Expression of a NGATHA1 Gene from Medicago truncatula Delays Flowering Time and Enhances Stress Tolerance
Abstract
1. Introduction
2. Results
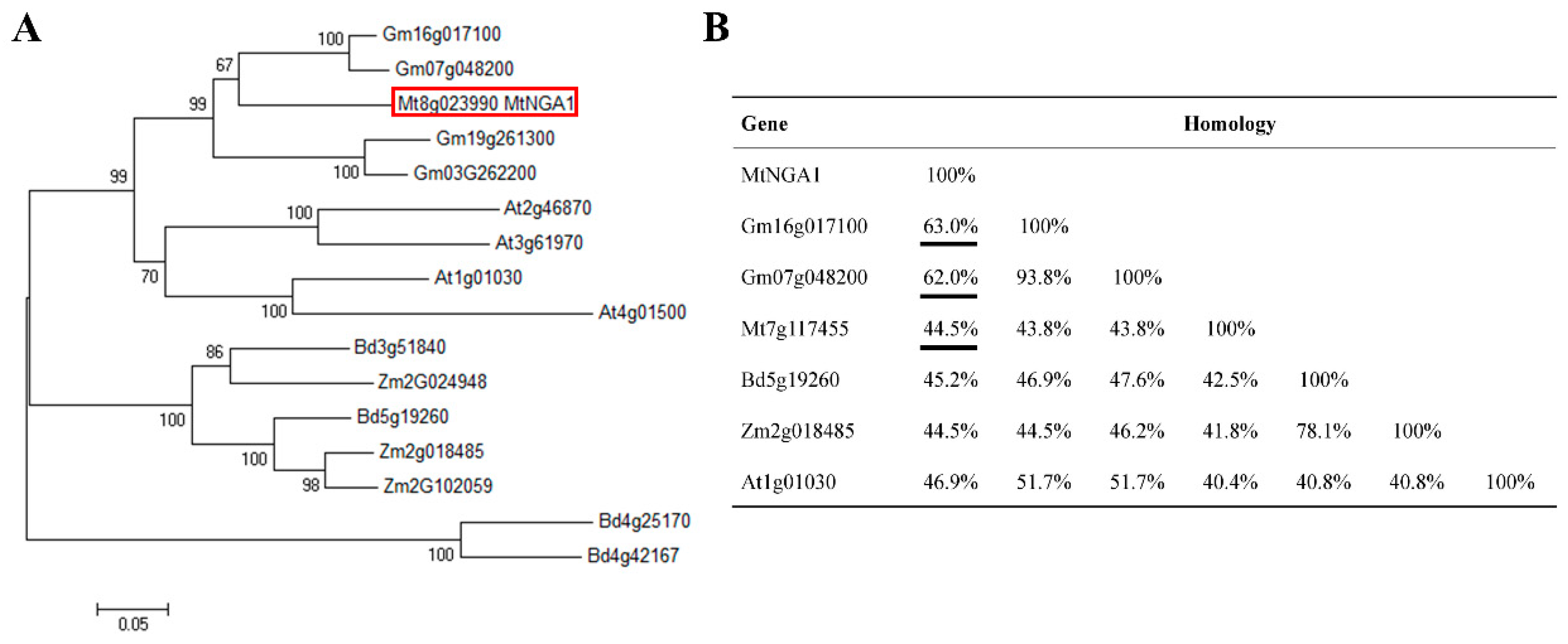
2.1. Identification and Homology Analysis of the MtNGA1 Gene in M. truncatula
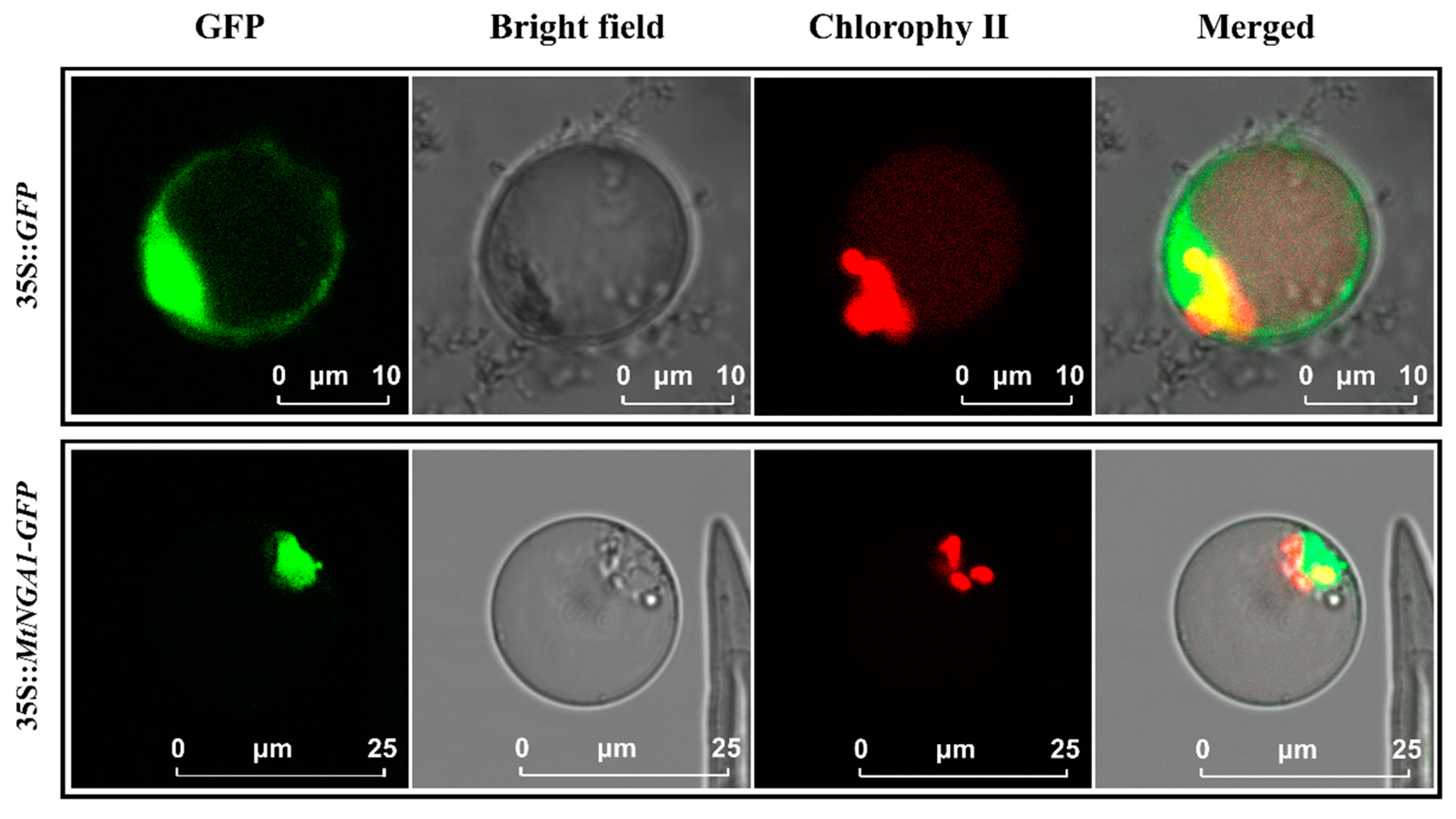
2.2. Subcellular Localization of MtNGA1
2.3. Promoter Analysis of MtNGA1
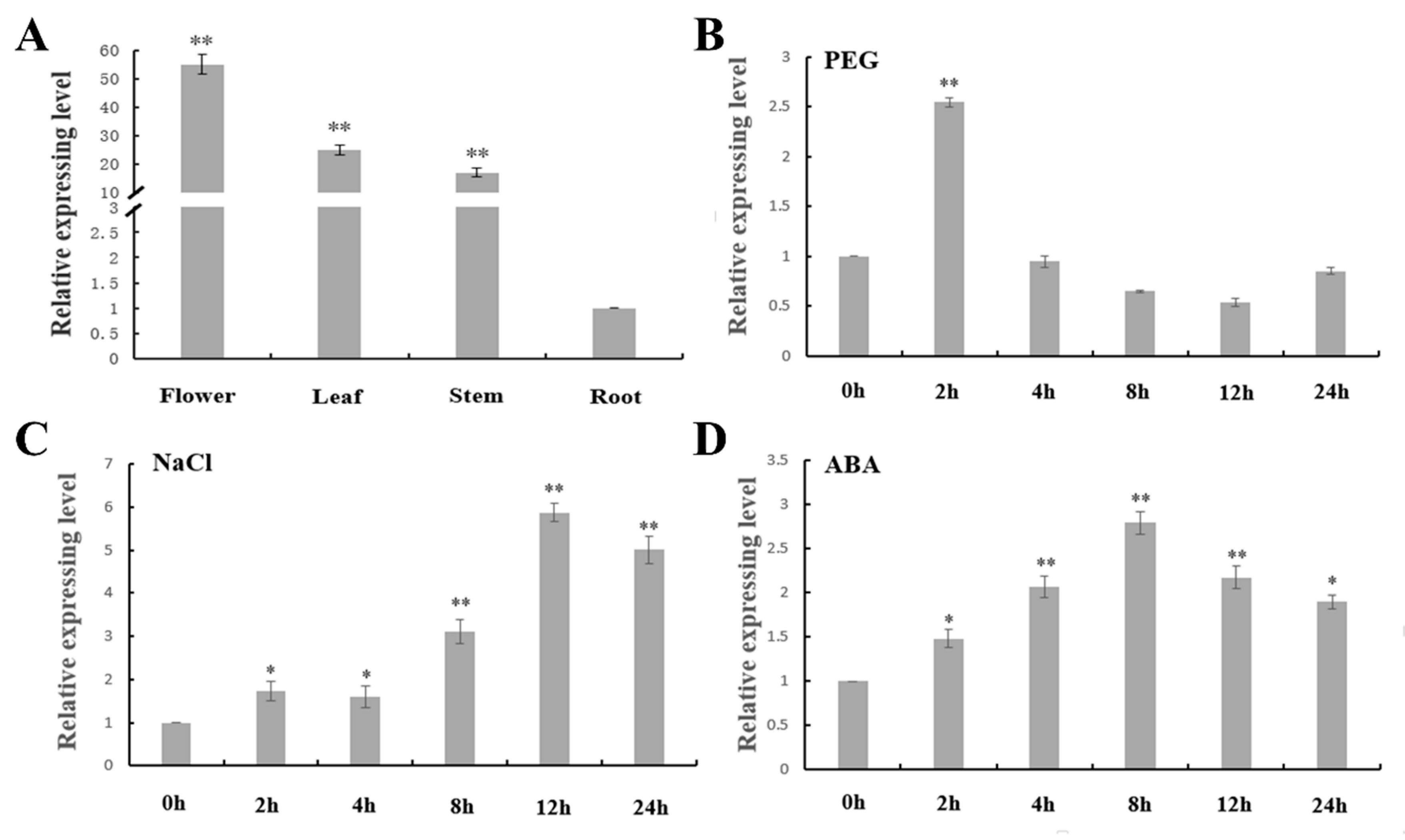
2.4. Expression Analysis of MtNGA1
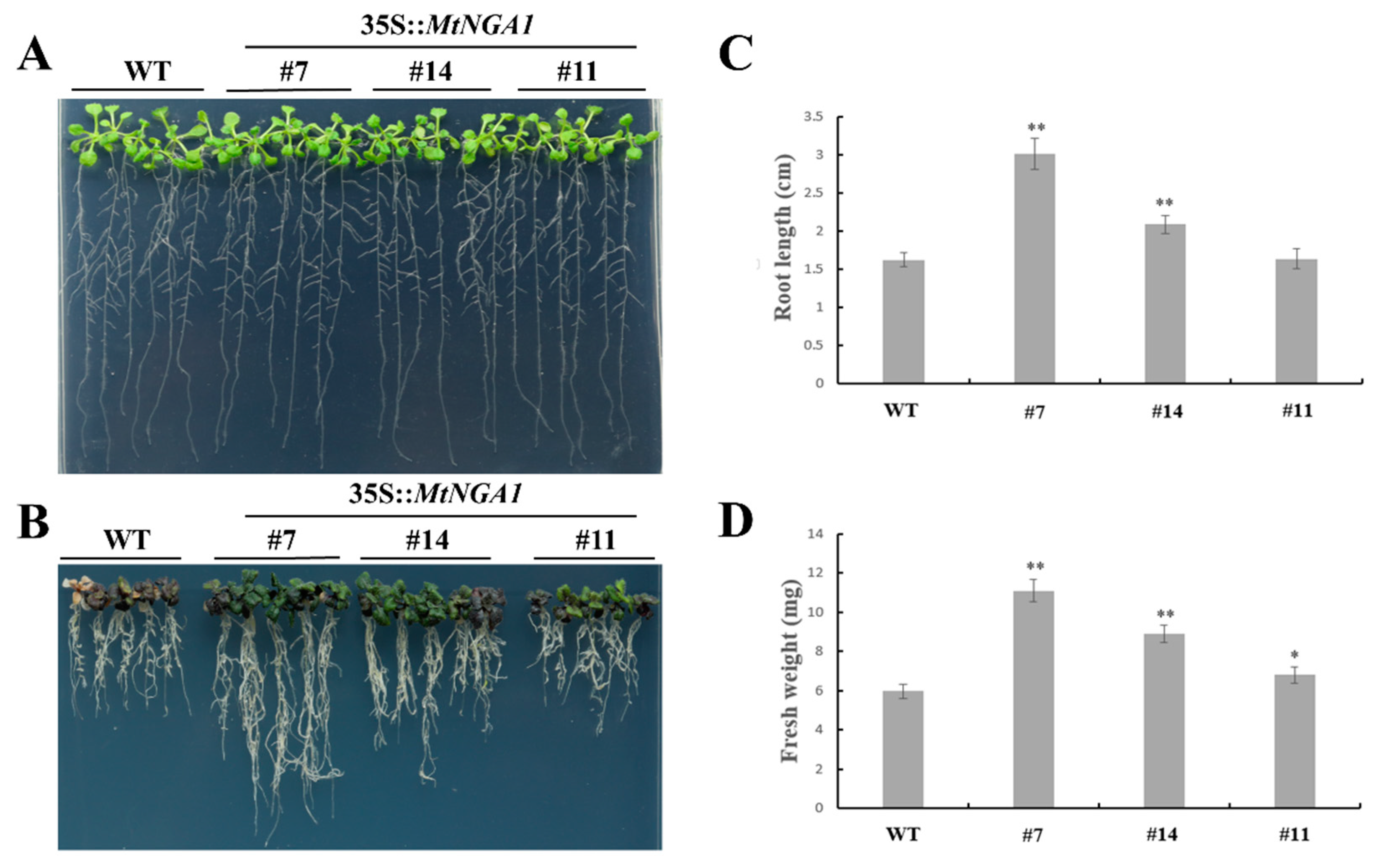
2.5. Overexpression of MtNGA1 Delayed Flowering Time and Reduced the Number of Arabidopsis Branches
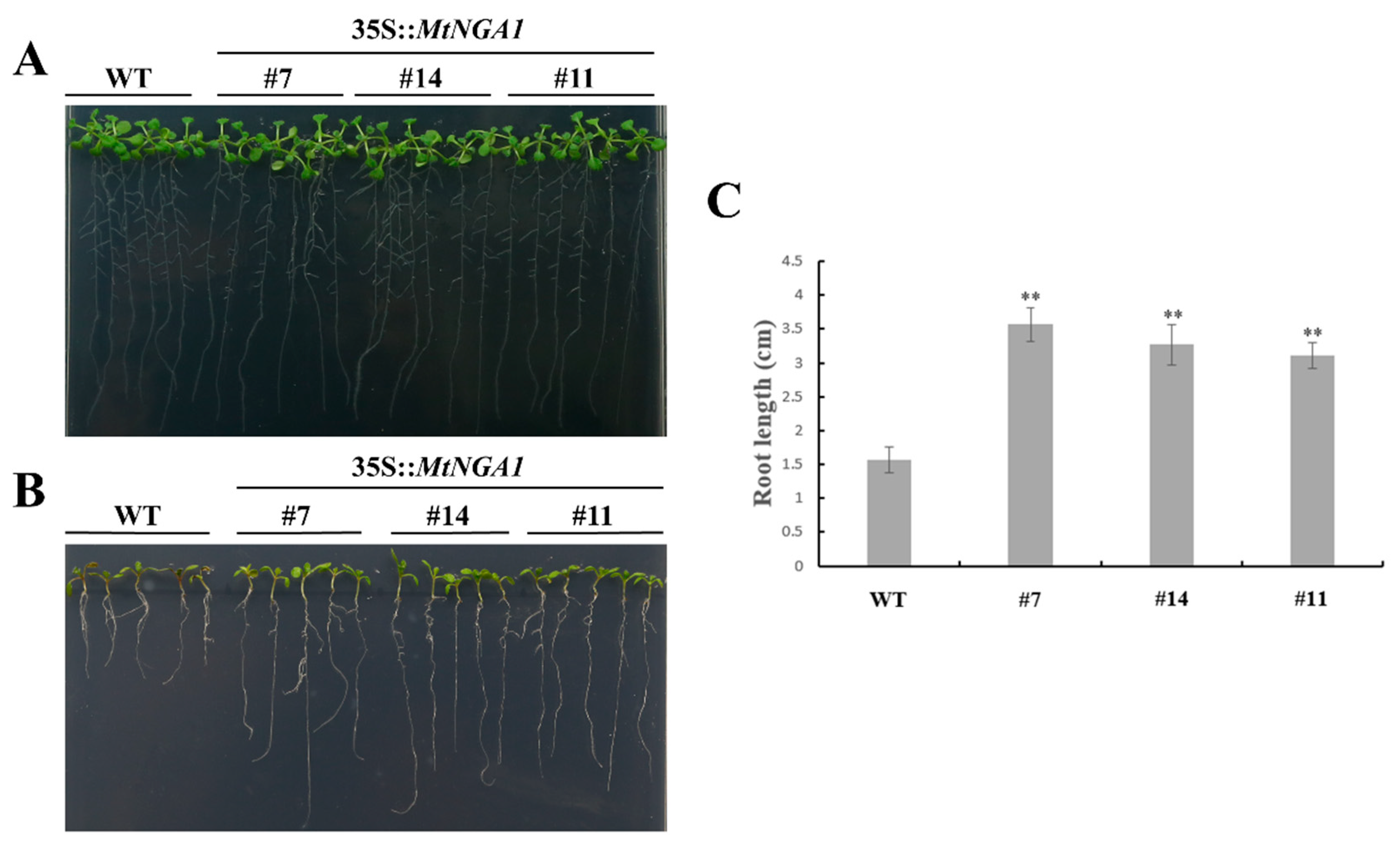
2.6. MtNGA1 Enhanced Resistance to Mannitol-Induced and NaCl Stresses
2.7. MtNGA1 Reduced the Sensitivity to Exogenous ABA
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Cloning of NGA1 from M. truncatula
4.3. Bioinformatics Analysis
4.4. Subcellular Localization Determination
4.5. Abiotic Stress and ABA Treatment
4.6. Quantitative Analysis
4.7. Obtaining Transgenic Plants
4.8. Stress Analysis of Transgenic Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Marella, H.H.; Sakata, Y.; Quatrano, R.S. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J. 2006, 46, 1032–1044. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Swaminathan, K.; Peterson, K.; Jack, T. The plant B3 superfamily. Trends Plant Sci. 2008, 13, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Sasnauskas, G.; Manakova, E.; Lapenas, K.; Kauneckaite, K.; Siksnys, V. DNA recognition by Arabidopsis transcription factors ABI3 and NGA1. FEBS J. 2018, 285, 4041–4059. [Google Scholar] [CrossRef] [PubMed]
- McCarty, D.R.; Hattori, T.; Carson, C.B.; Vasil, V.; Lazar, M.; Vasil, I.K. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 1991, 66, 895–905. [Google Scholar] [CrossRef]
- Suzuki, M.; Kao, C.Y.; McCarty, D.R. The Conserved B3 Domain of VIVIPAROUSI Has a Cooperative DNA Binding Activity. Plant Cell 1997, 9, 799–807. [Google Scholar] [PubMed]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Stone, S.L.; Kwong, L.W.; Yee, K.M.; Pelletier, J.; Lepiniec, L.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 2001, 98, 11806–11811. [Google Scholar] [CrossRef]
- Barreto, H.G.; Ságio, S.A.; Chalfun-Júnior, A.; Fevereiro, P.; Benedito, V.A. Transcriptional profiling of the AFL subfamily of B3-type transcription factors during the in vitro induction of somatic embryogenesis in the model legume Medicago truncatula. Plant Cell Tissue Organ Cult. (PCTOC) 2019, 139, 327–337. [Google Scholar] [CrossRef]
- Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; You, S.J.; Hsieh, T.H.; Chao, T.C.; Chan, M.T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011, 156, 213–227. [Google Scholar] [CrossRef]
- Min, H.; Zheng, J.; Wang, J. Maize ZmRAV1 contributes to salt and osmotic stress tolerance in transgenic Arabidopsis. J. Plant Biol. 2014, 57, 28–42. [Google Scholar] [CrossRef]
- Lee, S.C.; Choi, D.S.; Hwang, I.S.; Hwang, B.K. The pepper oxidoreductase CaOXR1 interacts with the transcription factor CaRAV1 and is required for salt and osmotic stress tolerance. Plant Mol. Biol. 2010, 73, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Cubas, P.; Jarillo, J.A.; Fernández-Calvín, B.; Salinas, J.; Martínez-Zapater, J.M. AtREM1, a Member of a New Family of B3 Domain-Containing Genes, Is Preferentially Expressed in Reproductive Meristems1. Plant Physiol. 2002, 128, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.H.; Chang, S.C.; Ko, J.; Song, J.T.; Kim, J.H. Overexpression of Brassica rapa NGATHA1 Gene Confers De-Etiolation Phenotype and Cytokinin Resistance on Arabidopsis thaliana. J. Plant Biol. 2011, 54, 119–125. [Google Scholar] [CrossRef]
- Alvarez, J.P.; Furumizu, C.; Efroni, I.; Eshed, Y.; Bowman, J.L. Active suppression of a leaf meristem orchestrates determinate leaf growth. eLife 2016, 5, e15023. [Google Scholar] [CrossRef]
- Lee, B.H.; Kwon, S.H.; Lee, S.J.; Park, S.K.; Song, J.T.; Lee, S.; Lee, M.M.; Hwang, Y.S.; Kim, J.H. The Arabidopsis thaliana NGATHA transcription factors negatively regulate cell proliferation of lateral organs. Plant Mol. Biol. 2015, 89, 529–538. [Google Scholar] [CrossRef]
- Kwon, S.H.; Lee, B.H.; Kim, E.Y.; Seo, Y.S.; Lee, S.; Kim, W.T.; Song, J.T.; Kim, J.H. Overexpression of a Brassica rapa NGATHA Gene in Arabidopsis thaliana Negatively Affects Cell Proliferation During Lateral Organ and Root Growth. Plant Cell Physiol. 2009, 50, 2162–2173. [Google Scholar] [CrossRef]
- Pfannebecker, K.C.; Lange, M.; Rupp, O.; Becker, A. Seed plant specific gene lineages involved in carpel development. Mol. Biol. Evol. 2017, 34, 925–942. [Google Scholar] [CrossRef]
- Lee, B.H.; Mai, T.T.; Song, J.T.; Kim, J.H. The Arabidopsis thaliana NGATHA1 transcription factor acts as a promoter of a general differentiation program and a carpel identity factor. J. Plant Biol. 2017, 60, 352–357. [Google Scholar] [CrossRef]
- Martinez-Fernandez, I.; Sanchis, S.; Marini, N.; Balanza, V.; Ballester, P.; Navarrete-Gomez, M.; Oliveira, A.C.; Colombo, L.; Ferrandiz, C. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front. Plant Sci. 2014, 5, 210. [Google Scholar]
- Alvarez, J.P.; Goldshmidt, A.; Efroni, I.; Bowman, J.L.; Eshed, Y. The NGATHA Distal Organ Development Genes Are Essential for Style Specification in Arabidopsis. Plant Cell 2009, 21, 1373–1393. [Google Scholar] [CrossRef] [PubMed]
- Trigueros, M.; Navarrete-Gómez, M.; Sato, S.; Christensen, S.K.; Pelaz, S.; Weigel, D.; Yanofsky, M.F.; Ferrándiz, C. The NGATHA Genes Direct Style Development in the Arabidopsis Gynoecium. Plant Cell 2009, 21, 1394–1409. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Liu, X.; Wang, R.; Zhang, G.; Yu, F. The overexpression of an Arabidopsis B3 transcription factor, ABS2/NGAL1, leads to the loss of flower petals. PLoS ONE 2012, 7, e49861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Du, L.; Xu, R.; Cui, R.; Hao, J.; Sun, C.; Li, Y. Transcription Factors SOD7/NGAL2 and DPA4/NGAL3 Act Redundantly to Regulate Seed Size by Directly Repressing KLU Expression in Arabidopsis thaliana. Plant Cell 2015, 27, 620–632. [Google Scholar] [CrossRef]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, Y.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.; Hong, X.; Agarwal, M.; Zhu, J. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Gene Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Kumar, G.M.; Mamidala, P.; Podile, A.R. Regulation of Polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics J. 2009, 6, 238–249. [Google Scholar]
- Fourquin, C.; Ferrándiz, C. The essential role of NGATHA genes in style and stigma specification is widely conserved across eudicots. New Phytol. 2014, 202, 1001–1013. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, S. Physiological and Proteomic Responses of Contrasting Alfalfa (Medicago sativa L.) Varieties to PEG-Induced Osmotic Stress. Front. Plant Sci. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Hohl, M.; Schopfer, P. Water relations of growing maize coleoptiles: Comparison between mannitol and polyethylene glycol 6000 as external osmotica for adjusting turgor pressure. Plant Physiol. 1991, 95, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Brady, R.L.; Banfield, M.J.; Winter, V.J.; Ahn, J.H.; Weigel, D.; Lee, J.H.; Henz, S.R.; Miller, D.; Yoo, S.Y. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Stirnberg, P.; van De Sande, K.; Leyser, H.O. MAX1and MAX2control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar]
- Duque, A.S.; López-Gómez, M.; Kráčmarová, J.; Gomes, C.; Araújo, S.S.; Lluch, C.; Fevereiro, P. Genetic engineering of polyamine metabolism changes Medicago truncatula responses to water deficit. Plant Cell Tissue Organ Cult. (PCTOC) 2016, 127, 681–690. [Google Scholar] [CrossRef]
- Dalal, V.K.; Tripathy, B.C. Modulation of chlorophyll biosynthesis by water stress in rice seedlings during chloroplast biogenesis. Plant Cell Environ. 2012, 35, 1685–1703. [Google Scholar] [CrossRef]
- Phung, T.; Jung, H.; Park, J.; Kim, J.; Back, K.; Jung, S. Porphyrin Biosynthesis Control under Water Stress: Sustained Porphyrin Status Correlates with Drought Tolerance in Transgenic Rice. Plant Physiol. 2011, 157, 1746–1764. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.Y.; Liao, L.L.; Liu, S.; Nie, M.M.; Li, J.; Zhang, L.D.; Ma, J.F.; Chen, Z.C. Magnesium Deficiency Triggers SGR–Mediated Chlorophyll Degradation for Magnesium Remobilization. Plant Physiol. 2019, 181, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Tasma, I.M.; Brendel, V.; Whitham, S.A.; Bhattacharyya, M.K. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 2008, 46, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Ohto, C.; Mizoguchi, T.; Shinozaki, K. A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1995, 92, 3903–3907. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Zhang, X.; Gao, J.; Wang, P.; Xu, X.; Liu, Z.; Shen, S.; Feng, B. A Buckwheat (Fagopyrum esculentum) DRE-Binding Transcription Factor Gene, FeDREB1, Enhances Freezing and Drought Tolerance of Transgenic Arabidopsis. Plant Mol. Biol. Rep. 2015, 33, 1510–1525. [Google Scholar] [CrossRef]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought Induction of Arabidopsis 9-cis-Epoxycarotenoid Dioxygenase Occurs in Vascular Parenchyma Cells1[W][OA]. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Kang, H.K.; Son, S.; Kim, S.; Nam, K.H. A Subset of Arabidopsis RAV Transcription Factors Modulates Drought and Salt Stress Responses Independent of ABA. Plant Cell Physiol. 2014, 55, 1892–1904. [Google Scholar] [CrossRef]
- Wenck AR, M.L. Large-scale protoplast isolation and regeneration of Arabidopsis thaliana. BIOTECHNIQUES 1995, (4), 640–643. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Wang, S.; Li, Y.; Yuan, J.; Xu, L.; Zhang, T.; Chao, Y.; Han, L. Expression of a NGATHA1 Gene from Medicago truncatula Delays Flowering Time and Enhances Stress Tolerance. Int. J. Mol. Sci. 2020, 21, 2384. https://doi.org/10.3390/ijms21072384
Guo T, Wang S, Li Y, Yuan J, Xu L, Zhang T, Chao Y, Han L. Expression of a NGATHA1 Gene from Medicago truncatula Delays Flowering Time and Enhances Stress Tolerance. International Journal of Molecular Sciences. 2020; 21(7):2384. https://doi.org/10.3390/ijms21072384
Chicago/Turabian StyleGuo, Tao, Shumin Wang, Yinruizhi Li, Jianbo Yuan, Lixin Xu, Tiejun Zhang, Yuehui Chao, and Liebao Han. 2020. "Expression of a NGATHA1 Gene from Medicago truncatula Delays Flowering Time and Enhances Stress Tolerance" International Journal of Molecular Sciences 21, no. 7: 2384. https://doi.org/10.3390/ijms21072384
APA StyleGuo, T., Wang, S., Li, Y., Yuan, J., Xu, L., Zhang, T., Chao, Y., & Han, L. (2020). Expression of a NGATHA1 Gene from Medicago truncatula Delays Flowering Time and Enhances Stress Tolerance. International Journal of Molecular Sciences, 21(7), 2384. https://doi.org/10.3390/ijms21072384




