Alpha-l-Locked Nucleic Acid-Modified Antisense Oligonucleotides Induce Efficient Splice Modulation In Vitro
Abstract
1. Introduction
2. Results
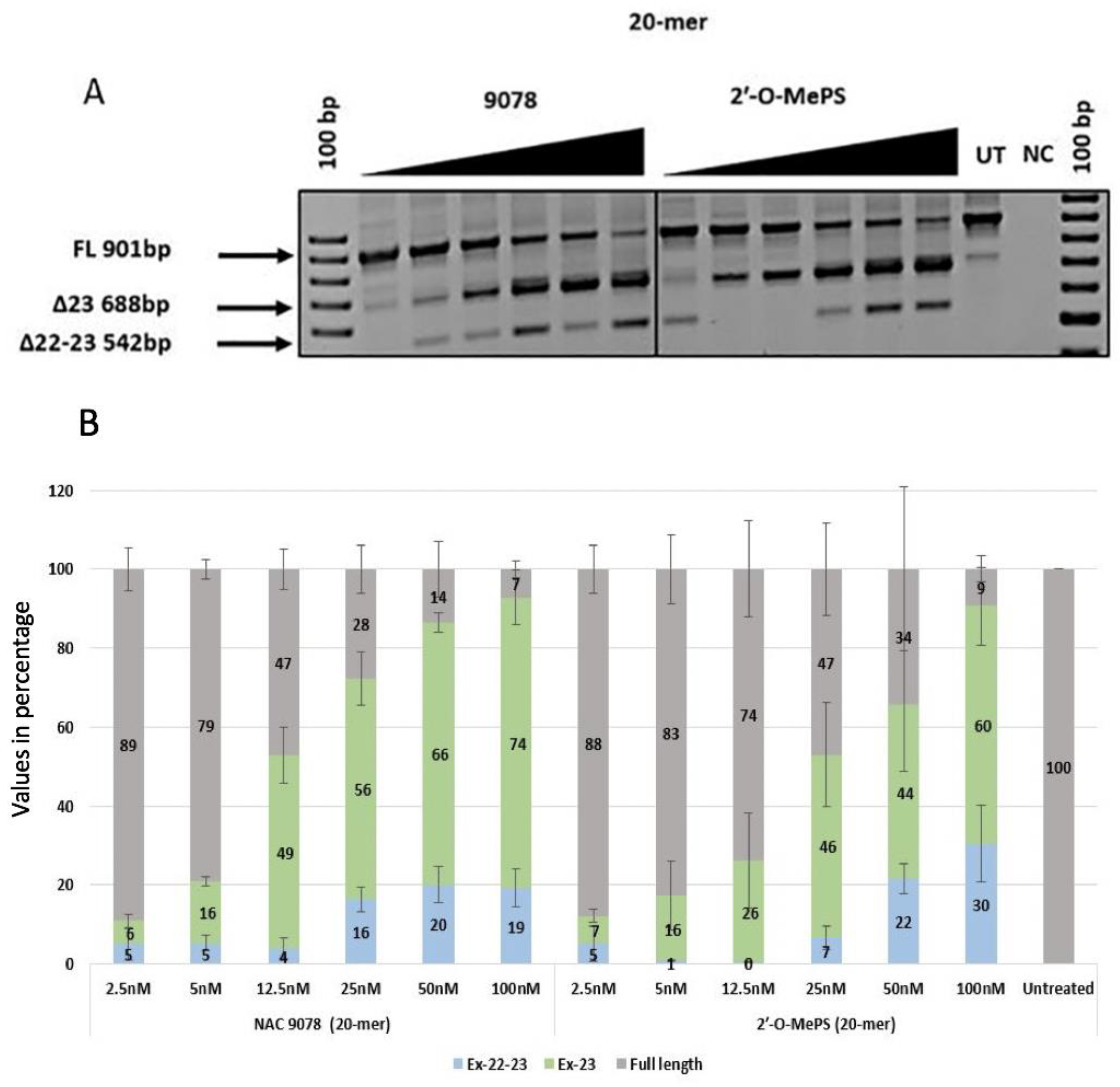
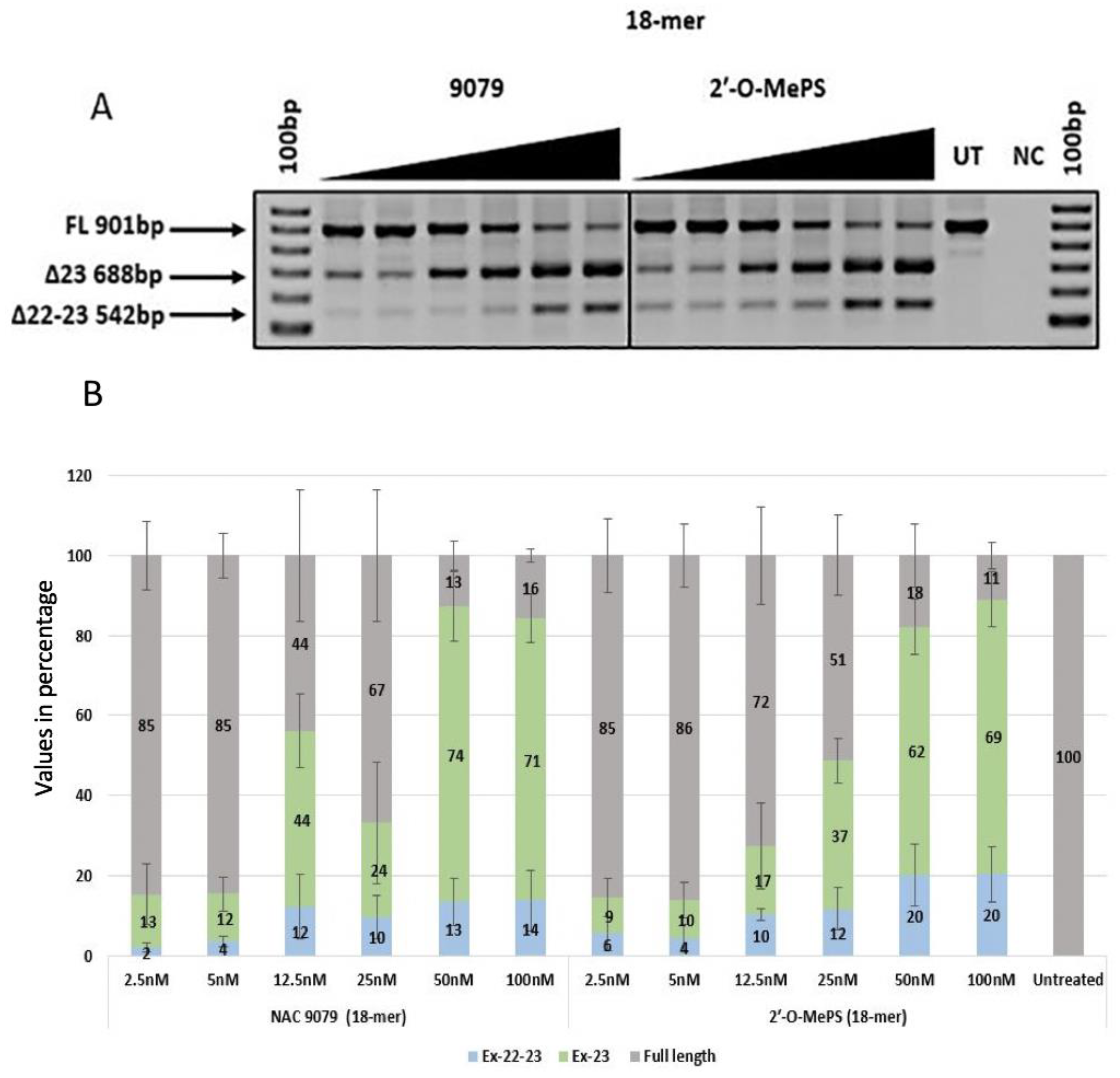
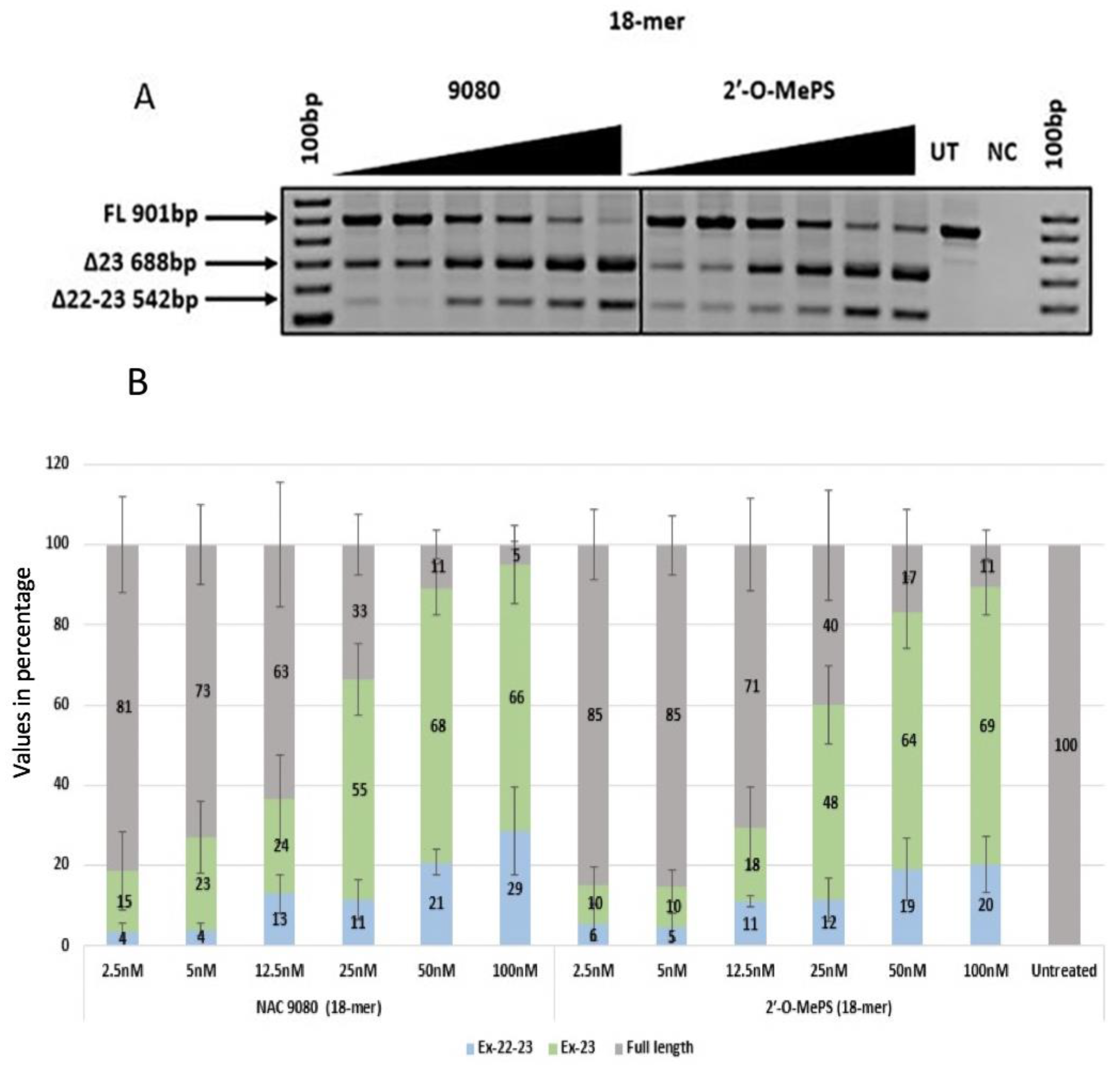
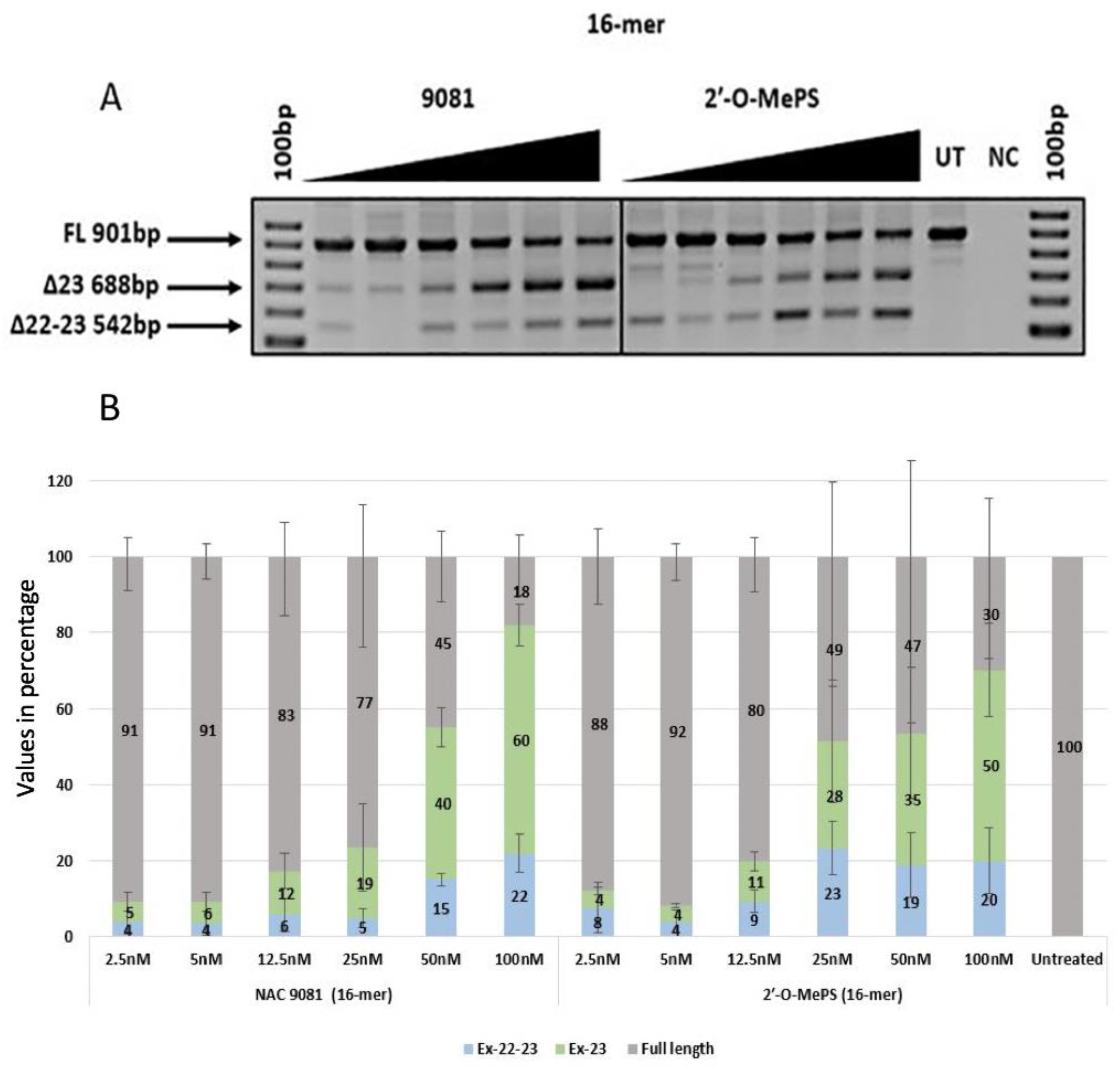
2.1. Evaluation of α-l-LNA AOs to Induce Exon-23 Skipping
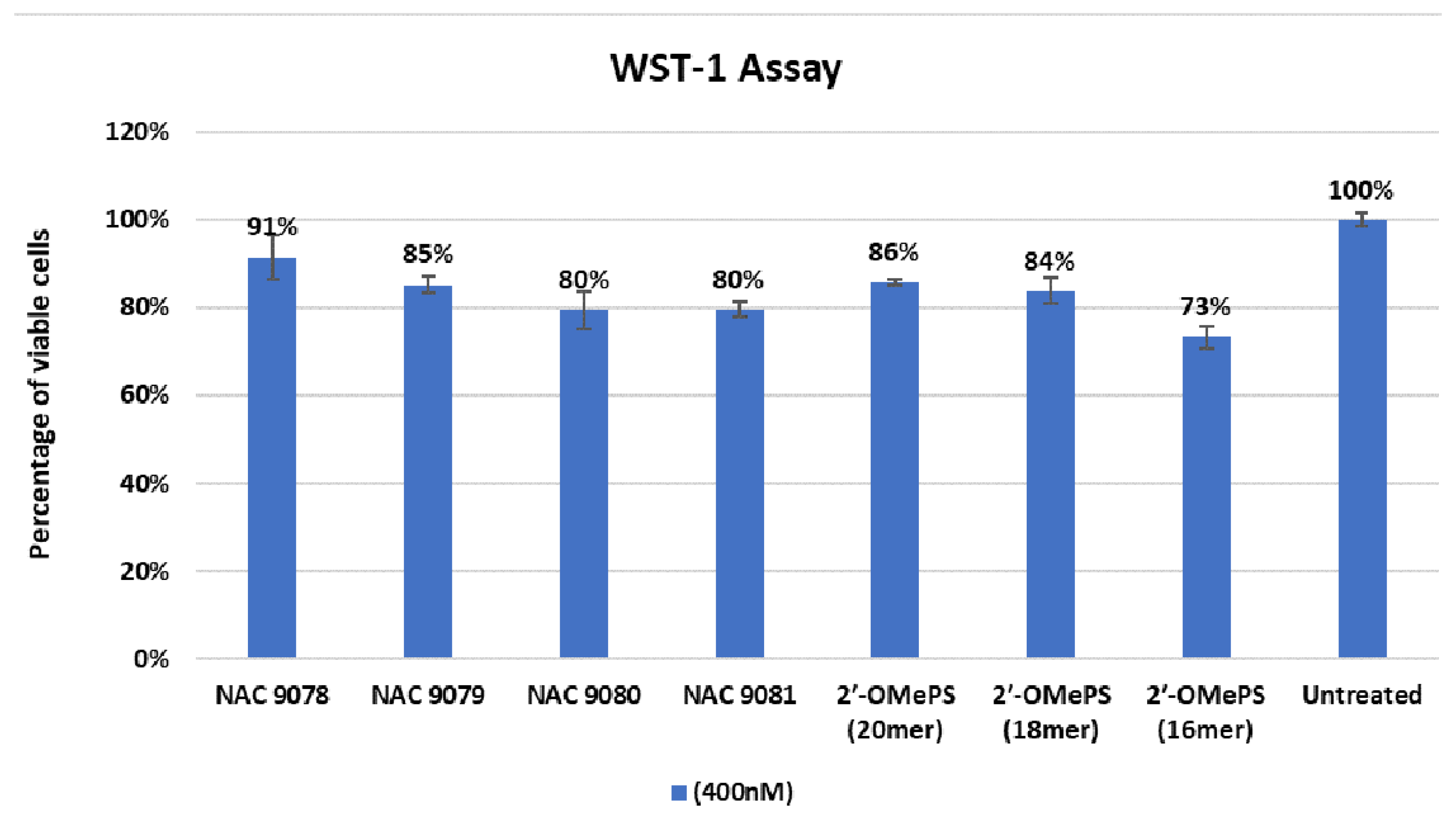
2.2. Evaluation of Cytotoxicity of AOs
3. Discussion
4. Materials and Methods
4.1. Melting Temperature Analysis
4.2. Cell Culture and Transfection
4.3. RNA Extraction and RT-PCR
4.4. Cell Viability and Cytotoxicity Assay
4.5. Evaluation of Efficiency of Exon Skipping
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AO | Antisense oligonucleotide |
| α-l-LNA | Alpha-L-locked nucleic acid |
| DMD | Duchenne muscular dystrophy |
References
- Fluiter, K.; Frieden, M.; Vreijling, J.; Rosenbohm, C.; De Wissel, M.B.; Christensen, S.M.; Koch, T.; Ørum, H.; Baas, F. On the in vitro and in vivo properties of four locked nucleic acid nucleotides incorporated into an anti-H-Ras antisense oligonucleotide. ChemBioChem 2005, 6, 1104–1109. [Google Scholar] [CrossRef] [PubMed]
- Rajwanshi, V.K.; Håkansson, A.E.; Sørensen, M.D.; Pitsch, S.; Singh, S.K.; Kumar, R.; Nielsen, P.; Wengel, J. The eight stereoisomers of LNA (locked nucleic acid): A remarkable family of strong RNA binding molecules. Angew. Chem. Int. Ed. Engl. 2000, 39, 1656–1659. [Google Scholar] [CrossRef]
- Jørgensen, A.S.; Hansen, H.H.; Vester, B.; Wengel, J. Improvement of a streptavidin-binding aptamer by LNA-and α-l-LNA-substitutions. Bioorg. Med. Chem. Lett. 2014, 24, 2273–2277. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Wengel, J.; Hrdlicka, P.J. 2’-N-(Pyren-1-yl) acetyl-2’-Amino-α-l-LNA: Synthesis and Detection of Single Nucleotide Mismatches in DNA and RNA Targets. ChemBioChem 2007, 8, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Madsen, A.S.; Østergaard, M.E.; Wengel, J.; Hrdlicka, P.J. Nucleic acid structural engineering using pyrene-functionalized 2’-amino-α-l-LNA monomers and abasic sites. J. Org. Chem. 2008, 73, 7060–7066. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.S.; Madsen, A.S.; Østergaard, M.E.; Sau, S.P.; Wengel, J.; Hrdlicka, P.J. Functionalized 2’-amino-α-l-LNA: Directed positioning of intercalators for DNA targeting. J. Org. Chem. 2008, 74, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yuan, F.; Zhou, C.; Plashkevych, O.; Chattopadhyaya, J. Free-radical ring closure to conformationally locked α-l-carba-LNAs and synthesis of their oligos: Nuclease stability, target RNA specificity, and elicitation of RNase H. J. Org. Chem. 2010, 75, 6122–6140. [Google Scholar] [CrossRef]
- Seth, P.P.; Allerson, C.R.; Berdeja, A.; Swayze, E.E. Replacing the 2’-oxygen with an exocyclic methylene group reverses the stabilization effects of α-l-LNA. Bioorg. Med. Chem. Lett. 2011, 21, 588–591. [Google Scholar] [CrossRef]
- Bao, T.L.; Veedu, R.N.; Fletcher, S.; Wilton, S.D. Antisense oligonucleotide development for the treatment of muscular dystrophies. Expert Opin. Orphan Drugs. 2016, 4, 139–152. [Google Scholar] [CrossRef]
- Mitrpant, C.; Fletcher, S.; Wilton, S.D. Personalised genetic intervention for Duchenne muscular dystrophy: Antisense oligomers and exon skipping. Curr. Mol. Pharmacol. 2009, 2, 110–121. [Google Scholar] [CrossRef]
- Fairclough, R.J.; Wood, M.J.; Davies, K.E. Therapy for Duchenne muscular dystrophy: Renewed optimism from genetic approaches. Nat. Rev. Genet. 2013, 14, 373. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krieg, A.M. Exon skipping therapy for Duchenne muscular dystrophy. Adv. Drug Deliv. Rev. 2015, 87, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Syed, Y.Y. Eteplirsen: First global approval. Drugs 2016, 76, 1699–1704. [Google Scholar] [CrossRef] [PubMed]
- Kyndrisa Approval Status. GSK and Prosensa Announce Primary Endpoint not Met in Phase III Study of Drisapersen in Patients with Duchenne Muscular Dystrophy; GSK: Brentford, UK, 2013. [Google Scholar]
- Dominski, Z.; Kole, R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 1993, 90, 8673–8677. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.T.; Rahimzadeh, K.; Shaikh, K.; Mohal, N.; Veedu, R.N. Synthesis of a morpholino nucleic acid (MNA)-uridine phosphoramidite, and exon skipping using MNA/2’-O-methyl mixmer antisense oligonucleotide. Molecules 2016, 21, 1582. [Google Scholar] [CrossRef]
- Le, B.T.; Adams, A.M.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Rational design of short locked nucleic acid-modified 2’-O-methyl antisense oligonucleotides for efficient exon-skipping in vitro. Mol. Ther. Nucleic Acids 2017, 9, 155–161. [Google Scholar] [CrossRef]
- Le, B.T.; Filichev, V.V.; Veedu, R.N. Investigation of twisted intercalating nucleic acid (TINA)-modified antisense oligonucleotides for splice modulation by induced exon-skipping in vitro. RSC Adv. 2016, 6, 95169–95172. [Google Scholar] [CrossRef]
- Le, B.T.; Murayama, K.; Shabanpoor, F.; Asanuma, H.; Veedu, R.N. Antisense oligonucleotide modified with serinol nucleic acid (SNA) induces exon skipping in mdx myotubes. RSC Adv. 2017, 7, 34049–34052. [Google Scholar] [CrossRef]
- Le, B.T.; Chen, S.; Abramov, M.; Herdewijn, P.; Veedu, R.N. Evaluation of anhydrohexitol nucleic acid, cyclohexenyl nucleic acid and D-altritol nucleic acid-modified 2’-O-methyl RNA mixmer antisense oligonucleotides for exon skipping in vitro. Chem. Commun. 2016, 52, 13467–13470. [Google Scholar] [CrossRef]
- Gebski, B.L.; Mann, C.J.; Fletcher, S.; Wilton, S.D. Morpholino antisense oligonucleotide induced dystrophin exon 23 skipping in mdx mouse muscle. Hum. Mol. Genet. 2003, 12, 1801–1811. [Google Scholar] [CrossRef]
- Le, B.T.; Hornum, M.; Sharma, P.K.; Nielsen, P.; Veedu, R.N. Nucleobase-modified antisense oligonucleotides containing 5-(phenyltriazol)-2’-deoxyuridine nucleotides induce exon-skipping in vitro. RSC Adv. 2017, 7, 54542–54545. [Google Scholar] [CrossRef]
- Chen, S.; Le, B.T.; Chakravarthy, M.; Kosbar, T.R.; Veedu, R.N. Systematic evaluation of 2’-Fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro. Sci. Rep. 2019, 9, 6078. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, S.; Honeyman, K.; Fall, A.M.; Harding, P.L.; Johnsen, R.D.; Wilton, S.D. Dystrophin expression in the mdx mouse after localised and systemic administration of a morpholino antisense oligonucleotide. J. Gene Med. 2006, 8, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Rando, T.A.; Blau, H.M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994, 125, 1275–1287. [Google Scholar] [CrossRef] [PubMed]
- Mann, C.J.; Honeyman, K.; Cheng, A.J.; Ly, T.; Lloyd, F.; Fletcher, S.; Morgan, J.E.; Patridge, T.A.; Wilton, S.D. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc. Natl. Acad. Sci. USA 2001, 98, 42–47. [Google Scholar] [CrossRef]
- Wang, T.; Larcher, L.M.; Ma, L.; Veedu, R.N. Systematic screening of commonly used commercial transfection reagents towards efficient transfection of single-stranded oligonucleotides. Molecules 2018, 23, 2564. [Google Scholar] [CrossRef]
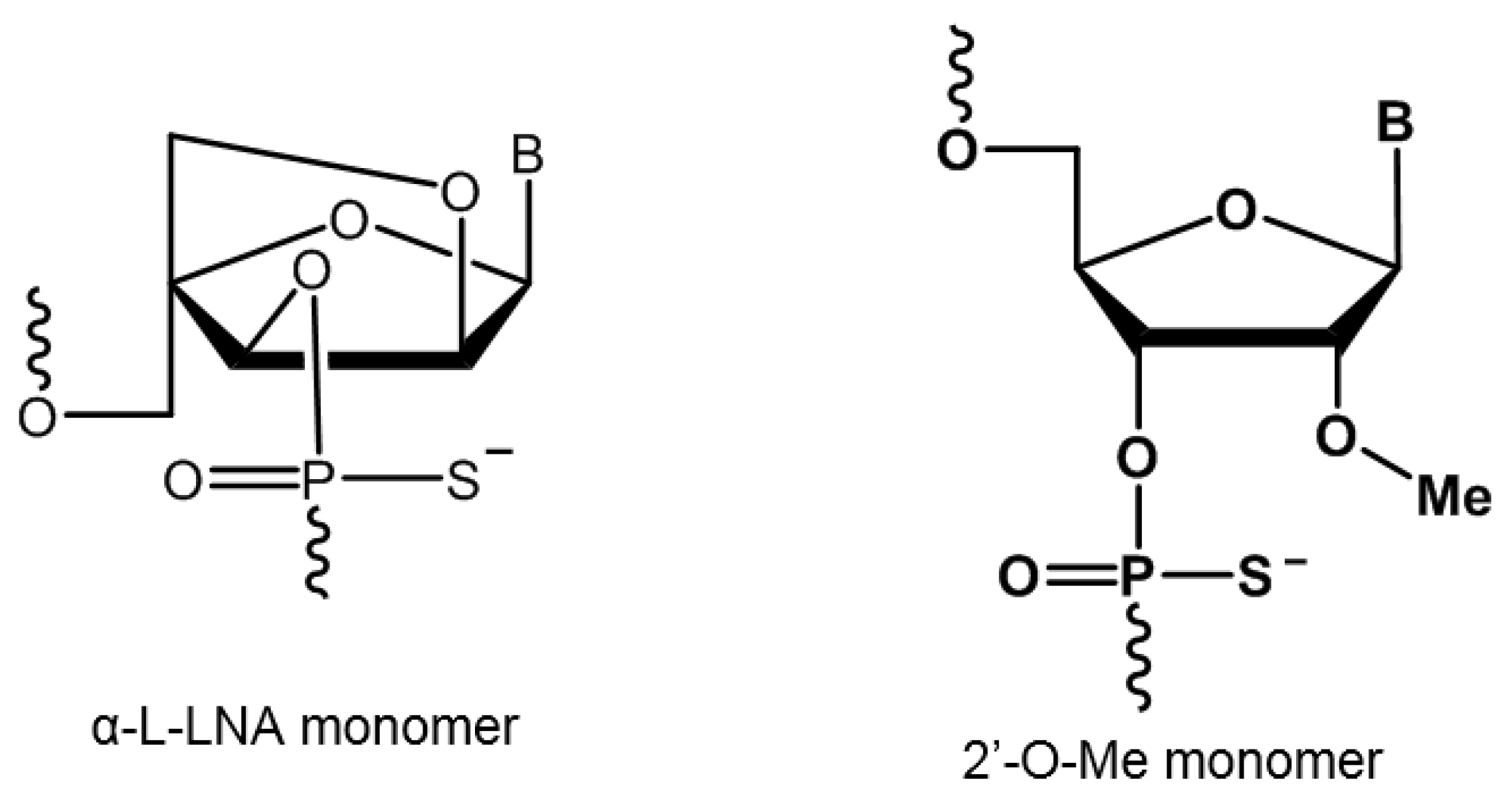

| AO Name | Sequence (5′-3′ direction) | Tm (°C) |
|---|---|---|
| NAC 9078 (20mer) | GGC CAA ACC UCG GCαT UAαC CαT | 65.2 |
| NAC 9079 (18mer) | GCC AAA CCU CGG αCUαT ACαC | 64.2 |
| NAC 9080 (18mer) | GαCC AAA αCCU αCGG CαTU AαCC | 68.8 |
| NAC 9081 (16mer) | CCA AAC CUC GGαC UαTA αC | 59.6 |
| 2′-O-MePS (20mer) | GGCCAAACCUCGGCUUACCU | 60.8 |
| 2′-O-MePS (18mer) | GCCAAACCUCGGCUUACC | 57.7 |
| 2′-O-MePS (16mer) | CCAAACCUCGGCUUAC | 53.1 |
| AO Name | Percentage of Exon 23 Skipping (%) | Percentage of Dual Exon 22/23 Skipping (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.5 nM | 5 nM | 12.5 nM | 25 nM | 50 nM | 100 nM | 2.5 nM | 5 nM | 12.5 nM | 25 nM | 50 nM | 100 nM | |
| NAC 9078 | 6 (2.93) | 16 (2.23) | 49 (12.41) | 56 (11.68) | 66 (4.21) | 74 (11.92) | 5 (6.95) | 5 (3.36) | 4 (4.51) | 16 (5.33) | 20 (8.13) | 19 (8.50) |
| NAC9079 | 13 (13.23) | 12 (7.51) | 44 (15.76) | 24 (26.30) | 74 (15.27) | 71 (10.53) | 2 (2.01) | 4 (2.27) | 12 (13.79) | 10 (9.26) | 13 (10.30) | 14 (12.94) |
| NAC 9080 | 15 (17.13) | 23 (15.42) | 24 (18.90) | 55 (15.52) | 68 (11.23) | 66 (17.12) | 4 (3.62) | 4 (2.53) | 13 (8.43) | 11 (8.79) | 21 (5.55) | 29 (18.86) |
| NAC 9081 | 5 (4.08) | 6 (4.35) | 12 (8.10) | 19 (19.71) | 40 (9.13) | 60 (9.29) | 4 (5.02) | 4 (5.26) | 6 (7.64) | 5 (4.37) | 15 (2.95) | 22 (8.75) |
| 2′-O-MePS (20mer) | 7 (3.10) | 16 (15.06) | 26 (20.88) | 46 (22.81) | 44 (29.25) | 60 (17.13) | 5 (7.68) | 1 (0.34) | 1 (0.18) | 7 (5.06) | 22 (6.65) | 30 (16.92) |
| 2′-O-MePS (18mer) | 10 (7.96) | 10 (6.83) | 18 (17.62) | 48 (16.86) | 64 (15.38) | 69 (12.22) | 6 (7.39) | 5 (5.99) | 11 (2.41) | 12 (9.07) | 19 (13.45) | 20 (11.89) |
| 2′-O-MePS (16mer) | 4 (1.79) | 4 (1.04) | 11 (4.32) | 28 (27.75) | 35 (29.98) | 50 (21.53) | 8 (11.34) | 4 (5.10) | 9 (5.06) | 23 (12.06) | 19 (14.58) | 20 (14.81) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raguraman, P.; Wang, T.; Ma, L.; Jørgensen, P.T.; Wengel, J.; Veedu, R.N. Alpha-l-Locked Nucleic Acid-Modified Antisense Oligonucleotides Induce Efficient Splice Modulation In Vitro. Int. J. Mol. Sci. 2020, 21, 2434. https://doi.org/10.3390/ijms21072434
Raguraman P, Wang T, Ma L, Jørgensen PT, Wengel J, Veedu RN. Alpha-l-Locked Nucleic Acid-Modified Antisense Oligonucleotides Induce Efficient Splice Modulation In Vitro. International Journal of Molecular Sciences. 2020; 21(7):2434. https://doi.org/10.3390/ijms21072434
Chicago/Turabian StyleRaguraman, Prithi, Tao Wang, Lixia Ma, Per Trolle Jørgensen, Jesper Wengel, and Rakesh N. Veedu. 2020. "Alpha-l-Locked Nucleic Acid-Modified Antisense Oligonucleotides Induce Efficient Splice Modulation In Vitro" International Journal of Molecular Sciences 21, no. 7: 2434. https://doi.org/10.3390/ijms21072434
APA StyleRaguraman, P., Wang, T., Ma, L., Jørgensen, P. T., Wengel, J., & Veedu, R. N. (2020). Alpha-l-Locked Nucleic Acid-Modified Antisense Oligonucleotides Induce Efficient Splice Modulation In Vitro. International Journal of Molecular Sciences, 21(7), 2434. https://doi.org/10.3390/ijms21072434





