirated. The leaves were placed in ½-strength MS medium for 3–5 days under dark culture. The dark-treated leaves were placed in a medium containing kanamycin for differentiation culture. When the shoots grew to 1 cm after 4–5 weeks, they were cut off and transferred to a resistant screening medium. The obtained transgenic plants were screened for DNA identification and qRT-PCR identification. The primers are in Table S3.
4.6. Subcellular Localization
The Agrobacterium was collected by centrifugation for 5 min at 5000 g. The cultures were resuspended in a medium containing 5 mM MES, 100 mM MgCl2 (pH 5.6), and 0.1 mM acetylsyringone [64]. The Agrobacterium cells were then injected into the abaxial surfaces of the tobacco leaves by using a needleless syringe. The plants were then cultured in a dark environment for 24 h. The transiently transformed tobacco leaves were fixed in sterile water and imaged with a Leica 20X or 40X water immersion objective, white light laser, and a hybrid detector with a resolution of 1024 × 1024 pixels. All images shown in this article are original images taken from the Leica SP8.
4.7. SEM Analysis of Poplar Leaves Trichomes
SEM (Hitachi S4800, Japan) was used to study the trichome density of 40-day-old WT and PtrMTA-overexpressing poplar leaves. We selected potted seedlings that had grown for 40 days and picked the sixteenth leaf of each plant. Then we cut out the middle area near the main vein of the leaf. Poplar leaves were trimmed to a size of 1 cm2. Samples were then immediately drop fixed in formalin–acetic acid–alcohol (FAA was composed of 5 mL 38% formaldehyde, 5 mL acetic acid and 90 mL 70% alcohol). The samples were dehydrated successively with 30%, 50%, 60%, 70%, 80%, and 90% ethanol for 30–40 min and were dehydrated overnight with anhydrous ethanol. A dryer was used for the dehydrated samples (Leica EM CPD300, Germany). Dry samples were sputter-coated with gold (110 s). Finally, the samples were imaged using a scanning electron microscope. We counted the number of trichomes in the 20 areas (1500 μm * 1000 μm) of leaves in WT and transgenic plants.
4.8. Drought-Tolerance Experiments
Three plants of the 3 transgenic lines and one WT poplar plant were grown in a greenhouse at 25 °C during the day and 22 °C at night. The greenhouse was illuminated for 16 h per day. All plants were grown in suitable pots with a diameter of 13.5 cm. All plants were subjected to a short-term drought treatment in which the soil RWC (relative water content) decreased from 70% [65,66]. The control plants were kept under the same conditions.
4.9. Measurement of MDA Content and SOD Activity
At 0 and 7 days, leaves that detached from drought-treated plants were measured to determine the MDA content and SOD activity. The MDA content and SOD activity of transgenic and WT plants were determined using a Solarbio Kit (Solarbio, China). MDA (nmol/g FW) = 5*(6.45*(A532–A600)-0.56*A450)/W (W: sample quality); SOD (U/g FW) = 11.4*inhibition percentage/(1-inhibition percentage)/W*F (W: sample quality; F: sample dilution multiple).
4.10. Determination of the m6A Level in RNA
RNA from transgenic and WT seedlings was extracted using an Adelaide RNA Kit (Adelaide China). The RNA concentrations of different samples were determined using a NanoPhotometer NP80 (Implen, Germany). The m6A level was determined using an Aderr kit (Epigentek, America). m6A (%) = (SampleOD-NcOD)/S/(PcOD-NcOD)/P*100% (Nc: Negative control; Pc: Positive control; S: RNA sample inputs, ng; P: Positive control inputs, ng).
5. Conclusions
This study shows that the m6A methyltransferase MTA from Populus trichocarpa was localized in the nucleus and co-localized with PtrFIP37 in the nucleus. Moreover, the overexpression of PtrMTA significantly affected the level of m6A and increased the density of trichomes. PtrMTA-overexpressing plants had a more developed root system. More importantly, the overexpression of PtrMTA can enhance the drought resistance of poplar. Therefore, our research not only enriched the study of the m6A function in Populus but also provided a basis for exploring the epigenetic molecular mechanism in woody plants.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/21/7/2462/s1, Figure S1. Phylogenetic tree of different MTA protein family members and amino acid sequence alignment of PtrMTA; Figure S2. MTA was highly conserved among various plant species; Figure S3. Growth status of wild type (WT) and PtrMTA-overexpressing plants under normal conditions; Table S1. Statistics on the number of trichomes in the 20 areas (1500 μm * 1000 μm) of leaves in wild type (WT) and transgenic plants; Table S2. Analysis of the PtrMTA promoter functional components; Table S3 Primers used in this study; Table S4 GenBank IDs of the PtrMTA homologs proteins.
Author Contributions
L.L. was the primary author of this study. Y.Z., Q.H., and Z.Q. participated in the experiments and modified the picture in the article. G.Z. and W.X. standardized the format of the references. T.Y. and G.W. offered some suggestions for the results section. R.L. designed the experiments and revised the manuscript for submission. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31970182, 31670182, 31761133009 and 31401149), the Fundamental Research Funds for the Central Universities (2019ZY29) and the State “13.5” Key Research Program of China (No. 2016YFD0600102). Compliance with ethical standards.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- Machnicka, M.A.; Milanowska, K.; Oglou, O.O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways-2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N-6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Bio. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.N.; Liu, J.Z.; He, C. RNA N-6-methyladenosine methylation in post-transcriptional gene expression regulation. Gene Dev. 2015, 29, 1343–1355. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Taylor, R.H. The methylation state of poly a-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Furuichi, Y.; Morgan, M.; Shatkin, A.J.; Jelinek, W.; Salditt-Georgieff, M.; Darnell, J.E. Methylated, blocked 5 termini in HeLa cell mRNA. Proc. Natl. Acad. Sci. USA 1975, 72, 1904–1908. [Google Scholar] [CrossRef]
- Wei, C.M.; Moss, B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977, 16, 1672–1676. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 1975, 255, 28–33. [Google Scholar] [CrossRef]
- Clancy, M.J.; Shambaugh, M.E.; Timpte, C.S.; Bokar, J.A. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002, 30, 4509–4518. [Google Scholar] [CrossRef]
- Levis, R.; Penman, S. 5′-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster. J. Mol. Biol. 1987, 120, 487–515. [Google Scholar] [CrossRef]
- Haugland, R.A.; Cline, M.G. Post-transcriptional modifications of oat coleoptile ribonucleic acids. 5′-Terminal capping and methylation of internal nucleosides in poly(A)-rich RNA. Eur. J. Biochem. 1980, 104, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Nicholsm, J.L. ‘Cap’ structures in maize poly(A)-containing RNA. BBA-Biomembranes 1979, 563, 490–495. [Google Scholar]
- Zhong, S.L.; Li, H.Y.; Bodi, Z.; Button, J.; Vespa, L.; Herzog, M.; Fray, R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 2008, 20, 1278–1288. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, T.D.; Lane, B.G. Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5′-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos. Can. J. Microbio. 1979, 57, 927–931. [Google Scholar]
- Saneyoshi, M.; Harada, F.; Nishimura, S. Isolation and characterization of N6-methyladenosine from escherichia coli valine transfer RNA. BBA-Biomembranes 1969, 190, 264–273. [Google Scholar] [CrossRef]
- Iwanami, Y.; Brown, G.M. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 1968, 126, 8–15. [Google Scholar] [CrossRef]
- Bringmann, P.; Luhrmann, R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987, 213, 309–315. [Google Scholar] [CrossRef]
- Liu, J.Z.; Yue, Y.N.; Han, D.L.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.F.; Yu, M.; Lu, Z.K.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5 ‘ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.L.; Zhao, J.C. N-6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. cell 2013, 49, 18–29. [Google Scholar]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature 2012, 485, 201–U284. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.K.; Gomez, A.; Hong, G.C.; Yue, Y.N.; Han, D.L.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.F.; et al. N-6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.K.; Han, D.L.; Ma, H.H.; Weng, X.C.; Chen, K.; Shi, H.L.; He, C. N-6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Xu, C.; Wang, X.; Liu, K.; Roundtree, I.A.; Tempel, W.; Li, Y.J.; Lu, Z.K.; He, C.; Min, J.R. Structural basis for selective binding of m(6)A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014, 10, 927–929. [Google Scholar] [CrossRef]
- Wei, L.H.; Songm, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.C.; Jia, G. The m(6)A Reader ECT2 controls trichome morphology by affecting mrna stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Scutenaire, J.; Deragon, J.M.; Jean, V.; Benhamed, M.; Raynaud, C.; Favory, J.J.; Merret, R.; Bousquet-Antonelli, C. The YTH domain protein ECT2 is an m(6)A Reader required for normal trichome branching in Arabidopsis. Plant Cell 2018, 30, 986–1005. [Google Scholar] [CrossRef]
- Arribas-Hernandez, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m(6)A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef] [PubMed]
- Bodi, Z.; Bottley, A.; Archer, N.; May, S.T.; Fray, R.G. Yeast m(6)A Methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wan, J.; Gao, X.W.; Zhang, X.Q.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef]
- Ke, S.D.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m(6)A residues are in the last exons, allowing the potential for 3 ‘ UTR regulation. Gene Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N-6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301. [Google Scholar] [CrossRef]
- Lence, T.; Akhtar, J.; Bayer, M.; Schmid, K.; Spindler, L.; Ho, C.H.; Kreim, N.; Andrade-Navarro, M.A.; Poeck, B.; Helm, M.; et al. m(6)A modulates neuronal functions and sex determination in drosophila. Nature 2016, 540, 242–247. [Google Scholar] [CrossRef]
- Luo, G.Z.; MacQueen, A.; Zheng, G.Q.; Duan, H.C.; Dore, L.C.; Lu, Z.K.; Liu, J.; Chen, K.; Jia, G.F.; Bergelson, J.; et al. Unique features of the m(6)A methylome in Arabidopsis thaliana. Nat. Commun. 2015, 5. [Google Scholar] [CrossRef]
- Vespa, L.; Vachon, G.; Berger, F.; Perazza, D.; Faure, J.D.; Herzog, M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2014, 134, 1283–1292. [Google Scholar] [CrossRef]
- Shen, L.S.; Liang, Z.; Gu, X.F.; Chen, Y.; Teo, Z.W.N.; Hou, X.L.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N-6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2018, 59, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Serna, L.; Martin, C. Trichomes: Different regulatory networks lead to convergent structures. Trends Plant Sci. 2006, 11, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Pan, J.B.; An, L.Z.; Gan, Y.B.; Feng, H.Y. The responses of trichome mutants to enhanced ultraviolet-B radiation in Arabidopsis thaliana. J. Photoch. Photobio. 2012, 113, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Sterky, F.; Bhalerao, R.R.; Unneberg, P.; Segerman, B.; Nilsson, P.; Brunner, A.M.; Charbonnel-Campaa, L.; Lindvall, J.J.; Tandre, K.; Strauss, S.H.; et al. A Populus EST resource for plant functional genomics. Proc. Natl. Acad. Sci. USA 2004, 138, 13951–13956. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.P.; Liu, S.; Dong, Y.; Zhao, Y.; Geng, A.K.; Xia, X.L.; Yin, W.L. PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 2016, 14, 849–860. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wang, H.L.; Li, H.G.; Su, Y.Y.; Li, S.; Yang, Y.L.; Feng, C.H.; Yin, W.L.; Xia, X.L. PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 2018, 16, 1514–1528. [Google Scholar] [CrossRef]
- Song, C.; Lu, L.; Guo, Y.; Xu, H.; Li, R. Efficient Agrobacterium-mediated transformation of the commercial hybrid Poplar Populus Alba x Populus glandulosa Uyeki. Int. J. Mol. Sci. 2019, 20, 2594. [Google Scholar] [CrossRef]
- Chen, T.; Hao, Y.J.; Zhang, Y.; Li, M.M.; Wang, M.; Han, W.F.; Wu, Y.S.; Lv, Y.; Hao, J.; Wang, L.B.; et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 2015, 16, 338. [Google Scholar] [CrossRef]
- Fukusumi, Y.; Naruse, C.; Asano, M. WTAP is required for differentiation of endoderm and mesoderm in the mouse embryo. Dev. Dynam. 2008, 237, 618–629. [Google Scholar] [CrossRef]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m(6)A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [PubMed]
- Martinez-Perez, M.; Aparicio, F.; Lopez-Gresa, M.P.; Belles, J.M.; Sanchez-Navarro, J.A.; Pallas, V. Arabidopsis m(6)A demethylase activity modulates viral infection of a plant virus and the m(6)A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. USA 2017, 114, 10755–10760. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N(6)-Methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.X.; Ye, Z.B. Trichomes as models for studying plant cell differentiation. Cell Mol. Life Sci. 2013, 70, 1937–1948. [Google Scholar] [CrossRef] [PubMed]
- McDowell, E.T.; Kapteyn, J.; Schmidt, A.; Li, C.; Kang, J.H.; Descour, A.; Shi, F.; Larson, M.; Schilmiller, A.; An, L.L.; et al. Comparative functional genomic analysis of solanum glandular trichome types. Plant physiol. 2011, 155, 524–539. [Google Scholar] [CrossRef] [PubMed]
- Riddick, E.W.; Simmons, A.M. Plant trichomes have mixed impacts on predatory insects. Pest Manag. Sci. 2014, 70, 1668. [Google Scholar] [CrossRef]
- Li, L.J.; Gu, W.R.; Meng, Y.; Wang, Y.L.; Mu, J.Y.; Li, J.; Wei, S. Physiological and biochemical mechanism of spermidine improving drought resistance in maize seedlings under drought stress. J. Ecol. 2018, 29, 554–564. [Google Scholar]
- Hochholdinger, F. Untapping root system architecture for crop improvement. J. Exp. Bot. 2016, 67, 4431–4433. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voss, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in roots: Uncovering form, function, and regulation. Plant physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef]
- Rogers, E.D.; Benfey, P.N. Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotech. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Karuppanapandian, T.; Moon, J.C.; Kim, C.; Manoharan, K.; Kim, W. Reactive oxygen species in plants: Their generation, signal transduction, and scavenging mechanisms. Aust. J. Crop Sci. 2011, 5, 709–725. [Google Scholar]
- Dietz, K.J.; Mittler, R.; Noctor, G. Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant physiol. 2016, 171, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Van, A.O.; Schwarzlander, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.J.; Jiang, P.P.; Qi, S.M.; Zhang, K.; He, Q.X.; Xu, C.Z.; Ding, Z.H.; Zhang, K.W.; Li, K.P. Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize type-ii h+-pyrophosphatase gene by deletion analysis in transgenic tobacco plants. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- de la Garma, J.G.; Fernandez-Garcia, N.; Bardisi, E.; Pallol, B.; Rubio-Asensio, J.S.; Bru, R.; Olmos, E. New insights into plant salt acclimation: The roles of vesicle trafficking and reactive oxygen species signalling in mitochondria and the endomembrane system. New Phytol. 2015, 205, 216–239. [Google Scholar] [CrossRef] [PubMed]
- Li, B.S.; Qin, Y.R.; Duan, H.; Yin, W.L.; Xia, X.L. Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J. Exp. Bot. 2011, 62, 3765–3779. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).